Abstract
In mammals, xanthine oxidoreductase is synthesized as a dehydrogenase (XDH) but can be readily converted to its oxidase form (XO) either by proteolysis or modification of cysteine residues. The crystal structures of bovine milk XDH and XO demonstrated that atoms in the highly charged active-site loop (Gln-423–Lys-433) around the FAD cofactor underwent large dislocations during the conversion, blocking the approach of the NAD+ substrate to FAD in the XO form as well as changing the electrostatic environment around FAD. Here we identify a unique cluster of amino acids that plays a dual role by forming the core of a relay system for the XDH/XO transition and by gating a solvent channel leading toward the FAD ring. A more detailed structural comparison and site-directed mutagenesis analysis experiments showed that Phe-549, Arg-335, Trp-336, and Arg-427 sit at the center of a relay system that transmits modifications of the linker peptide by cysteine oxidation or proteolytic cleavage to the active-site loop (Gln-423–Lys-433). The tight interactions of these residues are crucial in the stabilization of the XDH conformation and for keeping the solvent channel closed. Both oxidative and proteolytic generation of XO effectively leads to the removal of Phe-549 from the cluster causing a reorientation of the bulky side chain of Trp-336, which then in turn forces a dislocation of Arg-427, an amino acid located in the active-site loop. The conformational change also opens the gate for the solvent channel, making it easier for oxygen to reach the reduced FAD in XO.
Xanthine oxidoreductase (XOR) is a homodimer of molecular weight 290,000, and each subunit of the enzyme contains one molybdo-pterin (Mo-pt) cofactor, two distinct [2Fe-2S] centers, and one flavin adenine dinucleotide (FAD) cofactor (1, 2). The mammalian XORs catalyze the hydroxylation of hypoxanthine or xanthine at the Mo center, and reducing equivalents thus introduced into the enzymes are transferred via two [2Fe-2S] centers to FAD, where the reduction of NAD+ or molecular oxygen occurs (3). These enzymes are synthesized as the dehydrogenase form [xanthine dehydrogenase (XDH)] but can be readily converted to the oxidase form [xanthine oxidase (XO)] reversibly by oxidation of sulfhydryl residues or irreversibly by proteolysis (1, 2, 4–6). XDH shows a preference for NAD+ reduction at the FAD reaction site (although it still displays considerable reactivity with oxygen), whereas XO fails to react with NAD+ and exclusively uses dioxygen as its substrate, leading to formation of superoxide anion and hydrogen peroxide (1, 7). Previous investigations have suggested that the XDH/XO conversion is related to milk lipid secretion (8, 9) and is implicated in diseases characterized by oxygen radical-induced tissue damage such as postischemic reperfusion injury (10–13). Thus, the XDH/XO transition has attracted much attention from both basic and clinical researchers, not only because of the mechanistic interest in the different reactivity of FAD toward NAD+ or oxygen substrates (1, 7) but also because of the biological or pathological significance (14).
Based on studies with 6- and 8-mercaptoflavins as active-site probes, it has been suggested that the structural environment around the FAD reaction site is significantly different in XDH and XO, respectively. The pK values of the probes' ionizable substituents are >4 pH units higher in the XDH form than in both irreversibly (15) and reversibly (16, 17) generated forms of XO. The experiments also showed that FAD is quite accessible to solvent in XO but much less so in XDH (15, 17). The crystal structures of bovine milk XOR have been solved recently in both the XDH form at 2.1-Å resolution and in the pancreatin-cleaved XO form at 2.5-Å resolution, and the structural transition accompanying the XDH/XO conversion of the enzyme was demonstrated (18). Comparison of the bovine milk XOR crystal structures clearly showed that, during the XDH-to-XO conversion, several atoms in the highly charged active-site loop (Gln-423–Lys-433) around the FAD were dislocated by up to 20 Å, a quite remarkable distance. In the XO form, this loop blocks the approach of the pyridine ring of the NAD+ substrate to the isoalloxazine ring of the cofactor and changes the electrostatic environment around FAD. Thus, the movement of the active-site loop is considered to be the direct cause of conversion between XDH and XO (18).
Chemical modification studies have shown that Cys-535 and Cys-992 of rat liver XDH (19) or bovine XDH (20) are involved in the reversible conversion caused by modification of sulfhydryl residues by 1-fluoro-2,4-dinitrobenzene (19) or iodoacetamide (20). The crystal structure of XDH revealed that the FAD and Mo domains are connected by a linker consisting of amino acids 532–589, and this linker lies on the surface of the protein, spanning a distance of ≈70 Å. Corresponding electron density could not be found in the map of pancreatin-cleaved XO because of mobility shown by this stretch of amino acids in this form of the enzyme. Peptide bond cleavage of XDH with trypsin after Lys-551 or with pancreatin after Lys-569 is known to result in irreversible conversion from XDH to XO (18). Both Lys-551 and Lys-569 again are located on the linker connecting the FAD and Mo domains.
It is striking that the modification sites and the FAD reaction site, described above, are so far apart in the three-dimensional structure, and therefore the question naturally arises as to how these cysteine modifications or proteolytic cleavage sites can cause XDH/XO conversion. This article describes a cluster of amino acids that seems to play a dual role, one as the gate for a channel providing access for molecular oxygen to the FAD cofactor and another one as the core of the relay system coupling the events at modification sites with the dislocation of the active-site loop.
