Abstract
The segregating unit of mtDNA is a protein–DNA complex called the nucleoid. In an effort to understand how nucleoid proteins contribute to mtDNA organization and inheritance, we have developed an in organello formaldehyde crosslinking procedure to identify proteins associated with mtDNA. Using highly purified mitochondria, we observed a time-dependent crosslinking of protein to mtDNA as determined by sedimentation through isopycnic cesium chloride gradients. We detected ≈20 proteins crosslinked to mtDNA and identified 11, mostly by mass spectrometry. Among them is Abf2p, an abundant, high-mobility group protein that is known to function in nucleoid morphology, and in mtDNA transactions. In addition to several other proteins with known DNA binding properties or that function in mtDNA maintenance, we identified other mtDNA-associated proteins that were not anticipated, such as the molecular chaperone Hsp60p and a Krebs cycle protein, Kgd2p. Genetic experiments indicate that hsp60-ts mutants have a petite-inducing phenotype at the permissive temperature and that a kgd2Δ mutation increases the petite-inducing phenotype of an abf2Δ mutation. Crosslinking and DNA gel shift experiments show that Hsp60p binds to single-stranded DNA with high specificity for the template strand of a putative origin of mtDNA replication. These data identify bifunctional proteins that participate in the stability of ρ+ mtDNA.
Maintenance of respiratory function in eukaryotic cells requires accurate expression and faithful transmission of mtDNA. Most eukaroytic cells contain multiple copies of mtDNA that are organized as protein–DNA complexes called nucleoids. In haploid cells of Saccharomyces cerevisiae, there are some 10–20 nucleoids per cell that are easily visualized with DNA-specific dyes, such as 4′,6′-diamino-2-phenylindole. Data from many laboratories indicate that the nucleoid is the fundamental segregating unit of mtDNA. Therefore, to achieve a detailed understanding of mtDNA inheritance, nucleoid proteins must be identified and their roles investigated.
Nucleoids have been isolated from yeast by solubilizing mitochondria with detergents and purifying those complexes by sedimention (1–3). By this method, roughly 20 polypeptide species are enriched, but only a few of them have been identified. Known nucleoid proteins include Abf2p, an abundant, high-mobility group protein required for the stability of wild-type (ρ+) mtDNA in cells grown on fermentable carbon sources (4), as well as for efficient mtDNA recombination (5, 6), and Mgm101p, a protein possibly involved in mtDNA repair that is also required for the stability of ρ+ mtDNA (7, 8). Cells lacking Abf2p can maintain ρ+ mtDNA when grown on nonfermentable carbon sources, and based on morphological and biochemical criteria, the mtDNA is organized differently than in wild-type cells (2, 5). In addition, the sorting of mtDNA in zygotes derived from abf2Δ parents is altered in comparison to sorting in wild-type (ABF2) crosses (9).
Because of uncertainties about the stability of protein–mtDNA interactions during purification of these nucleoids, we developed a formaldehyde crosslinking procedure to identify proteins that are in close association with mtDNA in organello. Formaldehyde is membrane soluble and produces protein–protein, protein–RNA, and protein–DNA crosslinks (10). Formaldehyde crosslinks DNA-binding proteins (such as histones and transcription factors) to DNA in vivo and in vitro, whereas free protein is not crosslinked to DNA in vitro, even when present at high concentrations (11, 12). Specific crosslinks occur between primary amines that are within 2 Å of one another and are easily reversible (10). Although in vivo formaldehyde crosslinking is routinely used in conjunction with immunoprecipitation techniques to identify DNA sequences bound by specific proteins (9), it also has been used to recover proteins associated with kinetoplast DNA of trypanosomes (13).
We describe here the identification of 11 proteins (10 by mass spectrometry) that are formaldehyde crosslinked to mtDNA in organello. Some of these proteins have been shown previously to interact with mtDNA or to function in mtDNA maintenance, but others, such as the mitochondrial chaperone Hsp60p and the tricarboxylic acid (TCA) cycle protein Kgd2p, were not previously known to interact with mtDNA. Genetic experiments described here implicate both Hsp60p and Kgd2p in the stability of ρ+ mtDNA. Biochemical experiments show, surprisingly, that Hsp60p is a single-stranded (ss) DNA binding protein that binds specifically and with strand specificity to a putative origin of mtDNA replication.
Materials and Methods
Yeast Strains and Media.
Initial experiments used MATa and MATα forms of strain 14WW (ade2 leu2 trp1 ura3-52 cit1∷LEU2) with ρ+ mtDNA of strain D273-10B (2). Null alleles of ABF2 (abf2∷TRP1) and KGD2 (kgd2∷HIS3) were made in strains αW303-1B and aW303-1A (ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100) (14), respectively, using standard methods. Mutants of HSP60 were generated by random or site-specific mutagenesis of the cloned gene and transformed into αW303, and colonies were screened to identify temperature-sensitive mutants. Cells were grown in YP (1% yeast extract, 2% bactopeptone) or YNB + cas (0.67% yeast nitrogen base/1% casamino acids) medium containing 2% dextrose (D), 3% glycerol (Gly), or 2% ethanol (E).
Purification of Mitochondria and Formaldehyde Crosslinked Nucleoids.
