Abstract
The small heat-shock protein (sHSP) from Methanococcus jannaschii (Mj HSP16.5) forms a homomeric complex of 24 subunits and has an overall structure of a multiwindowed hollow sphere with an external diameter of ≈120 Å and an internal diameter of ≈65 Å with six square “windows” of ≈17 Å across and eight triangular windows of ≈30 Å across. This sHSP has been known to protect other proteins from thermal denaturation. Using purified single-chain monellin as a substrate and a series of methods such as protease digestion, antibody binding, and electron microscopy, we show that the substrates bind to Mj HSP16.5 at a high temperature (80°C) on the outside surface of the sphere and are prevented from forming insoluble substrate aggregates in vitro. Circular dichroism studies suggest that a very small, if any, conformational change occurs in sHSP even at 80°C, but substantial conformational changes of the substrate are required for complex formation at 80°C. Furthermore, deletion mutation studies of Mj HSP16.5 suggest that the N-terminal region of the protein has no structural role but may play an important kinetic role in the assembly of the sphere by “preassembly condensation” of multiple monomers before final assembly of the sphere.
Keywords: stress-induced protein
Small heat-shock proteins (sHSPs) are induced in cells under stress from all three domains of life and have chaperone activity (1–4). The proteins range in size from 12 to 43 kDa and are found as large complexes of 200–800 kDa. A sequence of ≈100 residues homologous to α-crystalline from the vertebrate eye lens is shared among all sHSPs. The sHSP from Methanococcus jannaschii (Mj HSP16.5, 147 residues) contains an α-crystalline domain composed of 90 residues (residues 46–135). Mj HSP16.5 forms a large homomeric complex of 24 subunits with a molecular mass of 400 kDa.
The crystal structure of Mj HSP16.5 revealed a hollow spherical complex of octahedral symmetry with eight trigonal and six square “windows” (5). The structure suggested that the first 32 residues are disordered and that they are inside the sphere. Haley et al. (6) have also shown with cryo-electron microscopy (cryo-EM) that Mj HSP16.5 has very regular and symmetric assemblies and strong octahedral symmetry as in the crystal structure. Cryo-EM reconstruction revealed a lower density inside the sphere presumably corresponding to the flexible N-terminal 32 residues that were not observed in the crystal structure.
In an attempt to understand the nature of complex formation and the mechanism of chaperone activity of Mj HSP16.5, we studied the complex formation between Mj HSP16.5 and single-chain monellin (SCM) using a series of biochemical and biophysical methods. Furthermore, the role of the N- and C-terminal regions outside the α-crystalline domain of the protein was also analyzed by deletion mutation studies.
Materials and Methods
Materials. SCM is an 11.1-kDa protein of high thermal stability (7). Proteinase K (EC 3.4.21.64) was obtained from Sigma. Recombinant Mj HSP16.5 was expressed and purified as described by Kim et al. (8).
Deletion Mutations. The mutants of Mj HSP16.5 lacking N- or C-terminal regions (N-5, N-12, N-31, or C-7) were constructed by oligonucleotide-directed mutagenesis by using the vector pET21a and N- or C-terminal mutagenic primers. In the N-5 deletion, methionine replaced the first six amino acids, resulting in shortening of the wild-type sequence by five amino acids. In the N-12 deletion, the first 13 amino acids were replaced by methionine. In the N-31 deletion, methionine replaced the first 32 amino acids, resulting in shortening of the wild-type sequence by 31 amino acids. In the C-7 deletion, the last seven amino acids were deleted from the C terminus.
PCR was performed by using appropriate primer pairs. The PCR products were cleaned by using Qiagen PCR cleanup kit (Qiagen, Valencia, CA) and eluted with 50 μl of water. A 0.1-μg DNA aliquot was digested with NdeI and BamHI and precipitated with ethanol. The resuspended PCR product (40 ng) was ligated with 40 ng of an NdeI- and BamHI-restricted pET21a at 16°C for 12 h. The ligation mix was digested with 10 units of NheI for 3 h at 37°C to reduce background and then precipitated with ethanol. The ligation products were electroporated into BL21(DE3)pSJS1244 and plated on LB plus ampicillin plates. The mutants were verified by DNA sequencing. Expression and purification were performed according to Kim et al. (8).
Gel Filtration. Analytical size-exclusion chromatography (SEC) was carried out as described (9). Mj HSP16.5 (90 μM) and SCM (90 μM) were mixed in buffer A (25 mM Hepes, pH 7.5/0.1 M NaCl) and analyzed on a TosoHaas (Montgomeryville, PA) TSK G4000SW column (7.5 × 30 cm) at a flow rate of 0.5 ml/min. The two proteins were also combined and heated at 80°C for 20 min and spun down to remove any precipitated protein, and the supernatant was run on the analytical SEC under the same conditions.
EM. The purified Mj HSP16.5 protein and Mj HSP16.5–monellin complex were applied to glow-discharged carbon-coated copper grids. After allowing the proteins to absorb for 2 min, the grids were rinsed on droplets of deionized water and stained with 2% (wt/vol) uranyl acetate. Specimens were examined in the JEOL 2010 at an accelerating voltage of 120 kV by using low-dose unit. Electron micrographs were recorded on Kodak film (SO163) at a magnification of ×37,000 (nominal magnification of ×40,000).