Experimental Procedures
Cloning of Bovine XDH cDNA and Site-Directed Mutagenesis. Total RNA was isolated from bovine liver tissue, and bovine XDH cDNA was obtained by RT-PCR with primers designed on the basis of the published cDNA sequence of bovine milk XDH (21). The cDNA fragment was cloned into a pBluescript II KS (-) vector (Stratagene). Site-directed mutagenesis was performed by using a QuikChange site-directed mutagenesis kit (Stratagene). The mutated double-stranded DNA was analyzed and transformed into Escherichia coli strain HB101 (Takara, Tokyo). The resultant double-stranded vectors were isolated and digested. Each resulting fragment was ligated into the baculovirus transfer vector pJVP10Z that had been digested previously with the same restriction enzyme and analyzed again.
Expression of Recombinant Enzymes and Purification of the Recombinant and Bovine Milk Enzymes. Cotransfection with Autographa californica nuclear polyhedrosis virus DNA and each newly constructed transfer vector was performed by using Bac Vector 2000 (Novagen) as described (22). Screening of the recombinant virus was carried out by plaque assay according to the manufacturer's manual. The baculovirus-Sf9 insect cell expression system was used for expression of the mutant enzymes (22). Each recombinant enzyme was purified according to the previously described method for rat recombinant XOR (22). First, the crude cell extract was partially purified by DEAE cellulose column chromatography to remove paramagnetic contaminants of the host insect cells. The enzyme preparation was used for spectral experiments of NADH reduction after passage through a hydroxyapatite column to remove monomeric species (22). The enzyme was purified further by passing it through a folate affinity column (23) for other experiments. Bovine milk XO was isolated from fresh bovine milk and purified according to the method reported (24) but using only the first folate affinity chromatography step.
DTT Treatment. Freshly purified milk XO or mutant enzymes were incubated with 5 mM DTT at pH 8.5 and 25°C for 1 h before being used for analysis.
Assay Methods. Enzyme activity was measured as reported (24). NAD-dependent activity was determined by following the absorbance change at 340 nm, and O2-dependent activity was determined at 295 nm without NAD+ by using a Hitachi (Tokyo) 3300 spectrophotometer equipped with a temperature-controlled cell. The concentration of mutant enzyme was quantified spectrophotometrically by using a molar absorption coefficient of 37,800 M-1·cm-1 at 450 nm (25). The activity-to-flavin ratio (AFR) was obtained at 25°C as described (26). In steady-state kinetics of xanthine-NAD activity, bovine milk XDH and the DTT-treated W336A mutant were incubated at 25°C in 0.1 M pyrophosphate buffer (pH 8.5) containing 0.2 mM EDTA, 0.15 mM xanthine, and various concentrations of NAD+, and the NAD-dependent activities were determined. In steady-state kinetics of xanthine-O2 activity, the same reaction conditions were used with various concentrations of O2 without NAD+. Before assays were started, each reaction mixture was bubbled with a different concentration of oxygen for 5 min at 25°C, and each enzyme sample was also treated under the same gas stream for 2 min at 25°C in a Chance-type cuvette (SLM Aminco).
Inhibition by NADH of Xanthine Oxidation Catalyzed by Bovine Milk XOR and the W336A Mutant. Twenty nanomolar milk XO, 150 nM milk XDH, 12 nM freshly purified W336A mutant, or 12 nM DTT-treated W336A mutant were assayed in the presence of various concentrations of NADH under aerobic conditions in 0.1 M pyrophosphate buffer (pH 8.5) containing 0.2 mM EDTA and 0.15 mM xanthine at pH 8.5 and 25°C. The AFR values of milk XO and the mutant enzyme were 111 and 33, respectively. The O2-dependent activities were determined and plotted as percentages of the initial O2-dependent activity for each type of enzyme.
Spectra During Reaction of NADH. Anaerobic enzyme samples of milk XOR (AFR = 143), 4.9 μM DTT-treated form, and the W336A mutant (AFR = 37), 5.1 μM DTT-treated form, in 0.1 mM pyrophosphate buffer (pH 7.5) containing 0.2 mM EDTA and 1/20 volume of NADH (10 mM) in the same buffer were prepared in an all-glass apparatus (27) by sequential evacuation and reequilibration with oxygen-free argon. They were mixed anaerobically, and the absorption spectra were taken by using a Beckman Coulter DU 7400 spectrophotometer.