Cells (a14WW ρ+) from a 22-liter culture were grown in YPGly medium (OD 1–2), harvested, washed in SCE (0.6 M sorbitol/0.3 M mannitol/20 mM K2HPO4/20 mM citric acid/1 mM EDTA), pH 5.8, containing 5 μl/ml β-mercaptoethanol, and digested with 0.5 mg Zymolyase 100T (ICN) per g of cells (wet weight) in SCE, pH 5.8. Cells were washed with SCE, pH 7.0, and were broken either osmotically (in 15% sucrose, 10 mM Tricine⋅KOH, 0.2 mM EDTA, pH 7.5) or with glass beads (in SCE, pH 7.0). Nuclei and cellular debris were removed by centrifugation (2,500 × g, 10 min) until no further material could be pelleted. Mitochondria then were pelleted (17,000 × g, 15 min), resuspended in 65% (wt/vol) sucrose in floatation buffer (FB) (10 mM Tricine⋅KOH/0.1 mM EDTA/50 mM NaCl, pH 7.5), overlayed with 15%, 43%, and 55% (wt/vol) sucrose steps and centrifuged at 40,000 × g for 1.5 h. Mitochondria were collected from the 43%/55% interface, diluted in 15% sucrose in FB, and pelleted as above. Mitochondria were resuspended (10 mg/ml) in FB containing 15% sucrose, adjusted to 40 mM Tris⋅HCl, pH 8.0, and 2.5 mM MgCl2 and treated with DNase I (1 unit/50 μg) for 1 h on ice. The reaction was terminated with 5 mM EDTA, pH 8.0; the mitochondria were pelleted, washed with resuspension buffer (RB) (0.5 M sucrose/20 mM Hepes, pH 7.6/2 mM EDTA/1 mM EGTA), and diluted to 2 mg/ml in crosslinking buffer (CB) [RB plus 50 mM Hepes/50 mM NaCl/7 mM β-mercaptoethanol/1 mM PMSF, 1× Protease Inhibitor mixture (Roche Molecular Biochemicals)/1 mM spermidine]. Formaldehyde was added to 1% and incubated at 4°C for 16 h with slow mixing. The reaction was quenched with 125 mM glycine, pH 7.0, and the mitochondria were pelleted, resuspended in CB plus 50 mM glycine, and lysed with 0.5% NP-40. The lysate was diluted 3-fold, and the nucleoid-enriched pellet was collected by centrifugation (110,000 × g, 1 h) through 20% sucrose (0.5% NP-40/50 mM Hepes, pH 7.6/50 mM NaCl). The pellet was resuspended in CB using a dounce homogenizer and treated with 1% sarkosyl and RNase A (50 μg/ml) for 1 h at room temperature. The mixture was adjusted to RI = 1.365 with CsCl (in TE, 1% sarkosyl), and 5–15 mg of the purified mitochondria was loaded into 11-ml tubes. Gradients were centrifuged (260,000 × g, 16 h, 25°C) in a fixed angle rotor and fractionated into either 0.5-ml or 1-ml fractions. Some experiments included a second CsCl gradient.
Protein Analysis.
SDS/PAGE gels and Western blots were carried out (2) by using 5 μl of each fraction (Fig. 1C) or after precipitation (0.3 M sodium acetate and 1 vol of isopropanol at room temperature with linear acrylamide added as a coprecipitate). Precipitates were resuspended in 3× SDS loading buffer (Bio-Rad) containing 5% β-mercaptoethanol and heated at 95°C for at least 30 min with intermittent vortexing. Rabbit antibodies were raised against a pMAL-Ilv5p fusion protein using standard methods. Polyclonal antisera against Abf2p, Ilv5p, Hsp60p, and Aco1p were used at 1/20,000; 1/5,000; 1/10,000; and 1/1,000 dilution, respectively. Gels were fixed overnight in 50% methanol and 10% glacial acetic acid, stained with Fast Stain (Zoion Biotech, Newton MA), destained with 10% glacial acetic acid, and dried by using a Novex drying kit.
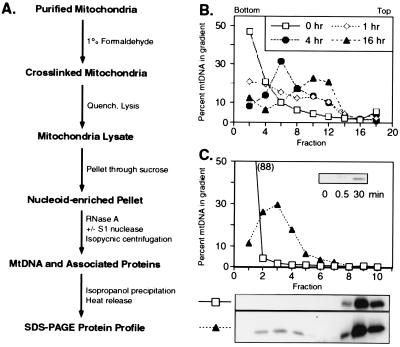
Figure 1.
Purification of in organello formaldehyde-fixed mtDNA–protein complexes. (A) Flow diagram of the formaldehyde crosslinking procedure. (B) Time course of in organello crosslinking. Mitochondria were treated for increasing lengths of time with 1% formaldehyde, quenched with glycine, and lysed with sarkosyl. The lysate was applied directly to CsCl gradients (RI = 1.37) and, after centrifugation, the distribution of mtDNA was determined by dot blots using a COXII probe. (C) Formaldehyde-dependent crosslinking of Abf2p to mtDNA. mtDNA, crosslinked for 16 h and prepared as described in A (▴), was separated from free (untreated) mtDNA (□) in CsCl gradients (RI = 1.365). In addition to the presence of free Abf2p at the top of the gradient (fractions 8–10), some Abf2p cosediments with mtDNA (peak = fraction 3) after formaldehyde treatment and is released by a 30-min heat treatment, but not by SDS alone or SDS and 0.5 min heating (Inset).
Protein Identification.