Proteinase K Susceptibility. Proteinase K was dissolved in 50 mM sodium phosphate, pH 7.5, at a concentration of 1 mg/ml and stored at -20°C. Mj HSP16.5 (1 μM) was mixed with SCM (1 μM) in 50 mM sodium phosphate, pH 7.5, at a molar ratio of 1:1 and incubated at 80°C for 20 min. Samples were centrifuged for 10 min at 10,000 × g, and the supernatants were mixed with varying amounts of proteinase K and allowed to incubate on ice for 10 min. Mj HSP16.5 and SCM alone were also treated with proteinase K. Reactions were terminated by the addition of 5 mM phenylmethylsulfonyl fluoride, and the proteins were run on a 12.5% Tricine SDS/PAGE gel followed by Coomassie blue R250 staining.
Western Blot. Proteins were run on a 0.8% agarose gel in native Laemmli buffer as described by Kim et al. (10). The proteins then were transferred onto nitrocellulose paper, and polyclonal antiserum against monellin was used to detect the presence of monellin in the complex formed by SCM and Mj HSP16.5 (11).
Aggregation Assay. Thermal protection of SCM by Mj HSP16.5 and N- and C-terminal deletion mutants were performed following the described procedures (9).
CD. For reproducibility of the results, CD studies were performed at different temperatures both on an Aviv 62DS spectrometer (Aviv Associates, Lakewood, NJ) equipped with a Peltier temperature controller and a Jobin-Yvon (Longjumeau, France) CD6 spectropolarimeter with water bath. Two results were consistent with each other. The experimental conditions for CD6 were as follows: Mj HSP16.5 and SCM were dissolved in 40 mM PBS/0.1 M NaCl, pH 7, at a concentration of 9.0 μM. After 1:1 mixing, the concentrations for both proteins became 4.5 μM. Spectra were measured with cells of 0.1-cm path lengths. For the CD spectrum, data points were taken every 1 nm. The spectra are the average of four scans. For temperature scans, the heating rate was 1°C/min, and equilibrium time was 10 s.
SDS/PAGE. SDS/PAGE was performed by using the discontinuous buffer system as described by Laemmli (12). Protein bands were stained with Coomassie blue R250.
Results
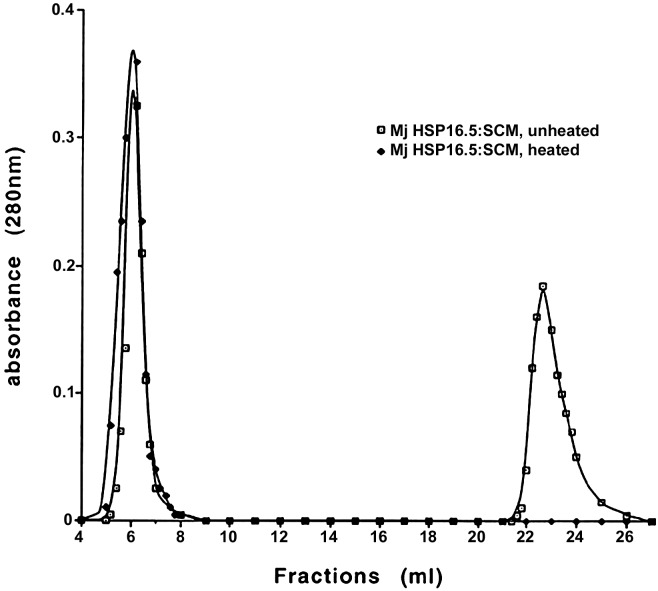
Complex Formation by Mj HSP16.5 and SCM as Detected by SEC and EM. Comparison of the elution profiles from SEC showed that incubation at room temperature of Mj HSP16.5 and SCM at a 1:1 molar ratio did not produce the complex as evident from the presence of two separate peaks, one corresponding to SCM and the other corresponding to Mj HSP16.5. If the two components were heated together at 80°C (a temperature at which SCM is almost fully denatured but Mj HSP16.5 is not) for 20 min and centrifuged, and the supernatant was then applied to the column, only one peak was detected with a slightly broader bandwidth (Fig. 1). This peak was collected and run on 15% SDS/PAGE, and both Mj HSP16.5 and SCM were detected (data not shown).
Fig. 1.
SEC was performed by using a TosoHaas TSK G4000SW column as described in Materials and Methods. Squares, Mj HSP16.5–SCM at a 1:1 molar ratio, unheated; diamonds, Mj HSP16.5–SCM at a 1:1 molar ratio was heated at 80°C for 20 min and centrifuged at 10,000 × g for 10 min, and the supernatant was applied to the sizing column.