Results and Discussion
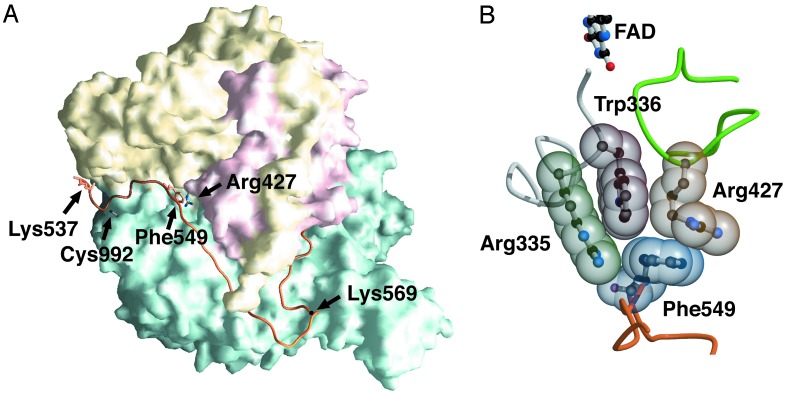
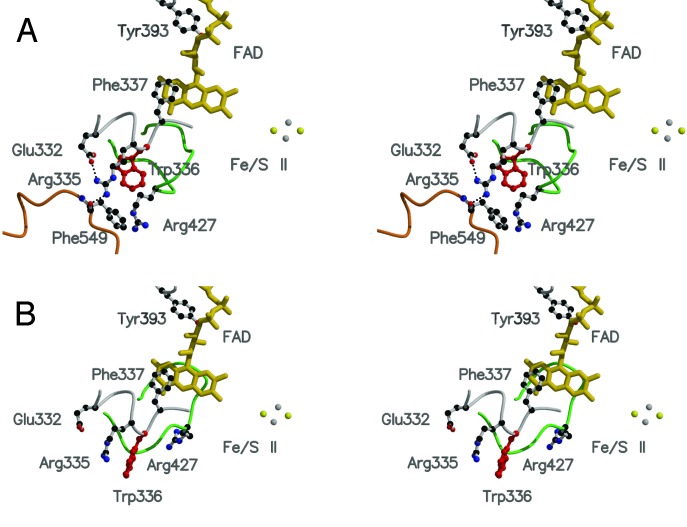
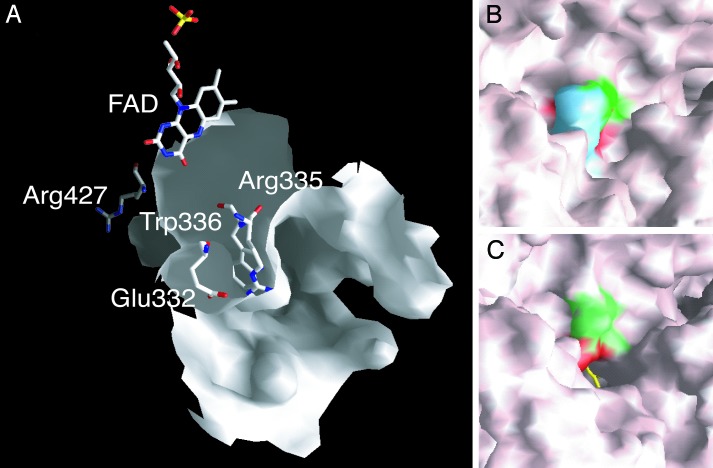
The Structure of a Tightly Packed Amino Acid Cluster Related with the XDH/XO Conversion. The modification sites responsible for reversible and irreversible conversion are located on the linker peptide (residues 532–589) connecting the Mo-pt and FAD domains, and the distances between the modification sites and the FAD reaction site are large (Fig. 1A). As described previously, modification of Cys-992 and Cys-535 or proteolysis along the linker (Lys-551 with trypsin or Lys-569 with pancreatin) causes parts of the active-site loop (Gln-423–Lys-433) to move by up to 20 Å to the front of the isoalloxazine ring of FAD, leading to the XDH/XO conversion (18). Further, a more detailed examination of the x-ray crystal structures indicates that the aromatic ring of Phe-549, located on the linker between FAD and the Mo-pt domain, in the XDH form makes close contact with the side chain of Arg-427, which is located on the loop undergoing the dramatic change in conformation (Fig. 1 A). The side chain of Trp-336 is located between those of Arg-335 and Arg-427 and is in van der Waals contact with the side chains of Arg-427 and Phe-549, forming a tightly packed cluster (Fig. 1B). In the XDH form, Arg-335 makes two hydrogen bonds with the carbonyl oxygens of Phe-549 and Glu-332; the bond distances are 2.6 and 2.7 Å, respectively (Fig. 2A). Accompanying the dislocation of the active-site loop, the orientation of Trp-336 is distinctly different between the XDH and XO forms (Fig. 2). It is also noted that the cluster exists at the bottom of the cavity (Fig. 3A) that accommodates the FAD cofactor and is closed tightly in the XDH form (Fig. 3B), whereas it is opened in the XO form (Fig. 3C); this results in increased solvent exposure of the isoalloxazine ring around the C(4)-C(6) positions (Fig. 3 A and C). Based on these structural differences, it seems likely that these residues play a key role in the XDH/XO conversion mechanism; we suggest that they not only form a relay system that couples the linker, which undergoes modification of cysteine residues or proteolytic cleavage, with the consequent change in conformation of the active-site loop but also a gate for a solvent channel leading toward the FAD cofactor. In particular, reorientation of the bulky moiety of Trp-336 is suggested to play a major role in the transition mechanism accompanying the effective removal of Phe-549 due to its large movement after proteolysis or oxidation of sulfhydryl residues. Differing solvent accessibilities in XDH and XO are consistent with the results reported by Massey et al. (15) from 6-SH-FAD active-site probe experiments showing that the 6-position of FAD is much more solvent-accessible in milk XO than in chicken liver XDH.
Fig. 1.
