Abstract
Overexpression of c-Myc is one of the most common alterations in human cancers, yet it is not clear how this transcription factor acts to promote malignant transformation. To understand the molecular targets of c-Myc function, we have used an unbiased genome-wide location-analysis approach to examine the genomic binding sites of c-Myc in Burkitt's lymphoma cells. We find that c-Myc together with its heterodimeric partner, Max, occupy >15% of gene promoters tested in these cancer cells. The DNA binding of c-Myc and Max correlates extensively with gene expression throughout the genome, a hallmark attribute of general transcription factors. The c-Myc/Max heterodimer complexes also colocalize with transcription factor IID in these cells, further supporting a general role for overexpressed c-Myc in global gene regulation. In addition, transcription of a majority of c-Myc target genes exhibits changes correlated with levels of c-myc mRNA in a diverse set of tissues and cell lines, supporting the conclusion that c-Myc regulates them. Taken together, these results suggest a general role for overexpressed c-Myc in global transcriptional regulation in some cancer cells and point toward molecular mechanisms for c-Myc function in malignant transformation.
The oncogene c-myc is frequently associated with human malignancies and plays a critical role in regulating cell proliferation, growth, apoptosis, and differentiation (1–6). Studies in rodent model systems have shown that overexpression of c-Myc can cause malignant transformation, and that sustained tumor growth depends on its continued expression (7–12). The molecular mechanisms by which c-Myc functions to effect tumorigenesis have been the subject of extensive research in the last several decades.
Several lines of evidence suggest that c-Myc may cause transformation through its function as a sequence-specific transcription activator. First, the c-Myc protein, along with the Max protein, can specifically recognize DNA sequences with a core motif of CACGTG (13, 14). The domain that is required for c-Myc DNA binding, the basic helix–loop–helix zipper domain, is essential for its oncogenic transformation (15). Second, c-Myc possesses an N-terminal transactivation domain. Deletions or mutations in this domain result in loss of c-Myc transformation (15). This model implies that c-Myc may cause transformation by activating a select set of genes that in turn play key roles in malignant transformation (3). Throughout the years, a large number of genes targeted by c-Myc regulation either directly or indirectly have been found. However, such “transformation” genes remain elusive (16).
Some evidence suggests that c-Myc may promote transformation through different mechanisms. First, the transcriptional activation potential of c-Myc does not always correlate with its ability to transform rodent fibroblast cells (4). For example, several studies showed that mutations in the Myc box II domain within c-Myc can abrogate its transformation capacity without affecting c-Myc activation of reporter gene constructs (17, 18). Second, c-Myc also acts as a transcriptional repressor. The mechanism of c-Myc-mediated repression is not entirely clear but in some cases may involve the association of c-Myc/Max with transcriptional activators (19–21). Because some of the genes normally repressed by c-Myc are key cell-cycle regulators, it is conceivable that c-Myc-mediated repression of these genes may also contribute to tumorigenesis (22).
More recently, it was shown that c-Myc could directly activate RNA polymerase (pol) III promoters (23). Because most pol III promoters lack the canonical c-Myc binding sites, it was demonstrated that c-Myc-regulated transcription from pol III promoters occurs through its association with the transcription factor IIIB (TFIIIB) complex, which is a pol III-specific general transcription factor. This suggests a potentially broad role for c-Myc in regulating gene expression and raises the possibility that c-Myc may use a similar mechanism in regulating pol II promoters.
Identifying the genomic binding sites of c-Myc in cancer cells should help resolve the long-standing questions regarding mechanisms of oncogenic transformation by c-Myc. As an initial effort to characterize c-Myc DNA binding in vivo, we used a genome-wide location-analysis approach that allows the determination of transcription factor-binding sites throughout the genome (24, 25). This method has allowed identification of the DNA-binding sites in the yeast genome for proteins involved in transcription, chromatin modification, and DNA replication (26–31). More recently, the same approach has been used to reveal the promoters directly bound and regulated by the E2F transcription factors in human cells (32, 33).
Here we report experiments designed to identify genomic binding sites of c-Myc in Burkitt's lymphoma cells, which express high levels of c-Myc due to a chromosomal translocation (34). We show that the overexpressed c-Myc binds to a large number of gene promoters in these cancer cells, amounting to nearly 15% of the loci tested. Furthermore, the DNA binding by c-Myc generally correlates with global gene transcription activities and largely coincides with binding of the general transcription factor TFIID throughout the genome. A majority of the c-Myc target genes we identified are expressed in other cell types in a manner correlated with c-myc mRNA levels. Taken together, these results suggest a rather general role for c-Myc in the regulation of genome expression in many cancer cells.
Methods
Design and Manufacturing of Human Promoter Microarrays. To conduct a comprehensive analysis of the genomic sites of c-Myc in human cells, we developed a DNA microarray (henceforth referred to as the hu6K array) that contained PCR products spanning the proximal promoters of 4,839 human genes chosen from the NCBI Refseq database. These genes were selected because their promoters were best-annotated, and there was a clear function associated with each gene. The DNA fragments have an average size of 900 bp and typically cover the sequence from 650 bp upstream to 250 bp downstream of the transcription start site in a gene. The choice of these regions is based on previous observations that human transcription factors frequently bind to proximal promoter sequences (32). The hu6K array also includes 729 coding sequence and 221 genomic regions >1 kb upstream of the transcription start site of a gene. The latter two categories of sequences serve as internal controls for the location analysis. The description of all the sequences on the hu6K array can be found in Array Description, which is published as supporting information on the PNAS web site, www.pnas.org.