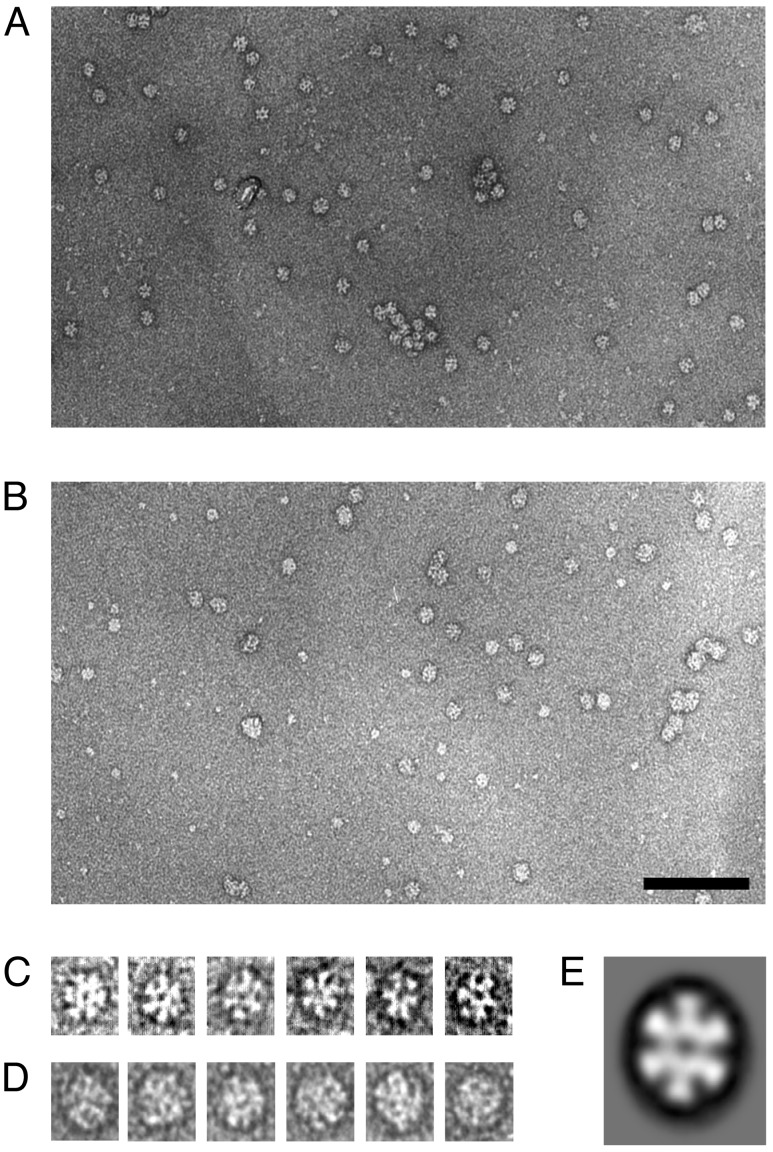
Negative-stain EM of Mj HSP16.5 at ambient temperature shows particle images of ≈13 nm in diameter and appear highly symmetric with symmetries consistent with octahedral geometry (Fig. 2 A and C). The hole in the center of the particle is probably due to the accessibility of stain through the windows observed in the crystal structure (5). In contrast, most of the particle images of Mj HSP16.5 complexed to SCM show a more irregular and increased size with a diameter of ≈16 nm, and the holes are now undetectable, suggesting that all or most of the windows are blocked by SCM, thus preventing the stain from entering the holes (Fig. 2 B and D). These observations are consistent with the notion that SCM binds to the outside surface of the sphere including the sites of the windows.
Fig. 2.
Electron micrographs and averaged particle images of Mj HSP16.5 and Mj HSP16.5 complexed with SCM, a substrate protein, prepared as described in Materials and Methods. (A) Mj HSP16.5. (B) Mj HSP16.5 in complex with SCM. (Scale bar, 100 nm.) (C and D) Gallery of single-particle images selected from a computer graphics display of the micrographs of A and B, respectively.
Susceptibility of SCM to Proteinase K When Bound to Mj HSP16.5. Mj HSP16.5 that had been incubated at 80°C before proteinase K digestion was only slightly susceptible to proteolysis (Fig. 3, lane 4). SCM, incubated at 60°C (the highest temperature to which SCM can be heated without denaturation and precipitation), was completely resistant to proteinase K digestion (Fig. 3, lane 5). When 1 μM SCM was incubated with 1 μM Mj HSP16.5 for 20 min at 80°C (conditions under which a complex is formed) and then subjected to proteinase K digestion for 10 min on ice, most of the SCM was digested by proteinase K, and only a small portion of Mj HSP16.5 was also susceptible to it (Fig. 3, lane 3). These data are also consistent with SCM being bound to Mj HSP16.5 in an unfolded conformation and also supports the idea that the substrate, SCM, is bound on the outside surface of Mj HSP16.5, because the window openings are too small for proteinase K to go inside the sphere.
Fig. 3.
