Abstract
An in vitro reconstitution system for the analysis of replication-coupled nucleosome assembly is described. In this “two-step system,” nucleosome assembly is performed in a separate reaction from DNA replication, wherein purified newly replicated DNA remains noncovalently marked for subsequent chromatin assembly factor-1 (CAF-1)-dependent nucleosome assembly. Because the nucleosome assembly is performed separately from the DNA replication step, this system is more versatile and biochemically tractable when compared with nucleosome assembly during simian virus 40 (SV40) DNA replication. The N-terminal domains of histones H3 and H4 play an important but redundant function in nucleosome assembly in the budding yeast, Saccharomyces cerevisiae. It had been proposed that at least one tail of histone H3 or H4 is required for replication-coupled nucleosome assembly. However, we demonstrate that the N-terminal domains of both histone H3 and H4 are dispensable for CAF-1-mediated formation of nucleosome cores onto newly replicated DNA in vitro. CAF-1 and each of its individual subunits stably bound to recombinant (H3.H4)2 tetramers lacking the N-terminal domains of both H3 and H4. Therefore, the N-terminal tails of histone H3 and H4 that contain the specific acetylation sites are not necessary for CAF-1-dependent nucleosome assembly onto replicated DNA. We suggest that the histone acetylation may be required for a CAF-1 independent pathway or function after deposition, by marking of newly replicated chromatin.
In S phase, preexisting chromatin structures are transiently disrupted, providing a window of opportunity for reprogramming gene expression. Therefore, mechanisms must exist to ensure that differences between cell types are maintained through DNA replication (1). During DNA replication, preexisting nucleosomes are segregated randomly onto the two progeny DNA molecules (2). Additional nucleosomes are formed from newly synthesized histone (H3.H4)2 tetramers, which subsequently associate with either preexisting or newly synthesized H2A.H2B dimers to form nucleosome cores (3, 4). The nucleosome cores are then organized into uniformly spaced arrays, a reaction that requires ATP hydrolysis and, in vitro, can be catalysed by a number of distinct proteins (5–7). The incorporation of histone H1 stabilizes the formation of higher-order structures in mature chromatin (8, 9).
De novo nucleosome formation onto nascent DNA in a human cell-free system for simian virus 40 (SV40) DNA replication requires chromatin assembly factor 1 (CAF-1). The CAF-1 protein consists of three polypeptide subunits: p150, p60, and p48 (10, 11). In human cells, CAF-1 acts as a core histone chaperone by forming a stable complex with newly synthesized histones H3 and H4 (10, 11), and targeting these histones to sites of DNA synthesis (12, 13). This preferential assembly of (H3.H4)2 tetramers onto replicating DNA is, at least in part, because of an interaction between CAF-1 and the proliferating cell nuclear antigen (PCNA) (14), an essential DNA polymerase processivity factor.
The histones associated with CAF-1 in human cells exhibit a heterogeneous pattern of posttranslational modification (11). Histone H4 is a mixture of molecules containing zero, one, two, or three acetyl groups at lysines 5, 8, or 12. Although histone H3 is also modified, the nature of this modification has not yet been determined. Interestingly, diacetylation of lysines 5 and 12 of H4 and acetylation of various residues of H3 have been observed in a number of different species (15). In addition, a dedicated enzyme that specifically acetylates lysines 5 and 12 of newly synthesized H4 before its incorporation into chromatin has been identified in a number of evolutionarily divergent organisms (16–18). The importance of either the N-terminal domain or these histone modifications for nucleosome assembly during S-phase has emerged from elegant molecular genetic experiments in Saccharomyces cerevisiae. Haploid yeast strains containing either truncated H3.H4 N-termini, or the triple point mutation K5G K8G K12G in H4 and an N-terminal deletion of H3, exhibited a defect in nucleosome assembly and accumulated nonviable cells arrested in G2 with a ≈3 N DNA content (19, 20). Thus, although redundant with each other, the N-terminal domains of H3 and H4 collectively play an essential role in maintaining cell viability, presumably through their site-specific acetylation. However, the precise biochemical function of the site-specific acetylation of newly synthesized H3 and H4 is currently unknown.
The human cell-free system for nucleosome assembly during SV40 DNA replication has proven particularly useful to identify CAF-1. However, its use for further analysis of replication-coupled nucleosome assembly has been limited. To overcome these limitations, we took advantage of the fact that newly replicated DNA remains marked as a preferential substrate for CAF-1-mediated nucleosome assembly (14). Here, this “two-step” system is described in detail and it is used to demonstrate that the N-terminal domains of both H3 and H4 were dispensable for preferential assembly of histones onto replicated DNA by CAF-1.