(A) Molecular surface of the XDH monomer divided into three major domains. The domains are iron/sulfur-center (residues 3–165; red), FAD (residues 226–531; yellow), and Mo-pt (residues 590–1,331; blue). The linker connecting the FAD domain with the Mo-pt domain (residues 537–589) is shown in orange. The positions of Cys-992 and Lys-537, the one still visible in the electron density map and closest to Cys-535, are shown as a stick model. The positions of Lys-551 and Lys-569, responsible for proteolysis, are also indicated. The positions of Phe-549, which is located on the linker connecting the Mo and FAD domains, and Arg-427, which is located on the sliding loop, are represented as stick models. (B) Space-filling model of the important cluster residues in the XDH form that are involved in the XDH/XO conversion.
Fig. 2.
The active-site loop and important residues involved in the XDH/XO conversion. Differences in active-site structure of the XDH (A) and XO (B) forms are shown. The active-site loop (Gln-423–Lys-433), which moves during the conversion from XDH to XO, is shown in green, and the linker connecting the Mo and FAD domains is shown in orange. Trp-336, which has different configurations in the XDH and XO forms, is shown as a red ball-and-stick model.
Fig. 3.
Molecular surface representation of the FAD cavity. (A) FAD and the amino acid cluster in its “open” conformation are shown in the cavity of the XO form as stick models colored by atom type. (B) The cluster in its “closed” conformation as part of the molecular surface of the XDH form. Phe-549 is shown in blue, Arg-335 is shown in green, and Trp-336 is shown in red. (C) Open conformation of the cluster as part of the molecular surface of the XO form; the accessible edge of the FAD ring is shown as a yellow stick model, and the surfaces corresponding to Arg-335 and Trp-336 are shown in green and red, respectively.
Mutation Analyses of the Cluster Residues. To confirm the roles of Arg-335, Trp-336, and Arg-427 in the XDH/XO conversion, we targeted these residues by site-specific mutagenesis in an effort to disrupt the amino acid cluster. As expressed and analyzed in the crude extract, the W336A mutant displayed mostly XO activity, whereas the wild-type enzyme was in the XDH form. The other mutants, R335A and R427Q, showed higher XO activity while still retaining significant XDH activity. All the recombinant enzymes were purified as the XO form not due to proteolysis as will be described below but due to cysteine oxidation. The absorption spectra of all three enzymes were indistinguishable (data not shown).
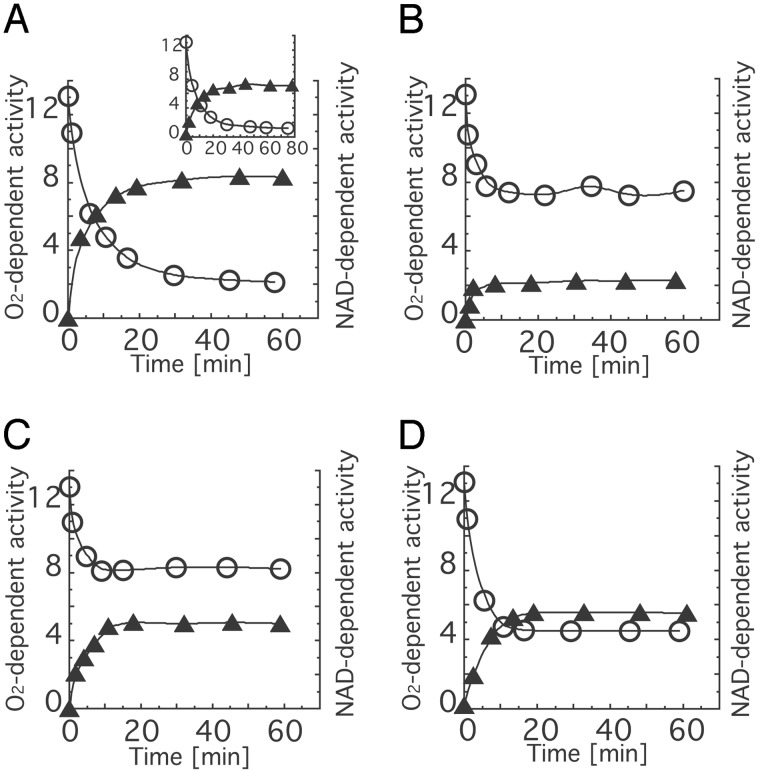
Because the finally purified enzymes were obtained as XO forms, we tried to reconvert them to their XDH forms by DTT treatment. Fig. 4 shows the time course of apparent conversion of the freshly purified recombinant wild-type enzyme and the mutant enzymes R335A, W336A, and R427Q during incubation with DTT. When the freshly purified recombinant wild-type XO was incubated with 5 mM DTT, most of the oxidase-type enzyme initially present was converted to the dehydrogenase type in the same manner as freshly purified bovine milk XOR (Fig. 4A). On the other hand, the W336A mutant showed only a slight increase of the NAD+-dependent activity and the O2-dependent activity remained high, in contrast to the wild-type enzyme after DTT treatment (Fig. 4B). The R427Q mutant also showed high O2-dependent activity after DTT treatment, to an extent similar to W336A, although the NAD+-dependent activity was comparable with that of the wild-type enzyme (Fig. 4C). The R335A mutant also showed higher O2-dependent activity and lower NAD+-dependent activity than wild-type enzyme but less pronounced than the W336A mutant (Fig. 4D).
Fig. 4.