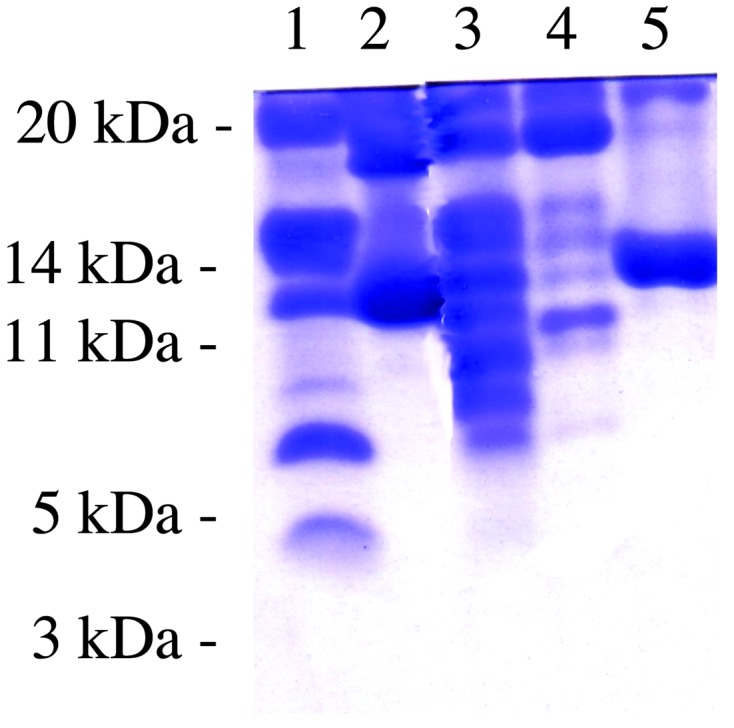
Proteinase K susceptibility. Lane 1, molecular mass standards; lane 2, SCM–Mj HSP16.5 at a 1:1 ratio, unheated; lane 3, SCM–Mj HSP16.5 at a 1:1 ratio, heated at 80°C for 20 min and centrifuged (supernatant was incubated with proteinase K as described in Materials and Methods); lane 4, Mj HSP16.5 was heated at 80°C for 20 min and then treated with proteinase K; lane 5, SCM was heated at 60°C for 20 min and then treated with proteinase K.
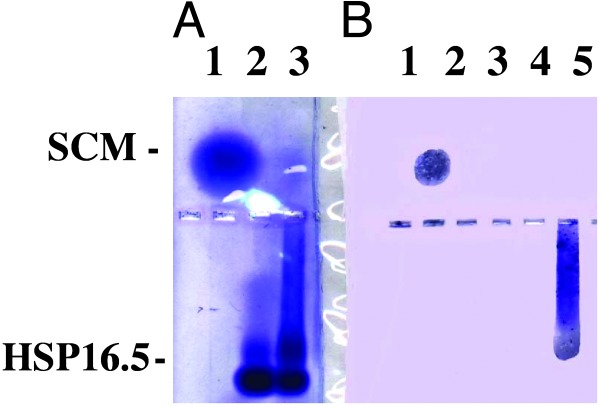
Western Blot of SCM–Mj HSP16.5 Complex. Complex between SCM and Mj HSP16.5 was formed by mixing the two components at a 1:1 molar ratio and heated at 80°C for 20 min. The solution was centrifuged, and the supernatant was loaded on a 0.8% native agarose gel along with uncomplexed SCM and Mj HSP16.5. These samples then were blotted onto nitrocellulose paper and probed with rabbit polyclonal anti-SCM antibody. As shown in Fig. 4, the antibody reacted against SCM alone (B, lane 1) and against the smeared area of the gel where the complex of SCM and Mj HSP16.5 is found, suggesting that SCM is on the outside surface of the sphere and accessible to the antibody (the windows are too small for the antibody to reach the inside of the sphere).
Fig. 4.
Western blot of SCM–Mj HSP16.5 complex. (A) A 0.8% native agarose gel was run as described in Materials and Methods. Lane 1, SCM, 5 μg; lane 2, Mj HSP16.5, 7.4 μg, was heated at 80°C for 20 min and centrifuged for 5 min at 14,000 × g, and the supernatant was loaded; lane 3, SCM–Mj HSP16.5 (1:1 molar ratio) was heated at 80°C for 20 min and centrifuged for 5 min at 14,000 × g, and the supernatant was loaded. (B) A portion of the gel from A was transferred onto nitrocellulose paper and probed with polyclonal antiserum raised against SCM. Lane 1, SCM, 5 μg; lane 3, Mj HSP16.5, 7.4 μg, was heated at 80°C for 20 min and centrifuged for 5 min at 14,000 × g, and the supernatant was loaded; lane 5, SCM–Mj HSP16.5 (1:1 molar ratio) was heated at 80°C for 20 min and centrifuged for 5 min at 14,000 × g, and the supernatant was loaded.