We designed oligonucleotide primers to amplify the genomic regions discussed above using the PCR. After PCR amplification, we purified each DNA fragment, verified the product by agarose gel electrophoresis, and then spotted the purified DNA to GAPSII glass slides (Corning) using a contact printer (Cartesian Technologies, Irvine, CA). After UV crosslinking, the glass slides were stored under vacuum until use.
Genome-Wide Location Analysis. A detailed protocol for genome-wide location analysis with mammalian factors can be found in Supporting Materials and Methods. Briefly, Daudi cells (a gift from William Sugden, University of Wisconsin, Madison) were grown in flasks with RPMI medium 1640 supplemented with 10% FBS/100 units/ml penicillin/100 μg/ml streptomycin to a density of 109 per liter at 37°C in 5% CO2. A total of 109 proliferating Daudi cells were fixed with formaldehyde, harvested, and disrupted by sonication. To enrich for target genes bound to a transcription factor, we immunoprecipitated the resulting chromatin fragments with polyclonal antibodies that specifically recognize c-Myc (sc-764), Max (sc-197), E2F1 (sc-193), TAFII250 (sc-735), or pol II (sc-9001), obtained from Santa Cruz Biotechnology. After reversal of crosslinks, the purified and enriched DNA was amplified by ligation-mediated PCR and subsequently labeled with the Cy5 fluorophore by random priming. For purposes of normalization, we also performed ligation-mediated PCR on DNA that was not enriched by immunoprecipitation and labeled the amplified sample with a second fluorescent dye, Cy3. Chromatin immunoprecipitation (ChIP)-enriched and nonenriched (total input) pools of DNA were mixed with Cot-1 DNA to suppress annealing of repetitive sequences and hybridized under stringent conditions to a hu6K array. DNA microarray hybridization was carried out as described in ref. 32. The microarray then was analyzed by using a GenenPix 4000B scanner (Axon Instruments, Foster City, CA).
Data Analysis. Analysis of microarray scanning images was performed according to published protocols (24) with modifications (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Data from independent replicate experiments were combined (24). An average enrichment ratio was calculated for each DNA species on the array from at least three replicate data sets. The binding of a factor to DNA was deemed significant if the average P value was <0.001. By using these criteria, no DNA achieved significance in control experiments where input DNA was compared with the same DNA. The Supporting Raw Data Set containing processed microarray data is published as supporting information on the PNAS web site.
Conventional ChIP. Confirmation of a select number of c-Myc/Max target sites by the conventional ChIP method was performed according to ref. 32. The primers used for amplification of genomic sequences corresponding to each binding site were MARS (5′-aagtgcgacttgccctaaaa-3′ and 5′-ccatgcagctgggactaca-3′), ATF4 (5′-ctcctttcgtgaggccataa-3′ and 5′-cgcgcagagaaaactacatct-3′), TTF2 (5′-ggtaaggggctagggtctca-3′ and 5′-cccagctcaagacctactcg-3′), CKN1 (5′-ggctctcacttttccctcct-3′ and 5′-catgggttggtctcaggatt-3′), and control (5′-tgggtttggtaggggacata-3′ and 5′-ctgggctctgctggctta-3′). The control PCR product lies 9,595 bp upstream of the transcription start site in the CAD gene.
Results
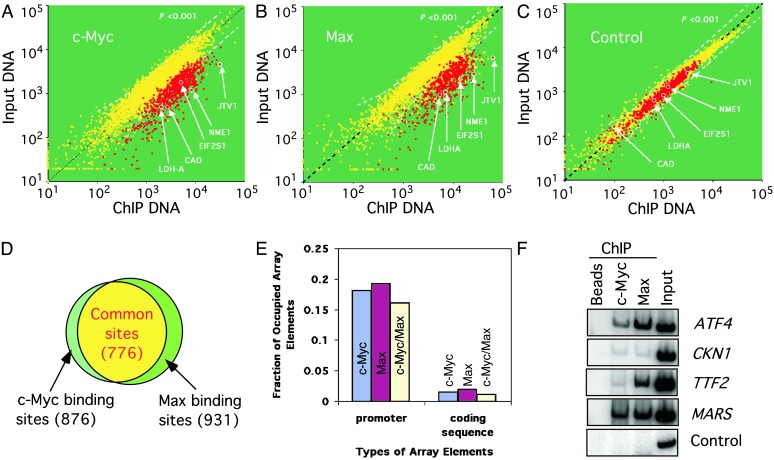
Widespread DNA Binding by c-Myc/Max in Burkitt's Lymphoma Cells. We first examined the promoter occupancy by c-Myc in Daudi cells, an established cell line originally derived from a Burkitt's lymphoma patient (34, 35). These cells express a high level of c-Myc and have been used extensively as an experimental system to study the role of c-Myc in tumorigenesis. We performed five independent c-Myc location-analysis experiments and averaged the results using an error model described in Supporting Materials and Methods (Fig. 1A). The analysis indicates that c-Myc is associated with the promoters of 876 genes based on a P value ≤0.001. Because c-Myc binds to DNA as a heterodimer with Max, we also performed five independent location-analysis experiments using antibodies that specifically recognize Max. A total of 931 gene promoters was found to be associated with Max in vivo (Fig. 1B). As a control, we carried out immunoprecipitation in the absence of specific antibodies. Only 32 gene promoters were enriched under such conditions, most likely because of their nonspecific interactions with the magnetic beads used in these experiments (Fig. 1C).