Time course of XDH/XO conversion of wild-type and mutant enzymes by incubation with dithiothreitol. Each protein was incubated with 5.0 mM DTT at pH 8.5 and 25°C. During the incubation, aliquots were withdrawn from the mixture to measure the enzyme activities. The activities were corrected for the measured value of AFR assuming that the AFR of fully active enzyme in XO and the freshly purified mutant enzymes was 200 (28). O2-dependent activity and NAD+-dependent activity are expressed as rate of urate formation [mol/mol per sec] and rate of NADH formation [mol/mol per sec], respectively. Open circles, O2-dependent activities; filled triangle, NAD+-dependent activities. Values for recombinant wild-type enzyme (A), bovine milk XOR (Insert), the W336A mutant (B), the R427Q mutant (C), and the R335A mutant (D) are shown.
Steady-State Kinetics for Bovine Milk XOR and the Mutant Enzymes R335A, W336A, and R427Q. Table 1 shows the Km and kcat values for O2 and NAD+ as substrates for each enzyme. The general properties of all the freshly purified mutant enzymes were similar to those of freshly purified wild-type XOR (XO) because the Km values for O2 and kcat values of xanthine-O2 activity were similar to those of the wild-type XO, and no NAD+-dependent activity was detected. However, DTT-treated mutant enzymes showed various properties that were in contrast to those of DTT-treated wild-type XOR (corresponds to XDH). Km values for O2 were decreased in all DTT-treated mutant enzymes, consistent with the interpretation that the cluster is the gate for solvent to the FAD and is opened by the mutation of the residues such that more oxygen can be available at the FAD site. On the other hand, although kcat values of xanthine-O2 activity remained quite high compared with wild-type XDH, their levels varied from mutant to mutant. Furthermore, although the Km values for NAD+ were close to those of milk XDH, the corresponding kcat values of xanthine-NAD+ activity were lower. The DTT-treated W336A mutant showed particularly pronounced changes; its kcat value for O2 was almost comparable with that of wild-type XO, i.e., five times as high as that of wild-type XDH, and the kcat value for NAD+ was the lowest one of all the mutant enzymes (≈27% of wild-type XDH). Because the cluster of the W336A mutant seemed to be disrupted most significantly, the properties of this enzyme were analyzed in more detail.
Table 1. Steady-state kinetic parameters for bovine milk XOR and mutant enzymes.
| Protein | Type | kcat X-O2, sec-1 | Km for O2, μM | kcat for NAD, sec-1 | Km for NAD, μM |
|---|---|---|---|---|---|
| Milk XOR | XO | 15.1 ± 0.2 | 37.7 ± 0.6 | Not detected | — |
| XDH | 2.5 ± 0.1 | 111.0 ± 5.3 | 12.3 ± 1.2 | 20.8 ± 2.2 | |
| W336A | Freshly purified | 15.1 ± 0.3 | 44.5 ± 0.8 | Not detected | — |
| DTT-treated | 11.5 ± 0.7 | 48.6 ± 3.0 | 3.3 ± 0.2 | 18.2 ± 0.2 | |
| R427Q | Freshly purified | 16.6 ± 1.0 | 73.5 ± 4.6 | Not detected | — |
| DTT-treated | 11.0 ± 0.2 | 66.6 ± 1.5 | 5.7 ± 0.4 | 28.5 ± 1.8 | |
| R335A | Freshly purified | 15.0 ± 0.4 | 42.9 ± 0.8 | Not detected | — |
| DTT-treated | 6.1 ± 0.1 | 40.6 ± 0.9 | 6.2 ± 0.3 | 23.5 ± 1.1 |
Apparent values with 0.15 mM xanthine, pH 8.5, at 25°C. The kcat values were corrected for the measured value of AFR of the XO form assuming an AFR value of 200 for both the sulfo form of the fully active XO enzyme and all freshly purified mutant enzymes (XO form) (28)
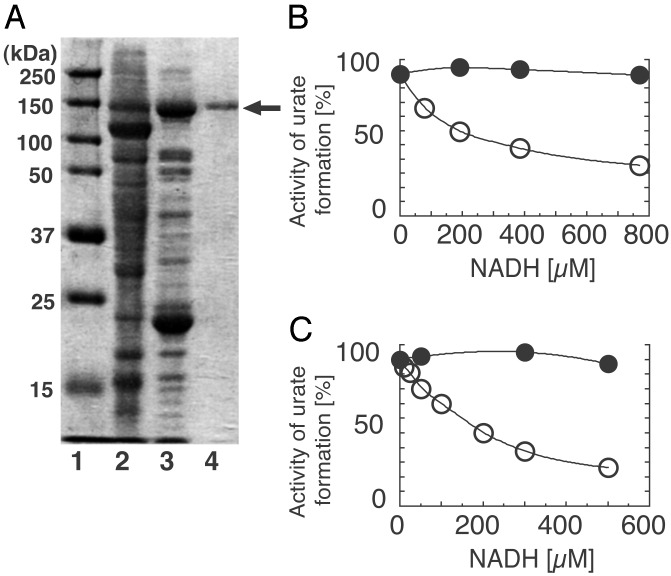
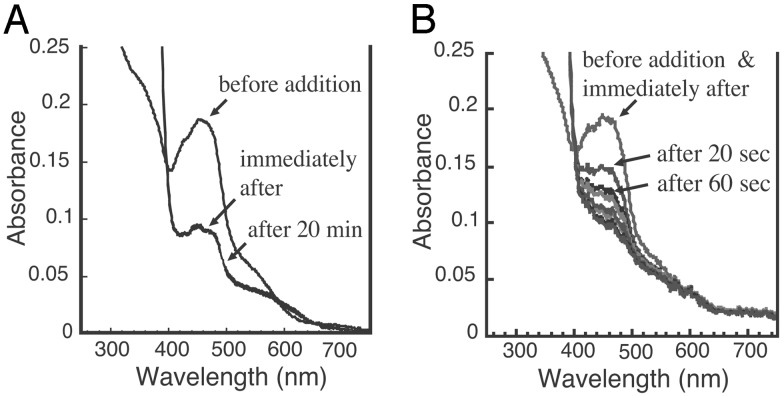
Properties of the W336A Mutant Enzyme Before and After DTT Treatment. Although the DTT-treated W336A mutant retained high oxidase activity, SDS/PAGE analysis (Fig. 5A) displayed only full-length enzyme, proving that this activity was not due to contamination with the irreversible, proteolytically generated oxidase form of the enzyme. No proteolysis was also observed with other mutants (data not shown). Another well established characteristic that distinguishes XDH from XO is the presence or absence of an NAD-binding site (1, 7, 17, 18, 24). When we compared the NADH inhibition of O2-dependent urate formation by the W336A mutant with that by bovine milk XOR we found that neither the wild-type nor the W336A enzyme were inhibited in the presence