Materials and Methods
Purified Proteins and Antibodies.
SV40 T antigen and DNA topoisomerases I and II were prepared as described previously (14). Recombinant human CAF-1, a complex of p150/p60, p150, and p60 were purified from baculovirus-infected Sf9 insect cells as described (10). The CAF-1 p60 (SS24) and p150 (SS1) monoclonal antibodies were described previously (21).
Affinity Purification of cH3.H4 and Preparation of H3/H4-Depleted S100 Extract.
Complexes containing newly synthesized and acetylated H3.H4 (cH3.H4) were partially purified from an extract from human 293 cells (S100 extract) by incubation for 2 h at 4°C with 1 ml of H4N antibody beads (22). The beads were washed with buffer A100 and eluted at 37°C with 2 ml of buffer A1000 containing 70 μg H4N peptide per ml twice. The elutes were pooled, dialyzed against buffer A50, and concentrated about 5-fold by dehydration in a bed of dry sucrose. H3 and H4 were immunodepleted as described previously (11). Buffer A is 25 mM Tris⋅HCl, pH 8.0, 10% glycerol, 1 mM Na2EDTA, 0.01% Nonidet-P40, and the number following buffer A is the NaCl concentration in millimolar. Each buffer also contained 5 mM sodium butyrate, 1 mM DTT, and a mixture of protease inhibitors (0.1 mM PMSF, 0.1 mM Prefabloc, 1 mM benzamidine· HCl, 1 μg/ml pepstatin A, 1 μg/ml leupeptin).
Preparation of Wild-Type and (H3.H4)2 Tetramers Lacking the N-Terminal Domains of H3 and/or H4.
Expression and purification of wild-type or mutant histones lacking the N-terminal domains of H3 and/or H4 was performed as described (23). H3ΔN lacked amino acid residues 1–23 and contained an S28C amino acid substitution introduced for x-ray crystallography; H4ΔN lacked residues 1–16, residue 1 being the first residue following the initiator methionine. Histone refolding and formation of (H3.H4)2 tetramers was also performed as described (23). To remove misfolded or monomeric H3 or H4 molecules, each recombinant (H3.H4)2 tetramer was purified by gel filtration through a Superdex-200 column in 2 M NaCl, 10 mM Tris⋅HCl (pH 7.5), 1 mM Na-EDTA, 5 mM 2-ME.
Nucleosome Assembly in Two-Step System.
SV40 DNA replication and semipurification of DNA by Miniprep Spun Column (Amersham Pharmacia) is described previously (14). The effluent (approximately 30 fmol of DNA as seen in Figs. 2A and 3 A and B, or 15 fmol of DNA as seen in Fig. 4B) was incubated with CAF-1, S100, or S100(−) extract, affinity-purified cH3.H4 or recombinant (H3.H4)2, and DNA topoisomerases I (13 ng) and II (100 ng). Nucleosome assembly was performed for 40 min at 37°C, followed by an additional 15 min after addition of 400 ng of histone H2A.H2B dimers purified from human 293 cell chromatin (24). For supercoiling assays, DNA samples were treated as described before (25).
Figure 2.
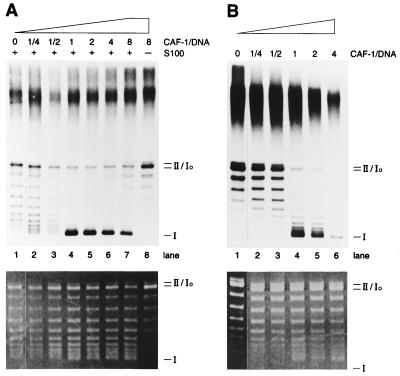
Biochemical analysis of CAF-1-dependent nucleosome assembly. (A) Nucleosome assembly in the two-step system. Replicated and semipurified DNA was subjected to supercoiling reactions with increasing amounts of CAF-1 either in the presence or the absence of S100 (56 μg), as indicated. (B) Nucleosome assembly during SV40 DNA replication. SV40 DNA replication reactions were performed with a large SV40 T-antigen and S100 (87.5 μg) in the presence of increasing amounts of CAF-1 for 45 min. (Upper) An autoradiograph and (Lower) an ethidium bromide-stained gel showing bulk DNA. Molar ratio between CAF-1 and total DNA (not replicated DNA) is indicated in A and B. Migration of relaxed, covalently closed circular DNA (form Io), nicked circular DNA (form II), and negatively supercoiled DNA (form I) are indicated.
Figure 3.