Fig. 1.
Genome-wide location analysis of c-Myc- and Max-bound promoters in Burkitt's lymphoma cells. Five independent experiments were performed to identify c-Myc or Max bound DNA in formaldehyde-crosslinked Daudi cells. (A) A scatter plot showing the fluorescent intensities for each spot on the hu6K promoter array in one of the c-Myc location-analysis experiments. A few previously known c-Myc target genes are highlighted. (B and C) Scatter plots corresponding to one of the Max location-analysis experiments, or the control experiment, in which ChIP was performed in the absence of primary antibodies. (D) Venn diagram comparing the DNA binding of c-Myc and Max. The data from five experiments were averaged (see Methods), and the genes with a P value <0.001 were counted as targets. Overall, 876 and 931 promoters are bound by c-Myc and Max, respectively. Marked with red in A–C are 776 promoters that are bound by both. (E) A comparison of the fraction of promoter or coding regions represented on the hu6K array that are bound by c-Myc, Max, or both in Daudi cells. (F) Conventional ChIP confirms the enrichment of c-Myc/Max target genes identified in the genome-wide location-analysis experiments. ChIP was performed with chromatin from Daudi cells by using the indicated primary antibodies, and enriched DNA was amplified with primers corresponding to a random select number of genes. As a negative control, magnetic beads lacking primary antibodies were used.
Several measures suggest that our location analysis is very robust and highly reliable. First, there is a significant overlap between the set of gene promoters bound by c-Myc and those bound by Max. A total of 776 gene promoters bound both proteins, accounting for 88% of the observed c-Myc binding sites and 83% of the observed Max binding sites (Fig. 1D). Second, consistent with previous knowledge, c-Myc and Max rarely bind to the coding sequences represented on the hu6K array. There is only 1.1% occupancy by c-Myc and Max for these regions compared with the 15% occupancy on gene promoters by both proteins (Fig. 1E). Assuming that observed binding of c-Myc and Max to coding sequences are false positives, we then estimate that the false-positive rates for the c-Myc/Max complex binding results is 7%, which is in line with previous estimates of false-positive rates of genome-wide location-analysis experiments (26). Third, we used the conventional ChIP method with a select set of c-Myc/Max target genes and confirmed all their in vivo protein–DNA interactions (Fig. 1F).
Based on this analysis, we estimate that at least 93% of the gene promoters identified as bound by c-Myc and Max in this study are true positives. Therefore, the c-Myc/Max complexes occupy at least 721 of 776 (93%) gene promoters, or nearly 15% of the 4,839 gene promoters examined, in Daudi cells. Because these genes are an unbiased selection of all human genes, c-Myc/Max complexes probably bind to close to 15% or 4,500 of the human genes in these Burkitt's lymphoma cells.
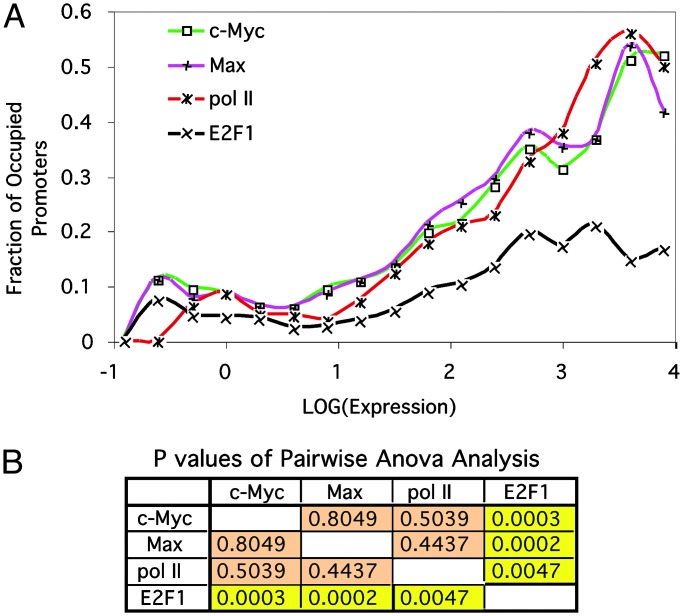
The DNA Binding of c-Myc and Max Corresponds to Genome Transcription Levels. In most cell types, only a small fraction of the genome is expressed. Because c-Myc binds to almost 15% of the human genes in Daudi cells and has a known function as a transcription activator, one prediction is that c-Myc DNA binding would correlate with global gene expression in these cells. To test this hypothesis, we analyzed the correlation of c-Myc or Max DNA binding and gene expression in Daudi cells. As a measure of the gene expression, we used previously reported transcriptome data obtained from the Daudi cells (36). We divided the total gene promoters represented by the hu6K array into 17 groups based on their transcription activity and plotted the fraction of c-Myc-bound promoters in each group with regard to transcription activities for genes in the group (Fig. 2A). The analysis indicates a strong correlation between c-Myc DNA binding and global gene transcription (ρ = 0.96). Nearly 50% of the highly active promoters bound c-Myc in Daudi cells, and the promoter occupancy gradually decreases in gene groups with lower promoter activities. Similarly, there is also a strong correlation (ρ = 0.95) between the DNA binding by Max and genome transcription activities (Fig. 2 A).