of NADH while in the XO form, whereas after DTT treatment both enzymes were subject to NADH inhibition (Fig. 5 B and C). These results argue that, after DTT treatment, competent NADH-binding sites exist in both mutant and wild type, a feature that is also confirmed by their NADH reducibility. Fig. 6 shows the reduction of the W336A mutant and of milk XDH with NADH under anaerobic conditions. The DTT-treated mutant enzyme was clearly reduced by NADH, but the observed rate of this reaction was significantly slower than is observed for wild-type XDH. Interference by the flexible loop with the proper access of the nicotinamide ring to the isoalloxazine ring of FAD presumably accounts for this effect, even though the ability of the enzyme to bind the pyridine dinucleotide is retained. It should be noted that the increase of the absorbance at 620 nm, which reflects the formation of flavin semiquinone (17, 30, 31), was much lower in the mutant enzyme, indicating that the flavin semiquinone is less stable in the DTT-treated mutant enzyme than in wild-type XDH. This is consistent with the fact that the DTT-treated mutant enzyme has a higher kcat value toward oxygen as described above, because it is known that fully reduced flavin reacts much faster with oxygen than does flavin semiquinone (30, 32). On the other hand, the non-DTT-treated mutant enzyme was not reduced at all within a few minutes by NADH but only after very prolonged incubation; similar results were obtained with the wild-type XO (data not shown). The results discussed in this paragraph point to structural and functional differences between W336A mutant and wild-type milk XO despite their common high O2-dependent oxidase activity.
Fig. 5.
(A) SDS/PAGE analysis of the W336A mutant. Lane 1, marker proteins (Bio-Rad) with molecular masses as indicated; lane 2, crude extract; lane 3, W336A mutant after DEAE cellulose chromatography; lane 4, purified W336A mutant used for assay. SDS/PAGE was performed according to the method of Laemmli (29) by using a 10% polyacrylamide gel. Inhibition of O2-dependent activity by NADH for milk XOR (B) and W336A mutant (C). Milk XOR and W336A mutant, both DTT-treated and untreated, were assayed aerobically with various concentrations of NADH added to the assay mixture as described in Experimental Procedures.O2-dependent activity was determined and plotted as the percentage of initial activity assayed without NADH. Filled circle, untreated enzyme; open circle, DTT-treated enzyme.
Fig. 6.
Changes of the absorption spectra of milk XDH and DTT-treated W336A mutant after mixing with NADH under anaerobic conditions. Milk XDH (4.9 μM, DTT-treated) (A) or5.1 μM DTT-treated W336A mutant (B) in0.1 M pyrophosphate buffer (pH 7.5) containing 0.2 mM EDTA was anaerobically mixed with 1/20 volume of NADH (10 mM) in the same buffer, and the absorption spectra were recorded at the times indicated. The initial spectra were taken within 5 sec of the addition of NADH.
General Discussion. In this article we define the amino acid cluster consisting of the residues Phe-549, Arg-335, Trp-336, and Arg-427 as the core of a relay system linking oxidative or proteolytic modifications occurring on the molecular surface of XOR to the large movement of an internal loop, which closely approaches the flavin reactive site and seems to be the one structural feature characterizing the transformation of the XDH to the XO form and vice versa. This cluster also plays the role of a gate guarding solvent access to the FAD cofactor. Mutagenesis experiments showed that the reverse XO-to-XDH conversion was disrupted the most in the W336A mutant, indicating that Trp-336 plays a major role in the stabilization of the XDH form likely by forming π-cation interactions with its arginine neighbors (33). In the Rhodobacter capsulatus XDH, which cannot be converted to an oxidase form, no such cluster exists; the residue corresponding to Trp-336 is not conserved but is replaced by an arginine residue, and the linker connecting the FAD and Mo domains is absent because the flavin and molybdenum domains reside in separate subunits. In addition, the conformation of the R. capsulatus enzyme seems to be stabilized in the XDH form by additional steric factors (34, 35).