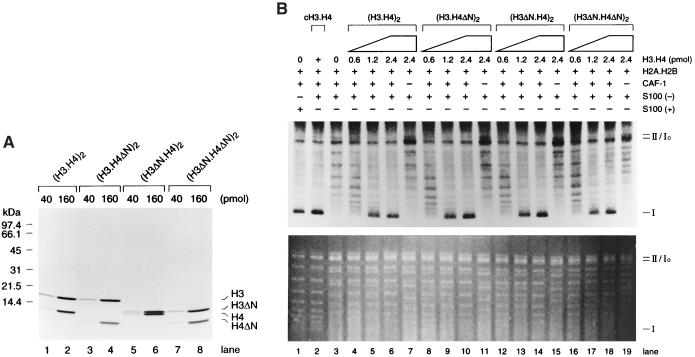
N termini of H3.H4 are not required for CAF-1-dependent nucleosome assembly. (A) A Coomassie blue-stained gel. Four different H3.H4 tetramers (40 or 160 pmol) were loaded on 18% polyacrylamide gel, and stained with Coomassie blue. (B) CAF-1-dependent nucleosome assembly with recombinant (H3.H4)2. A large-scale SV40 DNA replication was performed with H3.H4-depleted human cell extract S100(−). After separation of DNA, supercoiling assay was performed with CAF-1 (50 fmol), S100(−) (48 μg), and increasing amounts of each recombinant (H3.H4)2, as indicated by + or −. However, each recombinant (H3.H4)2 was preincubated at 4°C for 15 min with CAF-1, and subsequently with S100(−) at 37°C for 15 min. (Upper) An autoradiography and (Lower) an ethidium bromide-stained gel. Migrations of form I, Io, or II DNA are indicated as in Fig. 1.
Figure 4.
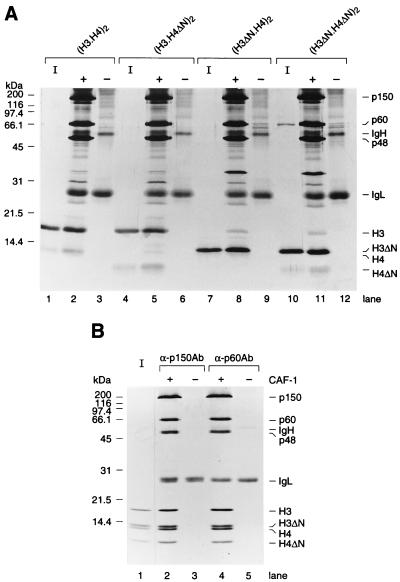
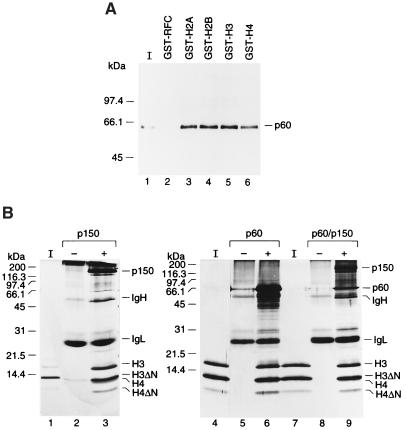
N termini of H3.H4 are not required for association between CAF-1 and H3.H4. (A) Binding between histone H3.H4 and CAF-1. A total of 60 pmol of either wild-type or mutant (H3.H4)2 tetramers was immunoprecipitated by anti-p60 antibody either in the presence or the absence of recombinant CAF-1 (30 pmol), as indicated by + or −. Precipitated proteins were loaded onto an 18% acrylamide gel, and analyzed by silver staining. (B) A competitive binding assay. A total of 300 pmol of wild-type or mutant (H3.H4)2 tetramers were immunoprecipitated by anti-CAF-1 antibodies (anti-p150 antibody for lanes 2 and 3 or anti-p60 antibody for lanes 4 and 5), either in the presence or the absence of CAF-1 (150 pmol), as indicated by + or −. Precipitated proteins were analyzed in an 18% polyacrylamide gel and stained with Coomassie blue. 12.5% of input histones were loaded in lanes I. Three subunits of CAF-1 (p150, p60, and p48), Ig heavy chain (IgH) and light chain (IgL), and each histone H3 and H4 are indicated.
Binding of CAF-1 to Recombinant (H3.H4)2 Tetramers.
The indicated amounts of purified (H3.H4)2 tetramers and recombinant CAF-1, p150, p60, or p150/p60 were incubated for 2 h at 4°C in buffer A400. After incubation with monoclonal antibody beads for 2 h, the beads were washed five times with 600 μl of buffer A400 per wash. Immunoprecipitated proteins were analyzed in SDS-18% polyacrylamide histone gels (26) and analyzed either by silver staining or Coomassie blue staining.