Fig. 2.
DNA binding of c-Myc and Max corresponds to genome transcription levels and resembles the behavior of a general transcription factor. (A) Line chart showing the correlation between gene-expression levels and promoter binding by c-Myc, Max, pol II, and E2F1 in Daudi cells. The 4,839 gene promoters in the hu6K arrays were divided into 17 groups based on their transcription activities (see Supporting Data Set for Fig. 2, which is published as supporting information on the PNAS web site). The lower and upper boundaries of log10-transformed gene-expression values for these groups are distributed evenly between -1 and 5. The fraction of promoters bound by each factor in every group is plotted with regard to the log10-transformed transcription activity for the group. (B) Table showing the pairwise comparison of genomic binding profiles of c-Myc, Max, pol II, and E2F1 with respect to genome expression. P values were calculated based on two-factor ANOVA analysis without replicate.
The extensive correlation between c-Myc's DNA binding and global gene transcription not only confirms our prediction of c-Myc function in Daudi cells but also reveals an extensive role for this oncogenic transcription factor in global gene regulation, a role that usually is associated with general transcription factors. To assess the similarity between the role of c-Myc in genome transcription and that of a general transcription factor, we performed genome-wide location-analysis experiments to identify the genomic binding sites of pol II in Daudi cells. As a control, we also examined the DNA binding of a typical transcriptional activator, E2F1, in these cells (32). In Fig. 2 A, the percentage of promoter occupancy by pol II and E2F1 is plotted with regard to levels of promoter activities in the 17 gene groups discussed above. Although the DNA binding for both proteins seems to correlate with genome transcription activities, the DNA-binding profile of E2F1 with respect to global gene expression is markedly different from that of pol II (P = 0.0047), c-Myc (P = 0.0003), and Max (P = 0.0002) when analyzed by using a two-factor ANOVA method (Fig. 2B). By contrast, the DNA-binding profile of pol II is indistinguishable from that of c-Myc (P = 0.50) and Max (P = 0.44) by using the same analysis. This result strongly suggests that c-Myc and Max act more like a general transcription factor than a sequence-specific transcription factor in Daudi cells.
c-Myc and Max Colocalize with TFIID on Gene Promoters. The similar DNA-binding profiles with respect to genome expression between c-Myc and pol II supports the model that c-Myc may play a general role in gene transcription in Daudi cells. To further confirm this and explore the mechanisms of its action in Daudi cells, we tested whether the DNA binding of c-Myc coincides with other general transcription factors. In particular, because c-Myc was shown to interact directly with TATA-box-binding protein (TBP) (37, 38), the core component of the general transcription factor TFIID, we tested whether c-Myc colocalizes with TFIID on gene promoters.
We performed three independent genome-wide location-analysis experiments to identify the gene promoters associated with TAFII250, the largest subunit of TFIID, in Daudi cells (39, 40). The analysis identified a total of 1,157 gene promoters occupied by TFIID (with a false-positive rate of 6% estimated from its observed DNA binding to coding sequences). In Fig. 3, the set of TFIID-bound promoters is compared with promoters bound by the c-Myc/Max complexes. A majority (603 of 1,157) of the promoters occupied by TFIID is also bound by c-Myc/Max, whereas almost all the c-Myc/Max-bound promoters also harbor TFIID in Daudi cells. The number of common binding sites between TFIID and c-Myc/Max (603) far exceeds a random overlap between two similar groups (185, with P << 0.001, χ2 test), suggesting that c-Myc functions together with TFIID in regulating global gene expression.
Fig. 3.

c-Myc and Max colocalize with TFIID in Burkitt's lymphoma cells. A Venn diagram compares the gene promoters bound by the c-Myc/Max complexes and that by TFIID in Daudi cells.