Chemical modification studies have implicated Cys-535 and Cys-992 of rat liver (19) or bovine XOR (20) in the formation of a disulfide bond during the reversible XDH/XO conversion. This interpretation has been challenged recently in a report describing gel chromatographic analyses of bovine enzyme in the XDH and proteolytically created XO forms (36). However, site-directed mutagenesis (Tomoko Nishino, H. Hori, T.M., and Takeshi Nishino, unpublished data) and x-ray crystallographic studies of the bovine XO form obtained by oxidation of XDH (B.T.E., K.O., Takeshi Nishino, and E.F.P., unpublished data) provide very strong evidence that these residues form a disulfide bond in the reversible conversion.
Recovery of the NADH-binding site by DTT treatment of the mutant enzymes can be explained by incomplete disruption of the cluster through mutational change. Another, more interesting cause, however, could be the involvement of another cysteine couple in the pyridine nucleotide-binding site, which does not involve Cys-992 and can be reduced by DTT. Supporting the latter possibility are reports that four cysteine residues are modified when bovine XDH is converted to XO by incubation with dithiodipyridine (17). In addition, Cys-992 and Cys-535 were labeled in the rapid phase of the 1-fluoro-2,4-dinitrobenzene modification reaction, whereas another cysteine couple might be involved in the slower phase of the reaction that was observed (19).
All mutant proteins were in the XO form after purification. In contrast to wild-type enzyme, however, DTT treatment of sulfhydryl residues, which had been oxidized during purification, resulted in only a partial increase in the level of NAD+-dependent activity, whereas the remaining oxidase activity was still as high as 77% of that of the untreated enzyme in the case of W336A mutant. The contributing factors for this are a lower Km value for oxygen and an increase in kcat. These findings can be explained, at least in part, by the opening of the cluster gate of the solvent channel, which provides an additional access route, thereby leading to higher oxygen concentrations at the reaction site [most likely the C(4a) position of the isoalloxazine ring]. Variations in the redox potential of flavin, i.e., destabilization of the semiquinone, are a possible explanation for the higher kcat value and could be caused by the movement of the active-site loop (Gln-423–Lys 433), which drastically modifies the electrostatic environment of the cofactor FAD (18). Persistent interference by the flexible loop with the proper access of the nicotinamide ring to the isoalloxazine ring of FAD presumably accounts for the lower kcat in xanthine-NAD+ activity as seen in the slower reduction of the W336A mutant with NADH, even though the ability of the enzyme to bind the pyridine dinucleotide is retained. The properties of the other mutants, R335A and R427Q, generally parallel those of the W336A mutant, although with smaller deviations from the milk enzyme behavior, indicating a less severe disruption of the cluster's function both as a solvent gate and a relay system. Different substitutions of this amino acid might be reflected in differences in flexibility in conformation of the active-site loop after DTT treatment, which then may cause a variation of kcat values for the substrates oxygen and NAD+.
In conclusion, the results presented in this article are consistent with the following interpretation. Residues Phe-549, Arg-335, Trp-336, and Arg-427 of bovine milk XOR form a cluster, the tight interactions of which are crucial in the stabilization of the XDH form of the enzyme. The cluster residues sit at the center of a relay system that transmits modifications of the linker peptide, caused by cysteine oxidation or proteolytic cleavage, to the active-site loop (Gln-423–Lys 433) causing a dramatic change in the conformation of the latter. Both modifications lead to the removal of Phe-549 from the cluster by either probably inducing a change in conformation (oxidation) or creating mobility (proteolysis), obvious in the electron density maps of bovine XO. The loss of the Phe-549–Trp-336 interaction leads to a reorientation of the bulky aromatic side chain of Trp-336, which then in turn forces a dislocation of Arg-427, an amino acid that is part of the active-site loop. Obviously, this is sufficient to cause a substantial change in the conformation of the whole loop (Gln-423–Lys 433) and accomplishes the conversion from the XDH to the XO form. At the same time it opens the gate to a solvent channel leading to the FAD, which will allow easier access for oxygen molecules to the XO active site.
Acknowledgments
We thank Prof. Tsutomu Araki for support and very valuable discussions and Ms. Yuko Kawaguchi for excellent technical assistance. This work was supported by Grants-in-Aid for Science Research in Priority Areas (to Takeshi Nishino and Tomoko Nishino), a Grant-in-Aid for Science Research from the Ministry of Education, Science, Sports, and Culture of Japan (to Takeshi Nishino), and a grant from the Canadian Institutes for Health Research and the Canada Research Chairs Program (to E.F.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: XOR, xanthine oxidoreductase; Mo-pt, molybdo-pterin; XDH, xanthine dehydrogenase; XO, xanthine oxidase; AFR, activity-to-flavin ratio.