Glutathione S-Transferase (GST)-Histones and GST Pull-Down Assays.
Expression and purification of GST-fused histones in Escherichia coli was performed as described (18, 27). Pull-down assays were performed in 400 μl of buffer A300, with 100 pmol of recombinant p60 and ≈5 μg of each GST-histone immobilized to glutathione-Sepharose beads. After incubation for 2 h at 4°C, beads were washed five times with 600 μl of buffer A300. The amount of p60 remaining bound to beads was determined by SDS/PAGE and immunoblotting.
Results
CAF-1 Assembles Nucleosomes onto Replicated DNA in a Rapid and Stoichiometric Manner.
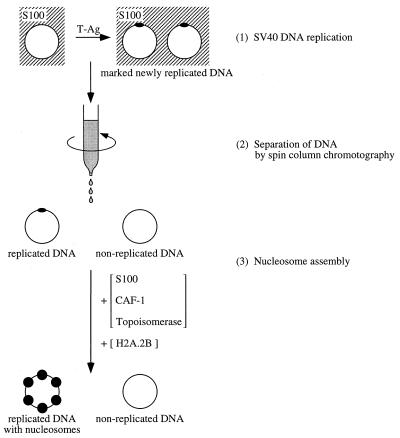
In a recent study, we showed that fully replicated DNA can be purified away from the bulk of free proteins in the S100 extract used for SV40 DNA replication by centrifugation through a gel filtration spin column. This purified replicated DNA remained a good substrate for CAF-1-mediated nucleosome assembly, whereas unreplicated DNA could not be assembled into nucleosomes. This observation allowed us to establish a two-step system for the analysis of replication-coupled nucleosome assembly (14, 28). In this two-step system, replication-coupled nucleosome assembly can be analyzed separately from DNA replication (Fig. 1). Although we do not know whether nucleosome assembly in vivo occurs concomitantly with the passage of replication fork or shortly after DNA synthesis, the nucleosome assembly by CAF-1 seen in our two-step system is clearly postreplicative. Indeed, no DNA synthesis was observed during the nucleosome assembly reaction when labeled dAMP was included in the assay (unpublished data, and ref. 14).
Figure 1.
Scheme of the two-step system for the analysis of replication-coupled nucleosome assembly. Newly replicated DNA purified by spin-column chromatography is competent for replication-dependent nucleosome assembly by CAF-1. In this vitro system, replication-dependent nucleosome assembly is analyzed in a completely separate reaction from the DNA replication reaction. The DNA replication products are first separated from the replication proteins, and then a second nucleosome assembly reaction is performed. To allow nucleosome assembly to proceed, the effluent from the spin column containing DNA is incubated with fresh S100 extract, CAF-1, and topoisomerase activities for various periods of time, followed by additional incubation with H2A.2B for 15 min. Because this is a reconstitution system, S100 extract can be replaced by H3.H4-depleted S100 extract and recombinant H3.H4 tetramers in some experiments (see Fig. 3B).
The amount of CAF-1 required for nucleosome assembly during SV40 DNA replication was compared with the amount necessary to achieve a similar level of nucleosome assembly onto isolated newly replicated DNA in the two-step system. The addition of identical amounts of CAF-1 changed the distribution of the labeled DNA from relaxed circular DNA (form Io) to supercoiled DNA (form I) to very similar extents in both reactions (Fig. 2A and B). Interestingly, in both systems, just a 2-fold increase in CAF-1 concentration resulted in a dramatic increase in supercoiled form I DNA as a result of nucleosome formation (Fig. 2 A and B, lanes 3 and 4). Because approximately 7% of the input DNA was replicated, we estimated that about one molecule of CAF-1 was necessary to form each nucleosome either during SV40 DNA replication (Fig. 2B, lane 4) or during their assembly onto replicated DNA (Fig. 2A, lane 4). Although this calculation was only an approximation, it clearly revealed that CAF-1 did not function in a catalytic manner to promote nucleosome formation in either of these assays.
Previous studies of the kinetics of nucleosome assembly during SV40 DNA replication showed that the appearance of form I DNA occurs after a lag of about 15–30 min (29). This corresponds roughly to the time taken for completely replicated circular DNA to appear in this system (30). A time course of nucleosome formation after the addition of CAF-1 and S100 extract to purified replicated DNA revealed that extensively supercoiled DNA (form I) was first observed between 5 and 10 min and was essentially complete after 15 min. The residual 5- to 10-min delay seen before the appearance of form I DNA could reflect either the time taken for the recruitment of CAF-1-histone complexes to the replicated DNA, the time necessary for the acquisition of nucleosome-constrained superhelical tension or the topoisomerase-mediated relaxation of superhelical tension in the internucleosomal DNA.