Standard methods (15) were used to prepare gel fragments for analysis in a PE Biosystems Voyager-DE matrix-assisted laser desorption ionization–time of flight mass spectrometer. Peptide mass values were searched against the S. cerevisiae NCBInr database by using the ms-fit routine (http://prospector.ucsf.edu). The number of peptides and percent coverage are as follows: Rim1p, seven peptides, 61% coverage; Abf2p, 14 peptides, 59% coverage; Mgm101p, 11 peptides, 49% coverage; Ilv5p, eight peptides, 45% coverage; Kgd2p, nine peptides, 34% coverage; Lpd1, six peptides, 18% coverage; Atp1p, 16 peptides, 43% coverage; Hsp60p, 15 peptides, 37% coverage; Ald4p, 19 peptides, 57% coverage; Aco1p, 19 peptides, 32% coverage. Hsp60p, Ilv5p, Aco1p, and Abf2p were confirmed and Hsp10p was identified by Western blotting. Tryptic peptides were purified on a prototype Waters Capillary HPLC system for partial amino acid sequence analysis by automated Edman degradation using a PE Biosystems Procise-HT Sequencer (Ald4p, VAFTGSTATG; Ilv5p, NSDLYGER, EFNSQ; Kgd2p, AQEPPVASNSF) or by LC/MS/MS using a ThermoQuest (San Jose, CA) LCQ-Deca ion trap mass spectrometer (Lpd1p, INVANFQK; Atp1p, AQPTEVSSILEER, GVSDEANLNETGR).
Construction and Expression of Hsp60p-6His.
PCR was used to introduce NcoI and XhoI sites in HSP60 (downstream of the amino terminal transit sequence and immediately before the termination codon, respectively) with primers 5′-GCCCCATGGCTTCCTCTCATAAAGAATTG-3′ and 5′-AATTTTGAATTAAGGCGGCTCGAGCATCATACCTGGCATTCC-3′. The 1.5-kb PCR product was cleaved with NcoI and XhoI (sites indicated) and cloned into pET-24d (Novagen) to provide a C-terminal 6-histidine tag. The recombinant Hsp60p-6His polypeptide lacks the amino-terminal mitochondrial targetting sequence but has an additional alanine residue before serine 22, which is the first amino acid of the mature Hsp60p (16). Hsp60p-6His was overexpressed in Escherichia coli BL21(DE3) and soluble extracts were prepared by lysozyme, sonication, and Ni2+ chromatography according to the manufacturer (Qiagen, Chatsworth, CA).
Electrophoretic Mobility Shift Assay.
Protein extracts were incubated with the DNA substrates (0.5 ng) in 10 mM Tris (pH 7.4), 4 mM MgCl2, 0.5 mM EDTA, and 50 mM KCl at 25°C for 30 min and resolved on a 3.5% (29.2:0.8) polyacrylamide gel in 0.25× TBE. All DNA fragments were end-labeled by using T4 polynucleotide kinase and [γ-32P]ATP, and include a 0.5-kb DraI–NdeI fragment from the ρ− hypersuppressive petite genome, HS40 (17) for ori5, a 670-bp fragment amplified from the ρ+ mtDNA of strain D273-10B using the primer pair 5′-CCTGACATAGAGACAATAATTAGAACTTC-3′ and 5′-TATATATATATATCTTATTTTATATATAC-3′ (18) for ori6, a 650-bp HindIII–NsiI fragment from the ρ− strain 5D-2–33 (19) for VAR1 and a 0.4-kb HincII–BamHI internal fragment of the 14S rRNA gene (rDNA). In some experiments, the above fragments were cloned into pGEM-3Zf(+) (Promega) and PCR products were made by using the T7 and SP6 primers. Probes were made ss by boiling double-stranded (ds) DNA probes, followed by quick cooling on ice.
Genetic Analysis.
The ρ+ strains kgd2Δ and abf2Δ were mated; diploids were selected, grown on solid presporulation medium, and sporulated in liquid medium plus supplements, and tetrads were dissected on yeast extract/peptone/dextrose (YPD) plates by using a Singer MSM (Singer Instruments, Sumerset, U.K.). To visualize the frequencies of petite spores, colonies from tetratype tetrads were picked, grown in YPD liquid medium for 8 h, and equal numbers of cells spotted onto YPD and 3-fold more cells onto yeast extract/peptone/ethanol (YPE) medium.
Results
In Organello Formaldehyde Crosslinking of Proteins to mtDNA.
To detect proteins that associate with mtDNA, we developed an in organello approach (Fig. 1A), whereby freshly isolated mitochondria from ρ+ cells grown on rich glycerol medium were treated with 1% formaldehyde for various lengths of time. After quenching the reaction, the mitochondria were lysed and applied directly to a CsCl gradient. After centrifugation, the distribution of mtDNA was analyzed by dot blots using a mtDNA COXII probe (Fig. 1B). To optimize resolution of crosslinked from free mtDNA, the CsCl gradient was adjusted to a refractive index of 1.37 so that mtDNA extracted from untreated mitochondria banded near the bottom of the gradient (Fig. 1B). Formaldehyde treatment resulted in a time-dependent shift of the DNA to lower density. After 16 h of treatment, nearly all of the mtDNA was found as a broad peak near the center of the gradient (fractions 8–12), well-resolved protein-free mtDNA (fractions 1–4), and from free protein (fractions 14–16). Shorter exposures of mitochondria to formaldehyde resulted in less efficient crosslinking with poorer resolution from protein-free mtDNA.