Deletion Mutations of Mj HSP16.5. Cryo-EM (6) as well as the x-ray structure of Mj HSP16.5 (9) show that the N-terminal region (≈27–32 residues) are seen as weak internal density in the cavity of the sphere and are disordered. The N-5 deletion mutant protein behaved like the wild-type protein in regards to protection of the Escherichia coli cell extract or SCM from heat denaturation as well as its behavior on SEC (Table 1). The C-7 deletion mutant, however, behaved like the wild-type protein in protecting the E. coli cell extract and SCM from heat denaturation, but on SEC it migrated as a tetramer. The N-12 deletion mutant behaved very differently from the other mutants. It did not protect the E. coli cell extract from denaturation. After expressing N-12, the cell pellet was sonicated and spun to remove the cell debris. The cleared lysate formed a gel that could be liquefied in the presence of 0.1 M NaCl or if the pH was raised to 11, but after storage of the liquefied cleared lysate at 4°C, the cleared lysate would form a gel again. The only way to purify this mutant was to treat it with 7 M urea. After dialyzing the urea out, this sample migrated as a tetramer on SEC and did not protect SCM from heat denaturation in vitro.
Table 1. Effect of deletion mutations on sHSP activity and size.
| Constructs of Mj HSP16.5* | Protection of E. coli cell extract in vivo† | Protection of SCM in vitro‡ | SEC size§, kDa |
|---|---|---|---|
| Wild type | 100% | 100% | 400 |
| N-5 | 100% | 100% | 400 |
| N-12 | No protection | No protection | 66 |
| N-31 | No protection | Not done | 669 (misfolded aggregate) 29 |
| C-7 | 100% | 100% | 66 |
Deletions were made as described in Materials and Methods
E. coli cells expressing each of the constructs were sonicated and spun, and the cell lysates were heated at 80°C for 20 min. The samples were centrifuged at 10,000 × g for 5 min, and the supernatants and pellets were analyzed by SDS/PAGE (9)
Thermal protection of SCM by purified Mj HSP16.5 protein from each of the constructs was performed as described by Kim et al. (9)
SEC was performed by using a TosoHaas TSK G4000SW column as described in Materials and Methods
Cells expressing the N-31 mutant did not lyse readily. A large part of expressed N-31 protein was found to be insoluble. The soluble fraction was purified further and found to migrate as two peaks: a large aggregate of 669 kDa and a peak of ≈29 kDa. The N-31 mutant did not provide heat protection of the E. coli cell extract.