CAF-1-Dependent Nucleosome Formation onto Replicated DNA from Core Histone Tetramers Lacking N-Terminal Domains of Both H3 and H4.
In human cells, the bulk of the CAF-1 protein is associated with acetylated H3/H4 (11) and molecular genetic experiments in S. cerevisiae strongly argue that site-specific acetylation of these histones is important for some aspect of nucleosome assembly (20). We therefore sought to determine whether the N-terminal domains of either H3 or H4 were essential for CAF-1-mediated nucleosome assembly onto replicated DNA by the two-step system. For this purpose, we purified core histone tetramers, either with or without the N-terminal domain(s) of H3 and H4, from recombinant histones expressed in E. coli (23). Fig. 3A shows a Coomassie brilliant blue-stained SDS-polyacrylamide gel containing equivalent amounts of each recombinant core (H3.H4)2 tetramer.
Both wild-type and mutant (H3.H4)2 tetramers lacking the N-terminal domains of H3 and/or H4 were mixed with purified CAF-1 to allow them to form complexes, and these complexes were added to a nucleosome assembly reaction containing replicated DNA and the H3/H4-depleted S100 extract [hereafter referred to as S100(−)]. As revealed by the lack of form I DNA, no extensive nucleosome formation took place in the absence of recombinant (H3.H4)2 tetramers (Fig. 3B, lane 3), indicating that the S100(−) extract did not contain residual histones H3/H4 in sufficient amounts to generate supercoiling of the replicated DNA. However, CAF-1-dependent nucleosome assembly, as monitored by preferential supercoiling of replicated DNA, was restored on addition of increasing amounts of any four distinct (H3.H4)2 tetramers (Fig. 3B, lanes 4–19). Importantly, the degree of supercoiling obtained with the different forms of the core histone tetramer, even (H3.H4)2 tetramers lacking the N-terminal domains of both H3 and H4, was very similar. In addition, the products obtained with either wild-type (H3.H4)2 tetramers or tetramers lacking the N-terminal domains of both H3 and H4 also gave rise to short nucleosomal ladders (primarily mono-, di-, and tri-nucleosomes) spaced by about 140 bp (data not shown) on treatment with micrococcal nuclease. These results demonstrated that the N-terminal domains of both H3 and H4 (including all of the known acetylation sites in H3 and H4) were dispensable for CAF-1-dependent nucleosome assembly onto replicated DNA.
The N-terminal Domains of H3 and H4 Are Not Required for Their Interaction with CAF-1.
The result shown in Fig. 3B suggested that CAF-1 binding to (H3.H4)2 tetramers did not require the presence of the N-terminal domains of either H3 or H4. To verify that this was the case, either wild type or mutant (H3.H4)2 tetramers lacking the N-terminal domains were incubated in the presence or absence of purified CAF-1. Histone binding to CAF-1 was detected by immunoprecipitation of CAF-1 with a monoclonal antibody against the p60 subunit. As shown in Fig. 4A, wild-type and mutant (H3.H4)2 tetramers were both precipitated by the CAF-1 p60 monoclonal antibody only in the presence of CAF-1. Because this gel was stained with silver, the H4 band is less prominent than the H3 band (Fig. 4A). This was most likely an artifact of our silver staining protocol, as opposed to reflecting a reduced efficiency of H4 precipitation relative to H3.
In an independent approach, equimolar amounts of wild-type and mutant (H3.H4)2 tetramers lacking the N-terminal domains of both H3 and H4 were incubated in the presence or absence of a substoichiometric amount of CAF-1 (1 mol CAF-1: 2 mol tetramer). The complexes formed were immunoprecipitated with monoclonal antibodies against two distinct subunits of the CAF-1 protein. Fig. 4B shows that equal amounts of both wild-type and mutant (H3.H4)2 tetramer were coprecipitated by different CAF-1 monoclonal antibodies only in the presence of CAF-1. Core histone tetramers were previously shown to be very stable even at relatively low ionic strength in the absence of DNA (31). Under the relatively mild conditions of our binding assays (50 mM Tris⋅HCl, pH 8.0, 400 mM NaCl), the original (H3.H4)2 tetramers were unlikely to dissociate into individual histones to reform mixed tetramers lacking only the N-terminal domain of either H3 or H4. Therefore, the result shown in Fig. 4B demonstrated that, under the conditions of our in vitro competitive binding assay, CAF-1 did not preferentially bind to tetramers containing wild-type histones, even when the mutant tetramers lacked the N-terminal domains of both H3 and H4.