Analysis of the distribution of crosslinked protein (derived from the nucleoid-enriched pellet, Fig. 1A) in the CsCl gradients (optimized for resolution from free protein) was initially done by Western blotting using antisera raised against recombinant Abf2p (2). Before SDS/PAGE, each fraction was treated with 1% sarkosyl in Tris buffer at 95°C for 30 min to reverse crosslinks. In the untreated control, all of the Abf2p migrates as free protein at the top of the CsCl gradient (Fig. 1C, fractions 8–10). In contrast, after formaldehyde treatment, about 5% of the total Abf2p applied to the gradient bands with mtDNA (fractions 2–4). Crosslinked Abf2p is released by a 30-min treatment at 95°C but not by treatment with 1% SDS or 1% SDS plus 30 s of heating (Fig. 1, Inset). These data indicate that Abf2p is covalently crosslinked to mtDNA by formaldehyde treatment of mitochondria and that the crosslinked protein–mtDNA complex can be separated from free protein by CsCl gradient centrifugation. The relative increase in the mtDNA to protein ratio is ≥350-fold.
Identification of Crosslinked Proteins.
Crosslinked mtDNA–protein complexes were prepared according to Fig. 1A, and about 20 polypeptides were resolved on SDS/PAGE gels stained with Coomassie brilliant blue (Fig. 2). Polypeptides were excised from gels, subjected to in-gel trypsinization, and identified by mass spectrometry (see Materials and Methods).
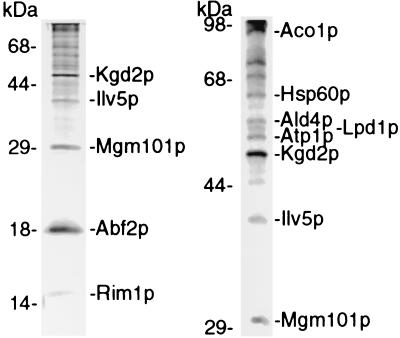
Figure 2.
Identification of polypeptides crosslinked with mtDNA. Heat-released polypeptides were fractionated through 15% (Left) and 9% (Right) polyacrylamide gels containing SDS, stained, excised, trypsinized, and subjected to mass spectrometry and/or sequence analysis to determine their identity. The position of molecular weight markers is shown on the left of each gel lane.
The proteins that we have identified are indicated in Fig. 2. Among the proteins that were expected to be crosslinked to mtDNA, we identified Abf2p (4), Rim1p [mitochondrial ssDNA binding protein (20)], and Mgm101p (7, 8). These proteins have been shown to bind mtDNA in vitro and in vivo by a combination of cosedimentation, fluorescence microscopy (green fluorescent protein-tagged proteins or indirect immunofluorescence), and direct DNA binding studies. Ilv5p also was identified as a crosslinked polypeptide. This mitochondrial matrix protein catalyzes a step in branched-chain amino acid biosynthesis (21), serves as a high copy suppressor of ρ+ mtDNA instability in a abf2Δ strain (5), is required for ρ+ mtDNA stability (5), and recently has been shown to influence nucleoid organization (22). These findings support the previous hypothesis that Ilv5p is a bifunctional protein (5).
Several mitochondrial proteins were identified in the crosslinked protein–mtDNA complex that were unanticipated. These included the mitochondrial chaperonin Hsp60p, Ald4p (an aldehyde dehydrogenase), three polypeptides of the tricarboxylic acid cycle, Kgd2p and Lpd1p (regulatory subunits of α-ketoglutarate dehydrogenase), Aco1p (aconitase), and Atp1p (the α-subunit of F1-ATPase), a protein associated with the inner mitochondrial membrane. Porin, an outer membrane protein, and Cox2p, an inner membrane protein, both of which are abundant mitochondrial proteins, were not detected in the crosslinked material (not shown). Because of their relative abundance among the proteins that are crosslinked, we examined in more detail the possible roles that Hsp60p and Kgd2p might play in mtDNA transactions.
Hsp60p Binds ssDNA in Mitochondria.
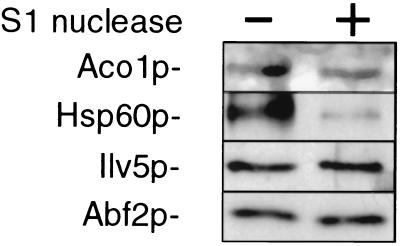
Hsp60p functions as an oligomer with Hsp10p and is required for proper folding of many polypeptides imported into mitochondria (23). It is of particular interest that Hsp60p was earlier identified as a ssDNA binding protein that stimulated yeast DNA polymerase II activity in vitro (24, 25). We reasoned that if Hsp60p binds to ssDNA in mitochondria, then removal of ssDNA from the crosslinked material by S1 nuclease treatment should result in a decreased recovery of Hsp60p. Western blot analysis was used to determine the levels of four proteins in the protein–mtDNA complex (Fig. 3). The level of Hsp60p is substantially reduced by S1 nuclease treatment, showing that it associates preferentially with ss mtDNA. Hsp10p is also present in the crosslinked material and was depleted after S1 nuclease treatment (data not shown), in contrast to Aco1p, Ilv5p, and Abf2p, which were unaffected by S1 nuclease treatment.
Figure 3.