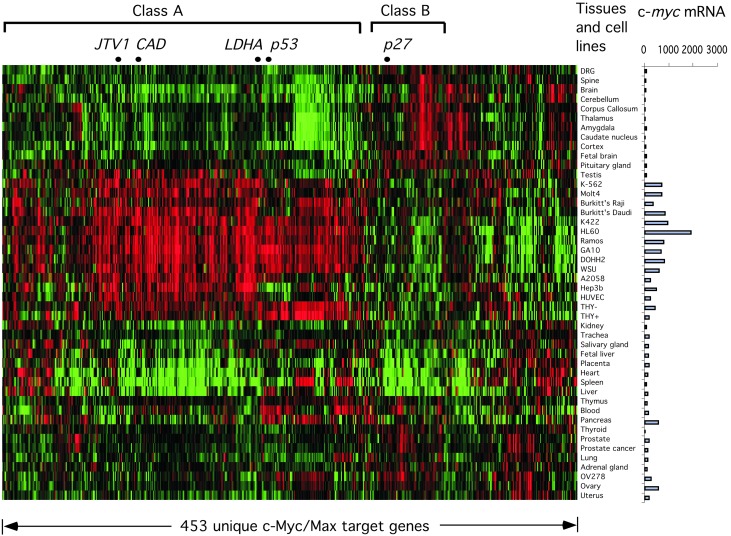
Expression of c-Myc/Max Target Genes Correlates with c-myc mRNA Levels in Diverse Tissues and Cell Lines. We next asked whether the promoters bound by c-Myc/Max are expressed in a c-Myc-dependent fashion. For this purpose, we compared the expression of these genes in a variety of human tissues and cell lines in which c-Myc transcript levels range from undetectable to very high (36). Among these c-Myc/Max targets, some genes were not included in the previous study, and many could not be reliably detected in the various samples. Therefore, these genes were excluded from this analysis. The remaining 453 c-Myc target genes were clustered based on their expression levels in these tissues and cell lines. As shown in Fig. 4, gene expression from a majority of the c-Myc/Max target promoters corresponds with c-myc mRNA levels (Class A). Genes in this category include known c-Myc target genes such as CAD, LDHA, p53, and JTV1. Interestingly, there is a small group of c-Myc/Max target genes with expression that inversely correlates with c-Myc mRNA levels (Class B). Among these genes is the gene encoding the cdk inhibitor p27, a known target of c-Myc repression. The identities of these genes can be found in Supporting Data Set for Fig. 4, which is published as supporting information on the PNAS web site.
Fig. 4.
Expression of c-Myc/Max target genes correlates with levels of c-myc transcript in human tissues and cell lines. (Left) Two-way clustering was performed on the expression data corresponding to 453 c-Myc/Max target genes in 46 human tissues and cell lines. The expression data for each gene were log-transformed and normalized such that the median log expression value is 0. The gene-expression values were represented by using a red-green color scheme, with red corresponding to higher-than-median expression values and green corresponding to lower-than-median expression values. The expression data corresponding to different genes are arranged from left to right, and different tissues are arranged from top to bottom. (Right) The c-myc mRNA levels in the corresponding samples, as measured by using Affymetrix GeneChip, are shown on a bar graph.
Discussion
A General Role for c-Myc in Global Transcriptional Regulation in Cancer Cells. Overexpression of the c-Myc gene has been associated with a large number of human malignancies (5). Although extensive research has been focused on the molecular function of this protein, it remains unknown how c-Myc promotes malignant transformation. In this study we attempted to identify the genomic binding sites for c-Myc and Max complexes in Burkitt's lymphoma cells. The results strongly suggest that c-Myc plays a general role in global gene regulation in these cells. First, we showed that the c-Myc/Max complexes bind to a large number of gene promoters in Burkitt's lymphoma cells, accounting for nearly 15% of genes tested. Second, DNA binding of c-Myc strongly correlates with transcription activities throughout the genome and resembles the genomic binding profile of pol II but not that of the sequence-specific transcription factor in the same cells. Third, c-Myc and Max extensively colocalize with the general transcription factor TFIID. Finally, expression from most c-Myc/Max target promoters corresponds to c-myc transcript levels in a diverse panel of human tissues and cell lines. Based on these observations, we propose a model whereby c-Myc plays a general role in the regulation of global gene expression in Burkitt's lymphoma cells. This model is consistent with the recent findings that the Drosophila dMyc/Max/Mnt network is involved in regulating ≈15% of the genome (41).
Program of Tumorigenesis in Cancer Cells. This extensive role of c-Myc in gene regulation might underlie tumorigenesis in cells that overexpress the oncogenic protein. Perhaps in primary cells or normal tissues, where c-Myc levels are low, the transcription factor only acts in a sequence-specific manner on a small number of genes and influences a limited spectrum of cellular behaviors. In Burkitt's lymphoma cells or other cancer cells expressing high levels of c-Myc, on the other hand, c-Myc may boost the global gene expression and influence a wide spectrum of cellular pathways by acting on a large number of genes. Many of the genes we identified as c-Myc/Max targets play key roles in signaling, cell-cycle regulation, DNA replication, protein biosynthesis, and energy metabolism (Table 1), and their increased expression as a result from c-Myc overexpression would be expected to lead to dramatic changes in multiple processes such as an increase in the overall rate of cell-mass accumulation and shortened cell cycle. Interestingly, some c-Myc/Max target genes we identified have been implicated previously in apoptosis. Activation of these genes, including p53, Daxx, and Dap3, most likely will sensitize cells to apoptotic stimuli and necessitate the accumulation of mutations in these pathways during expansion of the cell population. It is possible also that an increased rate of metabolism in Daudi cells may result in higher levels of reactive oxygen species and DNA damage, which may further sensitize cells for apoptotic signals (42). Subsequent loss of function of p53 or BLC2 then could lead to unchecked cell growth and malignant transformation (1).