In addition, trypsinized (H3.H4)2 tetramers, in which the N-terminal domains of both H3 and H4 are digested, and CAF-1 cosedimented as a complex during glycerol gradient sedimentation (data not shown). Thus, CAF-1 not only bound to trypsinized (H3.H4)2, but also the complex formed was essentially stable and static during glycerol gradient sedimentation at near physiological ionic strength (25 mM Tris⋅HCl, pH 8.0, 150 mM NaCl). We obtained similar results with either hyperacetylated or hypoacetylated (H3.H4)2 tetramers purified from human cells (data not shown).
All Three Subunits of CAF-1 Are Histone Binding Proteins.
We have argued that two acidic subunits of CAF-1, p150 and p48, have a high affinity for histones (10, 11). However, p60, a subunit of CAF-1 is also a histone binding protein. In Fig. 5A, recombinant p60 was mixed with excess but equal amounts of GST-histones and pulled down by glutathione beads. p60 was precipitated by all GST-histones almost equally well, indicating that p60 has a high affinity for four histones. This interaction is specific because p60 is a neutral protein with calculated pI of 7.27, and GST-RFC37 did not precipitate p60 at all.
Figure 5.
The N termini of H3.H4 are not required for association of p150 or p60 with H3.H4. (A) Binding of p60 to histones. A total of 50 pmol of recombinant p60 was incubated with excess (≈5 μg) GST-histones or GST-RFC (37-kDa subunit of human RFC) and pulled down with glutathione beads. Precipitated proteins were loaded on an 18% polyacrylamide gel and analyzed by Western blotting with anti-p60 monoclonal antibody. 25% of input histones (I) were loaded in lanes 1. (B) Competitive binding assay. A total of 50 pmol of wild-type or mutant (H3.H4)2 tetramers were immunoprecipitated by anti-CAF-1 antibodies (anti-p150 antibody for lanes 2, 3, 8, and 9 or anti-p60 antibody for lanes 5 and 6), either in the presence or absence of recombinant p150, p150/p60 (25 pmol), or p60 (50 pmol), as indicated by + or −. Precipitated proteins were resolved in an 18% polyacrylamide gel and silver stained. 12.5% of input histones (I) were loaded in lane 1. p150, p60, and p48, Ig heavy chain (IgH) and light chain (IgL), and individual histone H3 and H4 are indicated.
In the competitive binding assay, the need for the N-terminal domains of histones H3.H4 for the association with p150 and p60 was determined (Fig. 5B). Equimolar amounts of wild-type and mutant (H3.H4)2 tetramers lacking the N-terminal domains of both H3 and H4 were incubated in the presence or absence of a substoichiometric amount of p150, p60, or p150/p60. The complexes formed were immunoprecipitated with monoclonal antibodies against p150 or p60. Fig. 5B shows that p150, p60, and p150/p60 complex could precipitate equal amounts of both wild-type and mutant (H3.H4)2 tetramer. In a previous study, we showed that p48 binds to helix 1 in the histone-fold domain of H4, and binding of p48 to the helix 1 is largely independent of the proximal region (residues 1–12), which contains the deposition-related acetylation sites (18, 32). Here, we demonstrated that p150 and p60 can bind to histones H3.H4, and the N-terminal domains of H3 or H4 are not required for the binding.
CAF-1 was found to form a complex with newly synthesized histones H3.H4 in living cells, but not with histones H2A.2B (11). However, interestingly, CAF-1 can also bind to histone H2A.2B in vitro and remains associated with H3.H4 on replicated DNA even after the deposition of H3.H4, unlike other histone chaperones like NAP-1 (unpublished data). This observation suggests that CAF-1 on DNA may play some additional role in formation of chromatin than just assembly of histone H3.H4. We predict that CAF-1 might be involved in the deposition of histone H2A.2B dimmers to form a mature nucleosome, in a later deposition-related histone deacetylation reaction, or in recruiting of other chromatin components, like HP1 (33). Reconstitution of replication-coupled chromatin assembly with purified factors by our two step system in vitro and/or structural analysis of CAF-1 will contribute to further understanding of the mechanism of replication-coupled chromatin assembly.
Discussion
Previously, we showed that newly replicated DNA purified by centrifugation through a gel filtration spin column was competent for CAF-1-mediated nucleosome assembly, and that PCNA, a DNA polymerase clamp, functioned as a marker molecule for CAF-1 (14). This suggested that PCNA was involved in chromatin assembly during DNA replication in vivo by recruiting CAF-1 to the sites of DNA synthesis. In this report, we demonstrated that assembly of nucleosomes onto the isolated replicated DNA was facilitated by CAF-1 in a way similar to that seen during SV40 DNA replication.