References
- 1.Nishino, T. (1994) J. Biochem. 116 1-6. [DOI] [PubMed] [Google Scholar]
- 2.Amaya, Y., Yamazaki, K., Sato, M., Noda, K., Nishino, T. & Nishino, T. (1990) J. Biol. Chem. 265 14170-14175. [PubMed] [Google Scholar]
- 3.Komai, H., Massey, V. & Palmer, G. (1969) J. Biol. Chem. 244 1692-1700. [PubMed] [Google Scholar]
- 4.Della Corte, E. & Stripe, F. (1968) Biochem. J. 108 349-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stripe, F. & Della Corte, E. (1969) J. Biol. Chem. 244 3855-3863. [PubMed] [Google Scholar]
- 6.Della Corte, E. & Stripe, F. (1972) Biochem. J. 126 739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille, R. & Nishino, T. (1995) FASEB J. 9 995-1003. [PubMed] [Google Scholar]
- 8.McManaman, J. L., Neville, M. C. & Wright, R. M. (1999) Arch. Biochem. Biophys. 172 354-364. [DOI] [PubMed] [Google Scholar]
- 9.Vorbach, C., Scriven, A. & Capecchi, M. R. (2002) Genes Dev. 16 3223-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCord, J. M. (1985) N. Engl. J. Med. 312 159-163. [DOI] [PubMed] [Google Scholar]
- 11.Saugstad, O. D. (1988) Pediatr. Res. 23 143-150. [DOI] [PubMed] [Google Scholar]
- 12.Jarasch, E.-D., Grund, C., Bruder, G., Heid, H. W., Keenan, T. W. & Fanke, W. W. (1981) Cell 25 67-82. [DOI] [PubMed] [Google Scholar]
- 13.Roy, R. S. & McCord, J. M. (1982) Can. J. Physiol. Pharmacol. 60 1346-1352. [DOI] [PubMed] [Google Scholar]
- 14.Simmonds, A., Reiter, S. & Nishino, T. (1995) The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), 7th Ed., Vol. II, pp. 1781-1798. [Google Scholar]
- 15.Massey, V., Schopfer, L. M., Nishino, T. & Nishino, T. (1989) J. Biol. Chem. 264 10567-10578. [PubMed] [Google Scholar]
- 16.Saito, T., Nishino, T. & Massey, V. (1989) J. Biol. Chem. 264 15930-15935. [PubMed] [Google Scholar]
- 17.Hunt, J. & Massey, V. (1992) J. Biol. Chem. 267 21479-21485. [PubMed] [Google Scholar]
- 18.Enroth, C., Eger, B. T., Okamoto, K., Nishino, T., Nishino, T. & Pai, E. F. (2000) Proc. Natl. Acad. Sci. USA 97 10723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, T. & Nishino, T. (1997) J. Biol. Chem. 272 29859-29864. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen, J. T., Rasmussen, M. S. & Petersen, T. E. (2000) J. Dairy Sci. 83 499-506. [DOI] [PubMed] [Google Scholar]
- 21.Berglund, L., Rasmussen, J. T., Andersen, M. D., Rasmussen, M. S. & Petersen, T. E. (1996) J. Dairy Sci. 79 198-204. [DOI] [PubMed] [Google Scholar]
- 22.Nishino, T., Amaya, Y., Kawamoto, S., Kashima, Y., Okamoto, K. & Nishino, T. (2002) J. Biochem. 132 597-606. [DOI] [PubMed] [Google Scholar]
- 23.Nishino, T., Nishino, T. & Tsushima, K. (1981) FEBS Lett. 131 369-372. [DOI] [PubMed] [Google Scholar]
- 24.Saito, T. & Nishino, T. (1989) J. Biol. Chem. 264 10015-10022. [PubMed] [Google Scholar]
- 25.Massey, V., Brumby, P. E., Komai, H. & Palmer, G. (1969) J. Biol. Chem. 244 1682-1691. [PubMed] [Google Scholar]
- 26.Avis, P. G., Bergel, F. & Bray, R. C. (1955) J. Chem. Soc., 1100-1105.
- 27.Williams, C. H., Jr., Arscott, L. D., Matthews, R. G., Thorpe, C. & Wilkinson, K. D. (1979) Methods Enzymol. 62 185-198. [DOI] [PubMed] [Google Scholar]
- 28.Massey, V. & Edmondson, D. (1970) J. Biol. Chem. 245 6595-6598. [PubMed] [Google Scholar]
- 29.Laemmli, U. K. (1970) Nature 227 680-685. [DOI] [PubMed] [Google Scholar]
- 30.Schopfer, L. M., Massey, V. & Nishino, T. (1988) J. Biol. Chem. 263 13539-13543. [PubMed] [Google Scholar]
- 31.Harris, C. M. & Massey, V. (1997) J. Biol. Chem. 272 8370-8379. [DOI] [PubMed] [Google Scholar]
- 32.Nishino, T., Nishino, T., Schopfer, L. M. & Massey, V. (1989) J. Biol. Chem. 264 6075-6085. [PubMed] [Google Scholar]
- 33.Ma, J. C. & Dougherty, D. A. (1997) Chem. Rev. (Washington, D.C.) 97 1303-1324. [DOI] [PubMed] [Google Scholar]
- 34.Leimkuhler, S., Kern, M., Solomon, P. S., McEwan, A. G., Schwarz, G., Mendel, R. R. & Klipp, W. (1998) Mol. Microbiol. 27 853-869. [DOI] [PubMed] [Google Scholar]
- 35.Truglio, J. J., Theis, K., Leimkuhler, S., Rappa, R., Rajagopalan, K. V. & Kisker, C. (2002) Structure (London) 10 115-125. [DOI] [PubMed] [Google Scholar]
- 36.McManaman, J. L. & Bain, D. L. (2002) J. Biol. Chem. 277 21261-21268. [DOI] [PubMed] [Google Scholar]