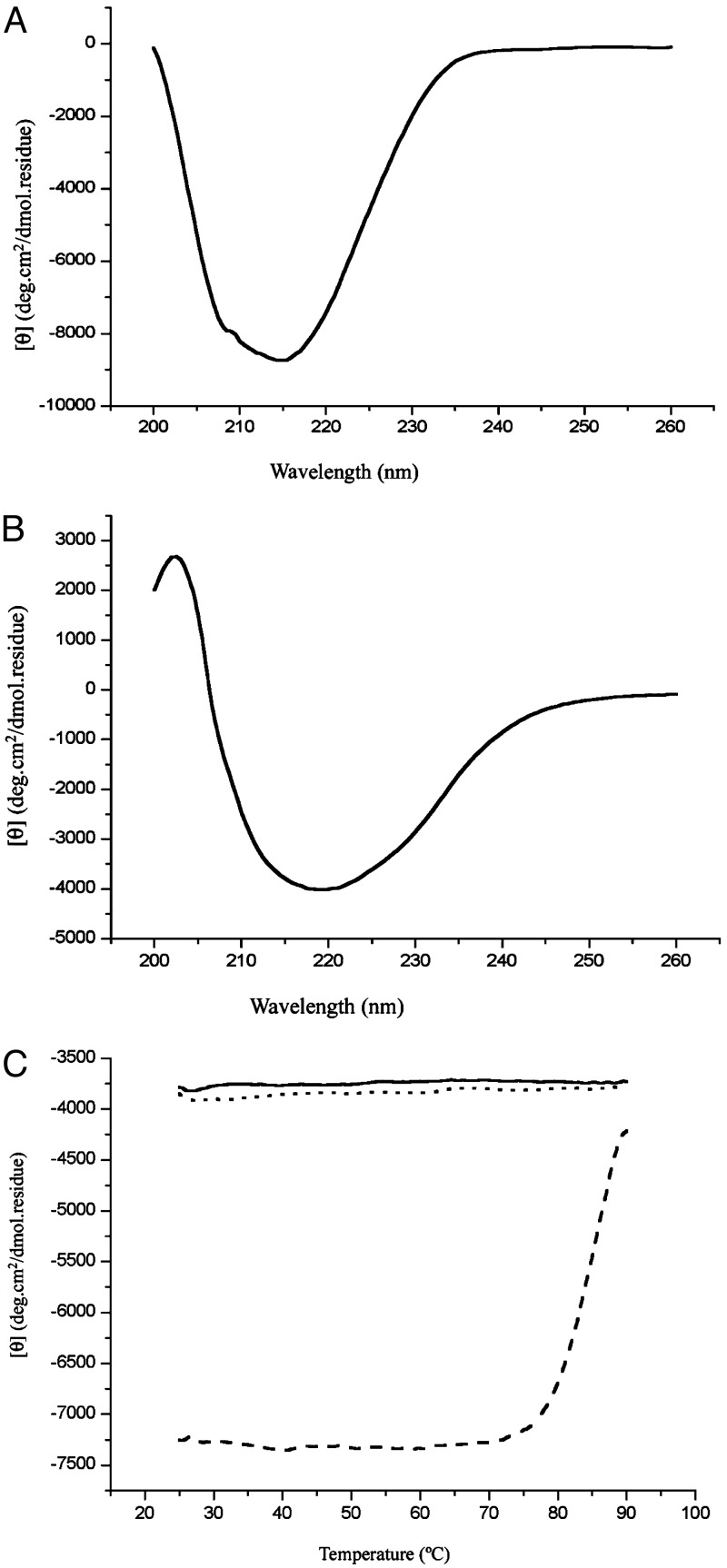
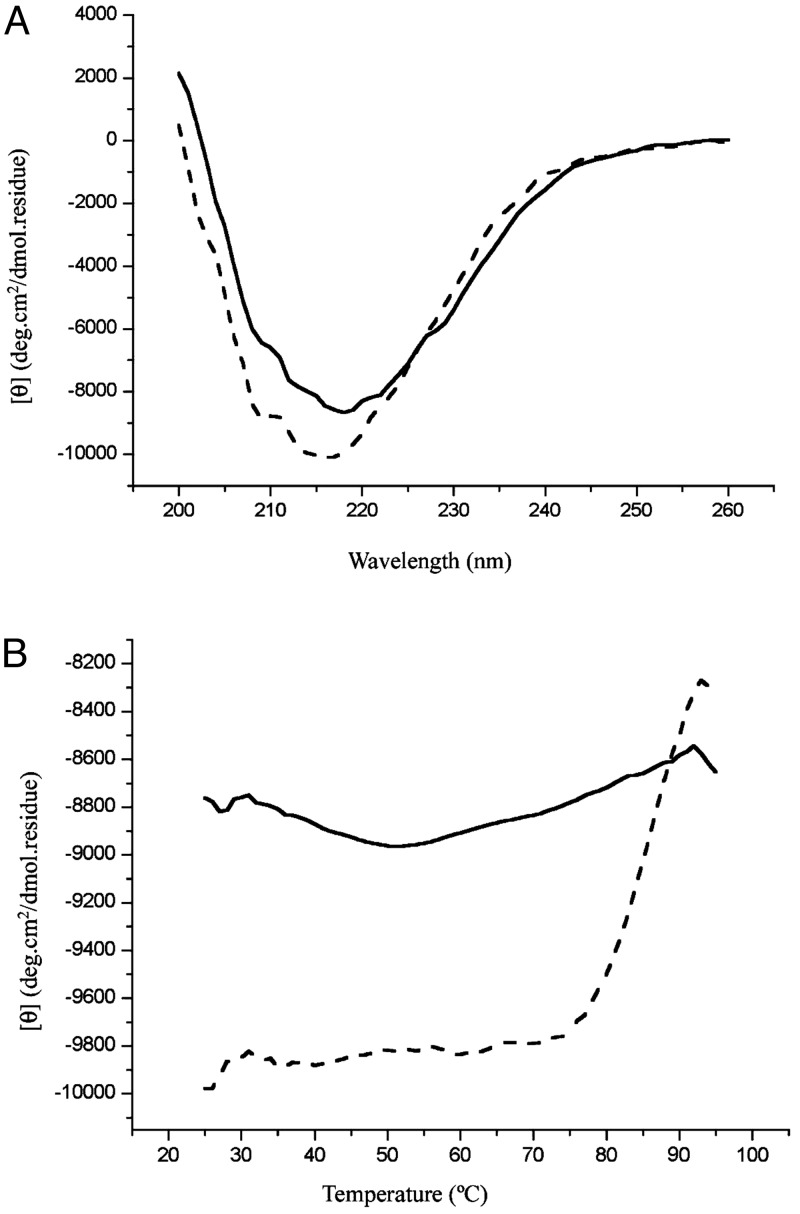
No Significant Conformational Changes of sHSP After Substrate Binding. EM studies of the apo complex and the SCM-bound complex showed that, although the SCM-bound complexes are larger than the apo complex in general and are more heterogeneous in size, the overall spherical shape is present in both. To assess potential conformational changes that may occur after substrate binding, we characterized Mj HSP16.5 and SCM individually as well as in complex by CD at different temperatures. First, thermal denaturation experiments were performed to qualitatively assess the conformational stability of the two proteins individually. Fig. 5A shows the CD wavelength scan of SCM, demonstrating that the structure is composed of mainly β-sheets at room temperature, which is consistent with the crystal structure of SCM having a five-stranded β-sheet and one α-helix (13). SCM starts melting at ≈70°C with a melting temperature of ≈78°C (Fig. 5C). The CD spectrum for Mj HSP16.5 also shows a structure consisting primarily of β-structure at room temperature (Fig. 5B), as expected from the crystal structure of Mj HSP16.5 (5). A heat-denaturation curve of Mj HSP16.5 monitored at 218 and 220 nm showed only background noise (Fig. 5C, solid and dotted lines, respectively). At a 1:1 molar ratio of Mj HSP16.5 and SCM, the CD spectra of the mixture (Fig. 6A, dotted line) were about the same as the sum of the two individual components at 25°C, indicating that at this temperature there are no detectable conformational changes after mixing, consistent with the fact that, at this temperature, the complex does not form (as mentioned earlier). The mixture preheated at 80°C then cooled to 25°C had a CD (Fig. 6A, solid line) similar to that of the sum of the CDs of the intact sHSP and denatured SCM. The heat-denaturation curve of the mixture of sHSP and SCM without preheating (Fig. 6B, dotted line) is very similar to that of SCM alone (Fig. 5C, dotted line), but the mixture preheated to form the complex had very little melting behavior (Fig. 6B, solid line), suggesting that SCM remains denatured once it is complexed as denatured SCM. Similar results were obtained at a 4:1 molar ratio of Mj HSP16.5–SCM (data not shown). All these results from the EM and SEC, protease-susceptibility, antibody-binding, and CD studies strongly imply that partially or fully denatured SCMs bind to sHSP, which experiences no or only small conformational changes.