The two-step system for the analysis of replication-coupled chromatin assembly presents two major advantages over the previous system, where CAF-1 was added during SV40 DNA replication. First, because the SV40 DNA replication step was performed separately from the nucleosome assembly step, the S100 extract could be readily fractionated to identify nucleosome assembly factors. In the past, it was necessary to purify numerous proteins required for reconstitution of SV40 DNA replication in vitro (34), to begin fractionating the S100 extract to identify nucleosome assembly factors. This approach was extremely labor-intensive. We are currently isolating these potential assembly factors using the two-step system. Our preliminary data suggest that the recently identified RCAF (35) is an active component in one of these fractions. Another advantage of the two-step system is that it greatly facilitates immunodepletion experiments. Indeed, the endogenous histone H3/H4 complexes present in the S100 extract could be immunodepleted and functionally replaced by complexes of CAF-1 and recombinant (H3.H4)2 tetramers lacking the N-terminal domains of H3 and/or H4.
Although recombinant core histones expressed in E. coli are not acetylated (36), they were rapidly acetylated during incubation at 37°C in the presence of the H3/H4-depleted S100(−) extract (unpublished data). Therefore, the fact that (H3.H4)2 tetramers lacking the N-terminal domains of both H3 and H4 were as active as tetramers containing full-length histones indicated that, once CAF-1 formed a stable complex with (H3.H4)2 tetramers, the N-terminal domains of H3 and H4 (and, by implication, their acetylation) were essentially dispensable for nucleosome assembly onto replicated DNA. This result stands in sharp contrast with the widespread view expressed in a number of literature reviews (37–40) that CAF-1 only uses acetylated (H3.H4)2 because it is bound to histones with a specific pattern of acetylation in vivo. Furthermore, we demonstrated that CAF-1 did not require the N-terminal domain of either H3 or H4 to bind to (H3.H4)2 tetramers in a complex.
We had reported previously that (H3.H4)2 tetramers isolated from chromatin could not replace the acetylated histones present in the S100 extract, because addition of chromatin-derived H3/H4 resulted in supercoiling of both replicating and nonreplicating DNA in a CAF-1-independent manner (25, 30). However, the precise biochemical reason for this was not clear. In retrospect, it seems likely that, under the low ionic strength conditions required for SV40 DNA replication, the addition of free, chromatin-derived (H3.H4)2 tetramers simply resulted in their nonspecific binding to DNA, which occurred independently of DNA replication. This interpretation is consistent with the fact that, unlike chromatin-derived H3/H4, the histones H3/H4 present in the S100 extract are physically associated with nonhistone chromosomal proteins that decrease their nonspecific affinity for DNA and prevent their binding to phosphocellulose (25). In this paper, we effectively bypassed this problem by preforming complexes of CAF-1 and (H3.H4)2 tetramers, which reduced the nonspecific affinity of H3/H4 for DNA. Although the experiments described here do not rule out the possibility that site-specific acetylation of either H3 or H4 may stimulate, or even be required for de novo nucleosome assembly during DNA replication in vivo, we propose that this histone acetylation may function after assembly, such as marking newly replicated chromatin.
A number of transcriptionally silent genes and heterochromatic loci are known to replicate late during S-phase (41). In Schizosaccharomyces pombe, the functional integrity of heterochromatin depends on maintenance of the histones in a generally hypoacetylated state (42, 43). Thus, if the pool of newly synthesized histones H3/H4 present in late S-phase tends to be substantially less acetylated than those in early S-phase, their assembly onto DNA by CAF-1 would ensure the maintenance of hypoacetylated chromatin at late-replicating loci. However, it is not yet clear whether the histones incorporated into nucleosomes during late S-phase are initially acetylated or not.
Furthermore, the ability of CAF-1 to promote assembly of core histone tetramers into nucleosomes irrespective of their state of acetylation could be physiologically relevant. CAF-1 mutants in S. cerevisiae show the mild UV sensitivity (44), suggesting a potential role of CAF-1 in DNA replication-independent and repair-associated nucleosome assembly in vivo. This was confirmed by the recruitment of CAF-1 to sites of DNA damage repair in UV-irradiated G1 or G2 human cells (13). In S. cerevisiae, a cell-free system for DNA replication-independent nucleosome assembly with extract outside S phase is inhibited by H3/H4 posttranslational modification (45). Therefore, the ability of CAF-1 to promote nucleosome assembly of unacetylated (H3.H4)2 tetramers, as reported here, may account for the involvement of CAF-1 in nucleosome assembly outside S phase in vivo.