S1 nuclease treatment of formaldehyde crosslinked mtDNA results in the decreased recovery of Hsp60p. The nucleoid-enriched pellet (Fig. 1A) was treated with or without S1 nuclease and mtDNA–protein complexes were purified. The relative abundance of representative polypeptides recovered in the crosslinked material was determined by Western blotting using antisera raised against Aco1p, Hsp60p, Ilv5p, and Abf2p.
Hsp60p Is a Sequence and Strand-Specific DNA Binding Protein.
To examine the binding characteristics of Hsp60p in more detail, a recombinant 6-histidine tagged form of Hsp60p was cloned, expressed in E. coli, and purified by Ni2+ chromatography (Fig. 4A). Based on Western blot analysis using an anti-Hsp60 polyclonal antibody (26), the purified Hsp60p-6His was free of GroEL, the Hsp60p homolog in E. coli (Fig. 4A). Electrophoretic mobility shift assays were performed to determine the binding specificity of Hsp60p-6His to a set of ds and ss mtDNA sequences (ori5, ori6, VAR1, and 14S rDNA). Ori5 is a putative origin of yeast mtDNA replication, whereas ori6 has been described as an inactive origin because of three short G + C-rich insertions (18). VAR1 DNA encodes a protein of the small mitochondrial ribosomal subunit and has a high A + T content, similar to that of ori5 and ori6. As shown in Fig. 4B, ds probes of these sequences were poor substrates for Hsp60p-6His binding. When these probes were made ss, the binding of Hsp60p-6His to each substrate was enhanced, indicating that Hsp60p-6His has a greater binding affinity for ssDNA than for dsDNA. Almost 50% of the total ss ori5 probe was shifted by the added protein, in contrast to only 6%, 15%, and 10% of the ori6, VAR1, and rDNA probes, respectively. Therefore, Hsp60p-6His exhibits both a general affinity for ssDNA as well as specificity for ori5 DNA among the four probes analyzed.
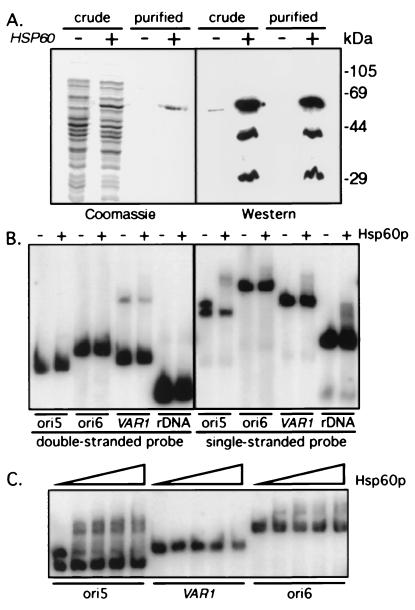
Figure 4.
Hsp60p-6His is a sequence-specific ssDNA binding protein. (A) Full-length HSP60 cloned into pET-24d (+) or vector alone (−) was induced in E. coli. Proteins present in the soluble extract (crude) or after Ni2+ column purification (purified) were fractionated through 10% polyacrylamide gels and either stained with Coomassie blue or subjected to Western blot analysis with anti-Hsp60p serum. Two polypeptides (44 and 29 kDa) copurified with Hsp60p-6His and presumably represent proteolytic products. (B) dsDNA or ssDNA probes (ori5, ori6, VAR1, and rDNA) were incubated in the presence (+) or absence (−) of 15 ng Hsp60p-6His and the protein–DNA complexes were separated from free DNA by electrophoresis through 3.5% polyacrylamide gels. Hsp60p-6His bound preferentially to one strand of ori5, in contrast to the absence of binding to the ds substrates and to the much reduced binding to the other ss substrates. (C) ssDNAs were prepared by boiling and quick cooling on ice and increasing concentrations of Hsp60p-6His (1.5, 3, 6, 12 ng) were added to the indicated probes, and protein–DNA complexes fractionated as above.
The fortuitous separation of the two strands of ori5 revealed that the binding of Hsp60p-6His to ori5 is strand-specific. Hybridization experiments (data not shown) with strand-specific probes indicate that Hsp60p-6His shifts the template strand of ori5. Similar experiments were performed with ss ori6 and VAR1 sequences. Only the template strand of ori6 is bound by Hsp60p-6His, but at a much lower level than is observed for ori5 DNA. In contrast, both strands of VAR1 are shifted with equally low efficiency, despite the similarity in A + T content with ori5 and ori6 DNA. These data indicate that Hsp60p-6His recognizes a template strand sequence or secondary structure that is in common to both ori5 and ori6 and that it also has a nonspecific ssDNA binding activity.
The experiments described above used a constant amount (15 ng) of Hsp60p-6His. At this concentration, all of the ssDNAs tested bound some Hsp60p-6His. However, the yield of bound product at a single protein concentration may not be representative of binding specificity. Therefore, for each of the probes used, we performed titration experiments using varying amounts of Hsp60p-6His (Fig. 4C). Even at the lowest protein concentration used (<2 ng), most of the template strand of ori5 is shifted. At this protein concentration, only a small amount of the ss ori6 DNA is bound, while no detectable shifted product is observed with ss VAR1 or rDNA probes. We conclude that Hsp60p-6His binds preferentially to the template strand of ori5 DNA.
HSP60 Point Mutants Affect Mitochondrial DNA Stability.