Fig. 5.
CD spectra for SCM and Mj HSP16.5. (A) CD spectra of SCM at 25°C. (B) Thermal melting of SCM. The CD signal at 218 nm is plotted as a function of temperature. (C) CD spectra of Mj HSP16.5 at 25°C. (D) Thermal melting of Mj HSP16.5. The CD signal at 218 nm is plotted as a solid line; at 220 nm it is plotted as a dashed line.
Fig. 6.
CD spectra before and after mixing SCM and Mj HSP16.5. (A) CD spectra of SCM and Mj HSP16.5 at a 1:1 molar ratio at 25°C. Dashed line, before mixing, where two solutions are separated by a divider; solid line, after mixing and heating. (B) Thermal melting of SCM–Mj HSP16.5 at a 1:1 molar ratio. Dashed line, before mixing; solid line, after mixing. The CD signal at 218 nm is plotted as a function of temperature.
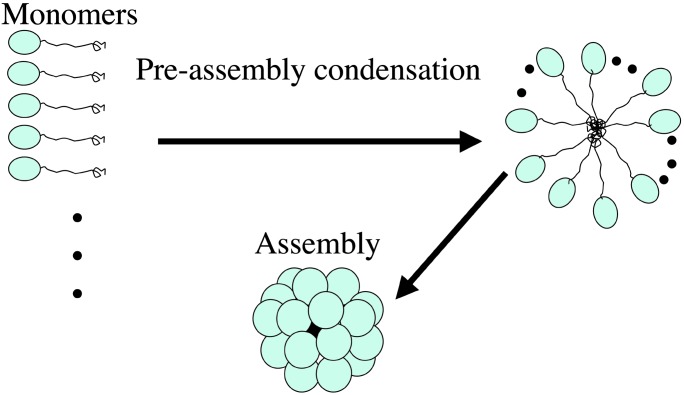
Role of the N- and C-Terminal Regions. All sHSPs contain the conserved α-crystalline domain and nonconserved N- and C-terminal regions. The crystal structure of Mj HSP16.5 revealed that the C-terminal region is heavily involved in additional stabilization of subunit interaction, suggesting that it may play a role in forming the types of multimeric complexes. The role of the N-terminal region is not clear from the structure, because it is disordered inside the cavity, suggesting that the N-terminal region does not play an important role in the architecture of the sphere. However, the mutant protein lacking the first 12 amino acids at the N terminus failed to assemble the sphere, suggesting that the N-terminal region still plays a role in the process of assembly. Because the N terminus seems to have no structural role (14), it may have a kinetic role in the assembly process of the sphere. One possible scenario is that, because the N-terminal disordered region has many hydrophobic residues, it may act as a hydrophobic tether of the folded α-crystalline domain, and many folded α-crystalline domains are brought together by the aggregation of many tethers. We propose that this “preassembly condensation” provides an entropically favorable situation for the kinetics of final assembly of the sphere (Fig. 7).
Fig. 7.
Model for assembly of Mj HSP16.5. The N-terminal region of Mj HSP16.5 acts as a hydrophobic tether of the folded α-crystalline domain. Many such tethered α-crystalline domains are brought together by the hydrophobic “glue” of the tethers to form a large multimer, a preassembly condensation process. With many subunits in proximity, the final assembly of the sphere is facilitated.
Conclusions
Initially, the hollow spherical structure of Mj HSP16.5 containing a cavity led us to propose a chaperone mechanism: During the assembly of the sphere in response to external stress, the sphere may trap inside the cavity some enzymes important for survival under stress and thus be protected from denaturation. These essential enzymes could still be functional, because most substrates and products could travel in and out of the cavity through the many windows found on the surface of the hollow sphere. However, some independent experimental evidence described here points to the fact that the substrates bind to the outside surface of the sphere. This conclusion suggests a different mechanism for preventing denaturation of substrate proteins by this sHSP.