Taken together, the data presented in this paper revealed that, once core histone tetramers were bound to CAF-1, the N-terminal domains of both H3 and H4 were essentially dispensable for nucleosome assembly onto replicated DNA. However, importantly, the studies in yeast indicated the redundant roles in nucleosome assembly of the site-specific acetylation of histone H4 and the N-terminal domain of H3 (20). Null mutations in any of the CAC genes encoding subunits of CAF-1 in S. cerevisiae grow normally without significant loss of genome integrity (46). This suggests the existence of at least one CAF-1-independent pathway for replication-coupled nucleosome assembly. One intriguing possibility is that some components of the CAF-1-independent pathway have high affinity to specifically acetylated nascent histones H3.H4 and facilitates their assembly onto newly synthesized DNA preferentially. Further work is needed to elucidate the biochemical function of the evolutionarily conserved posttranslational acetylation of newly synthesized H3 and H4 in nucleosome assembly.
Acknowledgments
We thank Drs. Karolin Lüger and Tim Richmond for their generous gift of purified recombinant H3 and H4, Patty Wendel for purified T antigen, and Dr. Viola Ellison for purified DNA topoisomerase II. K.S. is a research fellow of the Japanese Society for the Promotion of Science (JSPS). A.V. was a postdoctoral fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by Grant CA13106 from the National Institutes of Health.
Abbreviations
- CAF-1
chromatin assembly factor-1
- SV40
simian virus 40
- PCNA
proliferating nuclear antigen
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wolffe A P. J Cell Sci. 1991;99:201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- 2.Sogo J M, Stahl H, Koller T, Knippers R. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 3.Jackson V. Biochemistry. 1988;27:2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 4.Jackson V. Biochemistry. 1987;26:2315–2325. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- 5.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 6.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Nature (London) 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 7.LeRoy G, Orphanides G, Lane W S, Reinberg D. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 8.Graziano V, Gerchman S E, Ramakrishnan V. J Mol Biol. 1988;203:997–1007. doi: 10.1016/0022-2836(88)90124-6. [DOI] [PubMed] [Google Scholar]
- 9.Carruthers L M, Bednar J, Woodcock C L, Hansen J C. Biochemistry. 1998;37:14776–87. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman P D, Kobayashi R, Kessler N, Stillman B. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 11.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 12.Krude T. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 13.Martini E, Roche D M, Marheineke K, Verreault A, Almouzni G. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibahara K, Stillman B. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 15.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 17.Kolle D, Sarg B, Lindner H, Loidl P. FEBS Lett. 1998;421:109–114. doi: 10.1016/s0014-5793(97)01544-5. [DOI] [PubMed] [Google Scholar]
- 18.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 19.Ling X, Harkness T A, Schultz M C, Fisher-Adams G, Grunstein M. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 20.Ma X J, Wu J, Altheim B A, Schultz M C, Grunstein M. Proc Natl Acad Sci USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith S, Stillman B. J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Luger K, Rechsteiner T J, Richmond T J. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 24.Simon R H, Felsenfeld G. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith S, Stillman B. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas J O, Kornberg R D. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- 27.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 28.Fotedar R, Roberts J M. Proc Natl Acad Sci USA. 1989;86:6459–6463. doi: 10.1073/pnas.86.17.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith S, Stillman B. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 30.Stillman B. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 31.Baxevanis A D, Godfrey J E, Moudrianakis E N. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 32.Vermaak D, Wade P A, Jones P L, Shi Y B, Wolffe A P. Mol Cell Biol. 1999;19:5847–5860. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murzina N, Verreault A, Laue E, Stillman B. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 34.Waga S, Bauer G, Stillman B. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 35.Tyler J K, Adams C R, Chen S R, Kobayashi R, Kamakaka R T, Kadonaga J T. Nature (London) 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 36.Luger K, Rechsteiner T J, Flaus A J, Waye M M, Richmond T J. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 37.Roth S Y, Allis C D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 38.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 39.Adams C R, Kamakaka R T. Curr Opin Genet Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 40.Travers A. Curr Biol. 1999;9:R23–5. doi: 10.1016/s0960-9822(99)80037-2. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson J B, Gottschling D E. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grewal S I, Klar A J. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 43.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 44.Game J C, Kaufman P D. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altheim B A, Schultz M C. Proc Natl Acad Sci USA. 1999;96:1345–1350. doi: 10.1073/pnas.96.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]