Given the biochemical data presented, does Hsp60p play a role in mtDNA maintenance? Because HSP60 is an essential gene, we analyzed temperature-sensitive hsp60 mutants for effects on ρ+ mtDNA stability. These mutants were generated by random or site-directed mutagenesis and screened for temperature-sensitive alleles (26). Plasmid-borne mutant alleles were shuffled into a strain harboring a disrupted chromosomal copy of HSP60 that was maintained with the wild-type HSP60 gene on a 2-μm URA3 plasmid. While removing the wild-type copy of the gene by 5-fluororotic acid treatment on YPD plates, we noticed that a few of the mutant strains, although temperature-sensitive, could not be regrown on YPGly plates. This observation suggested that these mutations may interfere with maintenance of ρ+ mtDNA.
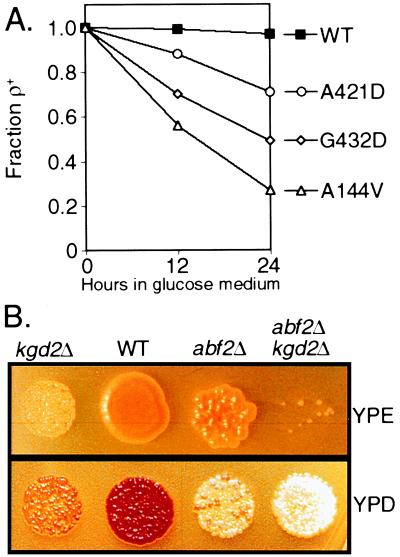
To determine whether Hsp60p functions in mtDNA stability, we analyzed three hsp60-ts ρ+ strains, A144V, G432D, and A421D (26), for elevated petite levels (Fig. 5A). The strains were grown for 24 h in YPD medium at the permissive temperature (25°C), and the percent of cells unable to grow on glycerol was determined. All three mutants generated gly− progeny cells at frequencies greater than wild type (yielding 72%, 49%, and 25% gly− cells, respectively, compared with 4% in the control strain). At 24 h, most of the cells expressing the hsp60-A144V allele had no mtDNA detectable by 4′,6′-diamino-2-phenylindole staining (data not shown). To determine whether these strains lost functional mtDNA because of inefficient mtDNA gene expression, bulk mitochondrial protein synthesis was measured by pulse labeling newly made proteins in the presence of cycloheximide (26). During the first 6 h of growth, all of these strains had wild-type levels of mitochondrial protein expression as well as normal levels of newly imported cytoplasmically synthesized mitochondrial proteins that appeared to be correctly folded (ref. 26; data not shown). These data indicate that, at the permissive temperature, the mutant Hsp60p-ts proteins function at or near wild-type capacity in mitochondrial protein synthesis and protein import, yet the stability of ρ+ mtDNA is reduced to allele-specific extents. Our evidence for a direct interaction of Hsp60p with mtDNA and the finding of sequence and strand-specific binding to a putative origin of mtDNA replication, plus the instability of ρ+ mtDNA, suggest a dual function for Hsp60p in mitochondria.
Figure 5.
Genetic analysis of HSP60 and KGD2. (A) Loss of functional ρ+ mtDNA in hsp60-ts mutants. Wild-type and three hsp60-ts mutant strains, listed on the right, were grown overnight in YPGly at 25°C and inoculated into YPD medium at 25°C. The fraction of ρ+ cells in the population present after 12 and 24 h growth was determined in a plating assay by their ability to grow on YPGly plates. (B) Synthetic petite formation between abf2Δ and kgd2Δ mutant strains. Sister spores, prepared as described in Materials and Methods, were plated in equivalent cell numbers on either YPD or YPE medium. Three-fold more cells were plated on YPE than on YPD medium. The loss of ρ+ mtDNA is demonstrated by the decreased number of cells that grow on YPE and the inability of these ade2− strains to develop a red color on YPD.
Phenotypic Analysis of kgd2Δ Strains.
Kgd2p, a subunit of α-ketoglutarate dehydrogenase, is recovered in the crosslinked material at nearly the same level as Abf2p. By analogy with Ilv5p and Hsp60p, we hypothesized that this protein is bifunctional and also would play a role in mtDNA stability. Because KGD2 is not an essential gene (27), we examined the phenotype of a KGD2 null allele singly and in combination with an abf2Δ mutation. The rationale for this approach is that subtle mtDNA phenotypes may be synthetically exacerbated when in combination with the abf2Δ allele. We generated a diploid yeast strain containing null alleles of KGD2 and ABF2, sporulated the diploid, germinated the haploid segregants on YPD medium, and assayed the ability of cells to turn red on YPD and to grow on YPE media, indicating that these cells were respiratory competent. As indicated in Fig. 5B, ρ+ mtDNA is as stable in kgd2Δ strains as in wild-type cells when germinated on YPD medium. By contrast, the abf2Δ kgd2Δ double mutant loses ρ+ mtDNA on dextrose much faster than wild-type, abf2Δ, or kgd2Δ single mutants. This mitochondrial dysfunction in abf2Δ kgd2Δ is caused by the loss of mtDNA rather than a metabolic defect because first, on mating the double mutant to an otherwise wild-type ρ0 strain, the diploid fails to grow on nonfermentable carbon sources and second, 4′,6′-diamino-2-phenylindole staining of the kgd2Δ abf2Δ strain indicates that these strains lose mtDNA and produce ρ0 cells (not shown). These data suggest that Kgd2p is also a bifunctional polypeptide, participating in both the TCA cycle and ρ+ mtDNA maintenance. These results are not simply a consequence of the loss of tricarboxylic acid cycle function, because a cit1Δ mutation, either singly or in combination with the abf2Δ mutation, has no effect on mtDNA stability (data not shown).