Table 1. Functional categories of selected c-Myc/Max target genes.
| Functional category | c-Myc/Max targets |
|---|---|
| Signal transduction | AKAP10, AKAP9, APPL, ARFRP1, ATF6, ATM, CD164, CD2AP, CD79B, CD97, COPS3, CORO1C, CR2, CREB1, CREBL2, CSF2RB, CUL5, DAP3, DAXX, DDXBP1, DPYSL3, FUS1, FZD5, GDAP1, GNA12, GNL1, GPRK2L, HCR, IFNAR1, IFNGR1, IL17C, ILK, ITGB3BP, LANCL1, LNPEP, LOC51657, MADD, MADH3, MAP2K5, MAP2K7, MAP3K5, MAPK7, MAPRE2, MD-1, MKLN1, MKNK1, MST1R, NFAT5, NFKBIB, NR1D1, NR6A1, P85SPR, PDK1, POR1, PREP, PRKAB1, PRKAB2, PRKCL2, PRKRA, PRKRIR, PTPN1, PTPN6, PTPRF, RAC2, RAGA, RAP2B, RGS16, RHEB2, RIPK2, RLN1, RNF7, RPS6KA2, RPS6KA5, RPS6KB1, RRAS, RXRB, SRD5A1, SRP54, SRP68, SRPR, TLE3, TRIP15, ZNF147, and ZNF259 |
| Cell cycle | 101F6, APC10, ATM, CCT7, CDC25B, CDC6, CDK6, CDKN1B, CUL5, DNAJA2, FRAP1, GAK, LATS1, LOC51723, MAPRE1, MCM5, MPHOSPH1, MPHOSPH6, P23, PA2G4, PCNA, PCTK1, PPP2CA, PSMD8, RBBP8, RBL1, SGT1, TOPBP1, TP53, and TSC2 |
| Cell growth/proliferation | APPL, BARD1, BLZF1, C8FW, CD164, CDC16, CDC25B, CDC6, CDK6, CDKN1B, CIAO1, CKS2, CUL5, DDX1, DNAJA2, ELL, FUS1, GNA12, HKLP2, ING1, LATS1, MAPRE1, MAPRE2, MD-1, MST1R, MT3, NR6A1, P23, PA2G4, PCNA, PRKRA, PRKRIR, SEI1, SKB1, SYK, TSPY, XIP, and ZNF259 |
| Cell death/apoptosis | ABS, APG12L, ASC, CRADD, CUL5, DAP3, DAXX, MAEA, MAP3K5, MD-1, PDCD8, PTPN6, RAGA, RIPK2, RNF7, SMAC, and TP53 |
| Transporter | ABCB6, AKR7A2, ATP5B, ATP5C1, ATP5G2, CACNB1, CLCN2, CLCN3, CLCN6, COX15, DEGS, H6PD, IDH3B, KCNH4, NUP153, NUP88, PPIA, PRDX5, SLC12A2, SLC1A4, SLC22A3, SLC22A4, SLC25A11, SLC26A4, SLC2A4, SLC31A1, SLC5A6, SLC7A1, SRD5A1, VDAC2, VDAC3, and XK |
| Cell adhesion | CD164, CD97, CNTN2, CORO1C, DGCR6, FLOT2, ILK, ITGB3BP, MAEA, MKLN1, P84, PTPRF, and SIP2-28 |
| DNA replication | CDC6, CHRAC17, MCM3, MCM5, PCNA, POLD4, PPIA, RAD9, TOPBP1, and XIP |
| DNA repair | ABH, ADPRTL2, ADPRTL3, ATM, DDB1, ERCC6, EXO1, FRAP1, G22P1, JTV1, MBD4, MSH2, PCNA, PIR51, PMS1, POLH, PRKDC, RAD50, RAD54L, RAD9, RBBP8, RECQL, RECQL5, SIP2-28, and TP53 |
| Protein biosynthesis | MARS, NACA, PET112L, PYCR1, RPL13, RPL15, RPL18, RPL19, RPL27A, RPL31, RPL37, RPL5, RPL8, RPL9, RPLP1, RPS13, RPS17, RPS19, RPS20, RPS21, RPS25, RPS26, RPS29, RPS5, RPS6, and SARS |
| Metabolism | ACAD8, ACOX3, AGPS, AKR7A2, ASAH, BCKDHA, BCKDHB, BTN2A1, CARKL, CEPT1, CH25H, CYBA, DPYSL3, EPM2A, GCLC, GMDS, GNPAT, IDH3B, LOC51172, LOC51601, LTA4H, MAT2A, MGAT2, MOCS2, MT3, MTHFD1, NAGA, NIFS, OAZ1, PDK1, PDK2, PDK3, PLA2G4B, PLA2G6, PMVK, PP, PRPS2, PRPSAP1, PYCR1, SCLY, SLC2A4, SLC7A1, SREBF2, SSB, ST6GALNACIV, TOPBP1, and ZNRD1 |
| Oncogenesis/tumor suppressor | ABCB6, AKR7A2, ARMET, BARD1, CUL5, DDX1, DLEU1, DLEU2, HMMR, ING1, LATS1, MADH3, MAPRE1, MEL, MSH2, MYBBP1A, PMS1, POV1, PPP2CA, PPP2R1B, PTP4A1, RPS19, TP53, TSC2, TSPY, and TTC4 |
| Proteasome | POH1, POH1, PSMA1, PSMA5, PSMB1, PSMB5, PSMB7, PSMC4, PSMC5, PSMD10, PSMD3, PSMD5, PSMD7, PSMD8, and PSME3 |
| Transcription factor/cofactor | ABT1, ATF4, ATF6, CALR, CBF2, CGBP, CIR, CREB1, CREBL2, CRSP6, CSDA, DDXBP1, GABPA, HCF-2, IRF3, LOC51042, LZTR1, MADH3, MAFF, MYCBP, NFAT5, NFKBIB, NR1D1, POU2F1, PSMC5, RBBP1, RBBP2, RFX5, RNF4, RXRB, SAP18, SAP30, SCML2, SP4, SUPT5H, TIF1, TMF1, TP53, TRAP150, TRIP13, YY1, ZNF136, ZNF142, ZNF147, ZNF174, ZNF192, ZNF274, ZNF35, and ZNF85 |
Models for c-Myc DNA-Binding Specificity in Cancer Cells. Our analysis of the sequences of the c-Myc-bound promoters in Burkitt's lymphoma cells indicates that the c-Myc recognition motif is not present in many of these target promoters. For example, only ≈26% of the promoters contain the CACGTG motif between 1,000 bp upstream of the transcription start site and the translation start site where it would be expected. Therefore, CACGTG motif-independent mechanisms may be used by c-Myc in its binding to gene promoters in Daudi cells. Two models could explain the widespread DNA binding of c-Myc in Burkitt's lymphoma cells.