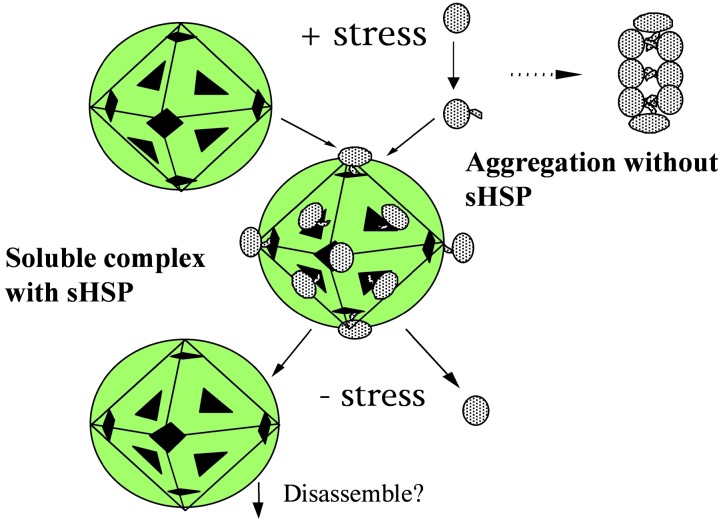
A mechanism we propose is presented schematically in Fig. 8. Most proteins in the cell become partially denatured under stress, exposing some hydrophobic surfaces, which then attract similar hydrophobic surfaces from the same protein or other proteins, resulting in aggregation and possibly precipitation of the proteins (15, 16). We propose that in the presence of sHSP, these partially denatured proteins are attracted to the surface of the sphere near the windows (because the amino acid residues on the inside surface as well as in the cavity are more hydrophobic than normal soluble proteins) and other partially hydrophobic patches and are prevented from aggregating among themselves. These attached proteins, as a part of the soluble complex, may be able to function during stress. As the stress is removed, the proteins renature and may fall off from the sphere because of the renaturation process or the disassembly of the sphere by a process yet to be discovered.
Fig. 8.
Schematic representation of the interaction of protein substrate with Mj HSP16.5. In the absence of sHSP, stress induces proteins to partially denature and expose hydrophobic regions that stick together to form protein aggregates and precipitation. In the presence of sHSP, these partially denatured proteins are attracted to the surface of the sphere and are prevented from aggregating to themselves. After release of stress, the substrate is released from the sphere and refolded.
Acknowledgments
We thank Ken Downing (Lawrence Berkeley National Laboratory) for scanning the EM negatives for an independent check. This work was supported by the director of the Office of Energy Research, Office of Biological and Environmental Research, of the U.S. Department of Energy under Contract DE-AC03-76SF0098 and National Institutes of Health Grant GM 62412 (to R.K. and S.-H.K.).
Abbreviations: sHSP, small heat-shock protein; Mj HSP16.5, sHSP from Methanococcus jannaschii; EM, electron microscopy; SCM, single-chain monellin; SEC, size-exclusion chromatography.
References
- 1.van den Ijssel, P., Norman, D. G. & Quinlan, R. A. (1999) Curr. Biol. 119 R103-R105. [DOI] [PubMed] [Google Scholar]
- 2.Saibil, H. (2000) Curr. Opin. Struct. Biol. 10 251-258. [DOI] [PubMed] [Google Scholar]
- 3.Clark, J. I. & Muchowski, Z. (2000) Curr. Opin. Struct. Biol. 10 52-59. [DOI] [PubMed] [Google Scholar]
- 4.Lee, G. J., Roseman, A. M., Saibil, H. R. & Vierling, E. (1997) EMBO J. 16 659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, K. K., Kim, R. & Kim, S.-H. (1998) Nature 394 595-599. [DOI] [PubMed] [Google Scholar]
- 6.Haley, D. A., Bova, M. P., Huang, Q. L., Mchaourab, H. S. & Stewart, P. L. (2000) J. Mol. Biol. 298 261-272. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S.-H., Kang, C.-H., Kim, R., Cho, J. M., Lee, Y. B. & Lee, T. K. (1989) Protein Eng. 2 571-575. [DOI] [PubMed] [Google Scholar]
- 8.Kim, K. K., Yokota, H. & Kim, S.-H. (1998) J. Struct. Biol. 121 76-80. [DOI] [PubMed] [Google Scholar]
- 9.Kim, R., Kim, K. K., Yokota, H. & Kim, S.-H. (1998) Proc. Natl. Acad. Sci. USA 95 9129-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, R., Yokota, H. & Kim, S.-H. (2000) Anal. Biochem. 282 147-149. [DOI] [PubMed] [Google Scholar]
- 11.Towbin, H., Staehelin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76 4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. (1970) Nature 227 680-685. [DOI] [PubMed] [Google Scholar]
- 13.Somoza, J. R., Jiang, F., Tong, L., Kang, C.-H., Cho, J. M. & Kim, S.-H. (1993) J. Mol. Biol. 234 390-404. [DOI] [PubMed] [Google Scholar]
- 14.Koteiche, H. A. & Mchaourab, H. S. (2002) FEBS Lett. 519 16-22. [DOI] [PubMed] [Google Scholar]
- 15.Yang, H., Huang, S., Dai, H., Gong, Y., Zheng, C. & Chang, Z. (1999) Protein Sci. 8 174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgio, M. R., Kim, C. J., Dow, C. C. & Koretz, J. F. (2000) Biochem. Biophys. Res. Commun. 268 426-432. [DOI] [PubMed] [Google Scholar]