Discussion
We have described a formaldehyde crosslinking procedure to purify proteins that interact with mtDNA. To minimize perturbation of nucleoids during isolation, as well as potential contamination with nuclear DNA binding proteins, our experiments have been carried out in organello with highly purified, DNase-treated mitochondria. Typically, in vivo crosslinking of proteins to nuclear DNA requires relatively short exposures to formaldehyde in contrast to the longer exposures used in our experiments. This difference may be caused, in part, by the highly oxidative environment of the mitochondria. Because most, if not all, of the Abf2p in mitochondria is bound to mtDNA in vivo (9, 28) we found that, despite these longer exposures to formaldehyde, no more than 5% of the total amount of Abf2p is recovered with mtDNA. This finding suggests that our method is biased against trapping casual interactions of proteins with mtDNA. Although we cannot rule out the adventitious presence of some proteins in the nucleoid preparations, the genetic analyses described in this paper allow us to determine whether any of the proteins we have identified affect mtDNA stability.
The application of mass spectrometry (and Western blotting) to the most abundant polypeptides resolved by SDS/PAGE allowed us to identify 11 polypeptide species, all of which are mitochondrial proteins. Although we estimate ≈20 polypeptide species in this preparation, not all of these proteins are necessarily DNA binding proteins as they may be present through protein–protein rather than protein–DNA interactions. In addition to Abf2p, Mgm101p, and Rim1p, which have been documented to interact with mtDNA, we identified Ilv5p, a bifunctional protein that participates in branched-chain amino acid biosynthesis (21) and mtDNA stability (5). Recent studies show that Ilv5p also plays a direct role in the parsing of mtDNA into nucleoids (22); thus, it was not surprising that Ilv5p was recovered as a major protein among those crosslinked to mtDNA. Other proteins identified in the current work, including Hsp60p, Kgd2p, Ald4p, and the α-subunit of the F1-ATPase, were not anticipated. Studies are presently underway to identify the remaining proteins in nucleoids using additional sensitive methods, such as ion trap mass spectrometry.
To begin assessing the significance of these proteins, we examined the roles of Hsp60p and Kgd2p in ρ+ mtDNA stability. We demonstrated that Hsp60p is crosslinked largely to ss mtDNA, consistent with an earlier report that Hsp60p binds preferentially to ssDNA (24, 25). We extended these observations using recombinant 6-His-tagged protein to show that Hsp60p-6His binds preferentially to the template strand of ori5, which is a putative origin of mtDNA replication. Moreover, we observed that, with limiting concentrations of protein, only two DNA–protein complexes (major and minor ones) were formed, suggesting that only one or two DNA binding sites are in the ori5 substrate (Fig. 4C). This is in contrast to the DNA binding of Abf2p, which forms multiple DNA–protein complexes when incubated with ds ori5 DNA (not shown), consistent with previous studies (29) that Abf2p binds nonspecifically to mtDNA.
A number of studies have shown that chaperones play an important role in prokaryotic DNA replication (30). Chaperones catalyze the folding/unfolding (or dissociation of aggregates) of activator proteins (such as E. coli DnaA) facilitating their binding at the origin of replication (oriC). In E. coli, for example, these functions are carried out either by the DnaK/DnaJ/GrpE or GroEL/GroES chaperonin systems (31). It is of particular interest that in yeast, null mutants of MDJ1 (the mitochondrial homolog of DnaJ) (32) and hsp60-ts mutants (Fig. 5A) are unable to maintain ρ+ mtDNA and are either petite or petite-inducing (when grown on glucose medium under permissive conditions). The remarkable specificity of Hsp60p binding to an active ori suggests that a role of Hsp60p in mtDNA inheritance may be at the level of mtDNA replication.
Although we did not observe any significant ρ+ mtDNA instability in kgd2Δ cells, there was a dramatic synthetic mtDNA instability phenotype in the kgd2Δ abf2Δ double mutant. It is noteworthy that although there is an increased instability of ρ+ mtDNA in ilv5Δ cells, that phenotype is not as severe as in abf2Δ cells, but is greatly exacerbated in the ilv5Δ abf2Δ double mutant (5). Taken together, these data suggest a relationship between the Kgd2p and Abf2p proteins in mtDNA inheritance, which may well extend to other proteins that we have identified.
These data also prompt us to speculate that the bifunctionality of mtDNA-associated proteins may be common. During the evolution of the eukaryotic cell, the lateral transfer of genes occurred from the endosymbiont genome (the progenitor of mtDNA) to the host genome (33). The persistence of a remnant of the endosymbiont genome (i.e., mtDNA) together with nuclear genome reduction, may have necessitated the evolution of mechanisms to retain and express mtDNA. One such mechanism would be the evolution of genes encoding both nuclear and mitochondrial isoforms of the same enzyme, examples of which include Pif1p (34), Trm1p (35), and Cdc9p (36). A second mechanism might require maintaining proteins that have acquired multiple functions. Recent evidence indicates that RPO41, encoding the mitochondrial RNA polymerase, functions in ρ+ mtDNA stability through a 180-aa N-terminal extension, which is independent of the transcription domain in the polypeptide (37). Our findings would suggest that the identified nucleoid proteins have evolved by the second mechanism, but it remains to be determined whether an individual subunit or the enzyme complex as a whole has evolved to maintain mtDNA. Further studies are in progress to identify and assess the function of the remaining nucleoid components.