In the first model, a high level of c-Myc protein may allow it to bind to sequences that are different from the CACGTG motif and are not occupied by c-Myc in normal cells. These weak c-Myc binding sites could be variants of the CACGTG motif or completely different sequences and occur more frequently in the genome than the canonical c-Myc binding sites. Although this model is simple and can explain the increased DNA binding of c-Myc in Daudi cells, it does not explain the mechanisms of specificity of c-Myc DNA binding or the strong correlation between c-Myc DNA binding and global transcription levels.
In the second model, c-Myc may bind to DNA through its association with other sequence-specific transcription factors or general transcription factors. Indeed, c-Myc has been shown previously to directly interact with TATA-box-binding protein (TBP) (37, 38), and we also demonstrate here that c-Myc DNA binding coincides with TFIID occupancy on gene promoters. Taken together, these results suggest that c-Myc may be recruited to gene promoters as part of general transcription complexes, especially in the absence of the canonical c-Myc binding sites. A similar mechanism has been implicated recently in c-Myc's activation of pol III promoters (23). In this case, c-Myc binds to TFIIIB, a pol III-specific general transcription factor, and directly activates pol III transcription.
In conclusion, by analyzing the in vivo binding of c-Myc and Max to ≈5,000 gene promoters, we observed that c-Myc is involved in the regulation of a surprisingly large number of gene promoters in Burkitt's lymphoma cells. Such global transcriptional regulatory function by c-Myc may be a key mechanism that this oncogenic protein uses to promote tumorigenesis.
Supplementary Material
Acknowledgments
We thank Dr. Richard Kolodner for the generous contribution toward the making of hu6K arrays; Dr. William Sugden for the Daudi cell line; Drs. Tae Hoon Kim and Frank Furnari for helpful discussions; and Ms. Leah Ortiz-Luis Barrera and Mr. Joseph Hegler for the initial bioinformatic analysis of c-Myc target-gene expression. We also thank Drs. Robert N. Eisenman and Herman J. Bussemaker for making their manuscript available to us before publication. This research was supported by the Ludwig Institute for Cancer Research, a Sidney Kimmel Foundation for Cancer Research Scholar Award (to B.R.), and National Institutes of Health Grant R01-GM60513 (to M.Q.Z.).
Abbreviations: pol, RNA polymerase; TF, transcription factor; hu6K, DNA microarray of ≈6,000 human genomic DNA fragments; ChIP, chromatin immunoprecipitation.