Acknowledgments
We thank Steve J. Afendis, Caroline R. Moomaw, and Betty Key for their excellent technical assistance. This work was supported by Grant GM33510 from the National Institutes of Health and by Grant I-6042 from the Robert A. Welch Foundation. Aco1p and Hsp10p antisera were kindly provided by Drs. C. Velot and S. Rospert, respectively.
Abbreviations
- ss
single-stranded
- ds
double-stranded
- YPD
yeast extract/peptone/dextrose
- YPE
yeast extract/peptone/ethanol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140063197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140063197
References
- 1.Miyakawa I, Sando N, Kawano K, Nakamura S, Kuroiwa T. J Cell Sci. 1987;88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- 2.Newman S M, Zelenaya-Troitskaya O, Perlman P S, Butow R A. Nucleic Acids Res. 1996;24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyakawa I, Fumoto S, Kuroiwa T, Sando N. Plant Cell Physiol. 1995;36:1179–1188. [PubMed] [Google Scholar]
- 4.Diffley J F, Stillman B. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troitskaya O, Perlman P S, Butow R A. EMBO J. 1995;14:3268–3276. doi: 10.1002/j.1460-2075.1995.tb07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacAlpine D M, Perlman P S, Butow R A. Proc Natl Acad Sci USA. 1998;95:6739–6743. doi: 10.1073/pnas.95.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X J, Guan M X, Clark-Walker G D. Nucleic Acids Res. 1993;21:3473–3477. doi: 10.1093/nar/21.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meeusen S, Tieu Q, Wong E, Weiss E, Schieltz D, Yates J R, Nunnari J. J Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto K, Perlman P S, Butow R A. J Cell Biol. 1998;142:613–623. doi: 10.1083/jcb.142.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlando V, Strutt H, Paro R. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 11.Solomon M J, Varshavsky A. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Ray D S. Proc Natl Acad Sci USA. 1993;90:1786–1789. doi: 10.1073/pnas.90.5.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przybyla-Zawislak B, Gadde D M, Ducharme K, McCammon M T. Genetics. 1999;152:153–166. doi: 10.1093/genetics/152.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen O N, Podtelejnikov A, Mann M. Rapid Commun Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Dubaquie Y, Schatz G, Rospert S. Methods Enzymol. 1998;290:193–202. doi: 10.1016/s0076-6879(98)90019-2. [DOI] [PubMed] [Google Scholar]
- 17.Parikh V S, Morgan M M, Scott R, Clements L S, Butow R A. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- 18.Baldacci G, Bernardi G. EMBO J. 1982;1:987–994. doi: 10.1002/j.1460-2075.1982.tb01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zassenhaus H P, Martin N C, Butow R A. J Biol Chem. 1984;259:6019–6027. [PubMed] [Google Scholar]
- 20.Van Dyck E, Foury F, Stillman B, Brill S J. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen J G L, Holmberg S. Nucleic Acids Res. 1986;14:9631–9651. doi: 10.1093/nar/14.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacAlpine D M, Perlman P S, Butow R A. EMBO J. 2000;19:767–775. doi: 10.1093/emboj/19.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubaquie Y, Looser R, Funfschilling U, Jeno P, Rospert S. EMBO J. 1998;17:5868–5876. doi: 10.1093/emboj/17.20.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown W C, Smiley J K, Campbell J L. Proc Natl Acad Sci USA. 1990;87:677–681. doi: 10.1073/pnas.87.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smiley J K, Brown W C, Campbell J L. Nucleic Acids Res. 1992;20:4913–4918. doi: 10.1093/nar/20.18.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallberg E M, Shu Y, Hallberg R L. Mol Cell Biol. 1993;13:3050–3057. doi: 10.1128/mcb.13.5.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repetto B, Tzagoloff A. Mol Cell Biol. 1990;10:4221–4232. doi: 10.1128/mcb.10.8.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelenaya-Troitskaya O, Newman S M, Okamoto K, Perlman P S, Butow R A. Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diffley J F X, Stillman B. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 30.Konieczny I, Zylicz M. Role of Bacterial Chaperones in DNA Replication. New York: Kluwer; 1999. [DOI] [PubMed] [Google Scholar]
- 31.Fayet O, Louarn J M, Georgopoulos C. Mol Gen Genet. 1986;202:435–445. doi: 10.1007/BF00333274. [DOI] [PubMed] [Google Scholar]
- 32.Duchniewicz M, Germaniuk A, Westermann B, Neupert W, Schwarz E, Marszalek J. Mol Cell Biol. 1999;19:8201–8210. doi: 10.1128/mcb.19.12.8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 34.Schulz V P, Zakian V A. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 35.Rose A M, Joyce P B, Hopper A K, Martin N C. Mol Cell Biol. 1992;12:5652–5658. doi: 10.1128/mcb.12.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer M, Rainey M, Pullen T, Stirling C J. Curr Biol. 1999;9:1085–1094. doi: 10.1016/s0960-9822(99)80477-1. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Shadel G S. Proc Natl Acad Sci USA. 1999;96:8046–8051. doi: 10.1073/pnas.96.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]