References
- 1.Pelengaris, S., Khan, M. & Evan, G. (2002) Nat. Rev. Cancer 2 764-776. [DOI] [PubMed] [Google Scholar]
- 2.Boxer, L. M. & Dang, C. V. (2001) Oncogene 20 5595-5610. [DOI] [PubMed] [Google Scholar]
- 3.Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. (2000) Annu. Rev. Cell Dev. Biol. 16 653-699. [DOI] [PubMed] [Google Scholar]
- 4.Cole, M. D. & McMahon, S. B. (1999) Oncogene 18 2916-2924. [DOI] [PubMed] [Google Scholar]
- 5.Nesbit, C. E., Tersak, J. M. & Prochownik, E. V. (1999) Oncogene 18 3004-3016. [DOI] [PubMed] [Google Scholar]
- 6.Dalla-Favera, R., Gelmann, E. P., Martinotti, S., Franchini, G., Papas, T. S., Gallo, R. C. & Wong-Staal, F. (1982) Proc. Natl. Acad. Sci. USA 79 6497-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams, J. M., Harris, A. W., Pinkert, C. A., Corcoran, L. M., Alexander, W. S., Cory, S., Palmiter, R. D. & Brinster, R. L. (1985) Nature 318 533-538. [DOI] [PubMed] [Google Scholar]
- 8.Leder, A., Pattengale, P. K., Kuo, A., Stewart, T. A. & Leder, P. (1986) Cell 45 485-495. [DOI] [PubMed] [Google Scholar]
- 9.Felsher, D. W. & Bishop, J. M. (1999) Mol. Cell 4 199-207. [DOI] [PubMed] [Google Scholar]
- 10.Felsher, D. W. & Bishop, J. M. (1999) Proc. Natl. Acad. Sci. USA 96 3940-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain, M., Arvanitis, C., Chu, K., Dewey, W., Leonhardt, E., Trinh, M., Sundberg, C. D., Bishop, J. M. & Felsher, D. W. (2002) Science 297 102-104. [DOI] [PubMed] [Google Scholar]
- 12.Pelengaris, S., Khan, M. & Evan, G. I. (2002) Cell 109 321-334. [DOI] [PubMed] [Google Scholar]
- 13.Blackwell, T. K., Kretzner, L., Blackwood, E. M., Eisenman, R. N. & Weintraub, H. (1990) Science 250 1149-1151. [DOI] [PubMed] [Google Scholar]
- 14.Blackwell, T. K., Huang, J., Ma, A., Kretzner, L., Alt, F. W., Eisenman, R. N. & Weintraub, H. (1993) Mol. Cell. Biol. 13 5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone, J., de Lange, T., Ramsay, G., Jakobovits, E., Bishop, J. M., Varmus, H. & Lee, W. (1987) Mol. Cell. Biol. 7 1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levens, D. (2002) Proc. Natl. Acad. Sci. USA 99 5757-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brough, D. E., Hofmann, T. J., Ellwood, K. B., Townley, R. A. & Cole, M. D. (1995) Mol. Cell. Biol. 15 1536-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bello-Fernandez, C., Packham, G. & Cleveland, J. L. (1993) Proc. Natl. Acad. Sci. USA 90 7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staller, P., Peukert, K., Kiermaier, A., Seoane, J., Lukas, J., Karsunky, H., Moroy, T., Bartek, J., Massague, J., Hanel, F. & Eilers, M. (2001) Nat. Cell Biol. 3 392-399. [DOI] [PubMed] [Google Scholar]
- 20.Seoane, J., Pouponnot, C., Staller, P., Schader, M., Eilers, M. & Massague, J. (2001) Nat. Cell Biol. 3 400-408. [DOI] [PubMed] [Google Scholar]
- 21.Gartel, A. L., Ye, X., Goufman, E., Shianov, P., Hay, N., Najmabadi, F. & Tyner, A. L. (2001) Proc. Natl. Acad. Sci. USA 98 4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gartel, A. L. & Shchors, K. (2003) Exp. Cell Res. 283 17-21. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Roman, N., Grandori, C., Eisenman, R. N. & White, R. J. (2003) Nature 421 290-294. [DOI] [PubMed] [Google Scholar]
- 24.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290 2306-2309. [DOI] [PubMed] [Google Scholar]
- 25.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409 533-538. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298 799-804. [DOI] [PubMed] [Google Scholar]
- 27.Robyr, D., Suka, Y., Xenarios, I., Kurdistani, S. K., Wang, A., Suka, N. & Grunstein, M. (2002) Cell 109 437-446. [DOI] [PubMed] [Google Scholar]
- 28.Kurdistani, S. K., Robyr, D., Tavazoie, S. & Grunstein, M. (2002) Nat. Genet. 31 248-254. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., Kouzarides, T. & Schreiber, S. L. (2002) Proc. Natl. Acad. Sci. USA 99 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28 327-334. [DOI] [PubMed] [Google Scholar]
- 31.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P. & Aparicio, O. M. (2001) Science 294 2357-2360. [DOI] [PubMed] [Google Scholar]
- 32.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A. & Dynlacht, B. D. (2002) Genes Dev. 16 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein, E., Klein, G., Nadkarni, J. S., Nadkarni, J. J., Wigzell, H. & Clifford, P. (1968) Cancer Res. 28 1300-1310. [PubMed] [Google Scholar]
- 35.Nilsson, K., Giovanella, B. C., Stehlin, J. S. & Klein, G. (1977) Int. J. Cancer 19 337-344. [DOI] [PubMed] [Google Scholar]
- 36.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hateboer, G., Timmers, H. T., Rustgi, A. K., Billaud, M., van't Veer, L. J. & Bernards, R. (1993) Proc. Natl. Acad. Sci. USA 90 8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maheswaran, S., Lee, H. & Sonenshein, G. E. (1994) Mol. Cell. Biol. 14 1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruppert, S., Wang, E. H. & Tjian, R. (1993) Nature 362 175-179. [DOI] [PubMed] [Google Scholar]
- 40.Hisatake, K., Hasegawa, S., Takada, R., Nakatani, Y., Horikoshi, M. & Roeder, R. G. (1993) Nature 362 179-181. [DOI] [PubMed] [Google Scholar]
- 41.Orian, A., van Steensel, B., Delrow, J., Bussemaker, H. J., Li, L., Sawado, T., Williams, E., Loo, L. W. M., Cowley, S. M., Yost, C., et al. (2003) Genes Dev., 17 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vafa, O., Wade, M., Kern, S., Beeche, M., Pandita, T. K., Hampton, G. M. & Wahl, G. M. (2002) Mol. Cell 9 1031-1044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.