Abstract
Methionine sulfoxide reductase A (MsrA) maintains the function of many proteins by reversing oxidation of methionine residues. Lack of this repair mechanism very likely increases aging-related disease susceptibility. In Saccharomyces cerevisiae, disruption of the msrA gene increases free and protein-bound methionine sulfoxide and decreases cell viability. Although the underlying mechanisms in the induction of the msrA gene are still unknown, a transcriptional regulation may be involved. Hence, a search of nuclear proteins regulating the msrA gene is a major target of the experiments reported in this article. Using protein purification combined with MS, we discovered that calcium phospholipid-binding protein (CPBP), a homologue of elongation factor-1γ, is a component of a complex that binds to the msrA promoter. By measuring CPBP cooperative binding to the msrA promoter, we have mapped the CPBP binding site to a 39-bp sequence at the 3′ end of the promoter. In a mutant yeast strain lacking the CPBP-encoding gene, the ability to overexpress msrA mRNA and MsrA protein was impaired and MsrA catalytic activity was greatly reduced, suggesting that CPBP may enhance msrA gene expression.
Oxidative damage of nucleic acids, lipids, and proteins is associated frequently with an aging-related pathology including metabolic diseases and a decline of cognitive function (see ref. 1 for review). An age-related increase of protein oxidation was reported in Drosophila melanogaster (2). Interestingly, flies that were resistant to oxidative damage exhibited a lifespan prolongation (3, 4). Many proteins whose biological activity depends on methionine residues are particularly sensitive to oxidation (5). Unlike other oxidized amino acid residues, methionine sulfoxide can be reduced to methionine with the thioredoxin-dependent methionine sulfoxide reductase (Msr), types A and B (6, 7). This enzymatic reduction occurs ubiquitously and may protect cells against oxidative damage (8, 9). The msrA gene of Saccharomyces cerevisiae is up-regulated by oxidative and other environmental stresses (Saccharomyces Genome Database, www.yeastgenome.org). MsrA seems to play an important role in the adaptive response of yeast to oxidative stress. In fact, deletion of the msrA gene results in extensive oxidative damage followed by cell death (10, 11).
Because the promoter region of the mammalian msrA gene still is unexplored, we used a yeast model to study the transcriptional regulation of msrA. An upstream noncoding DNA sequence harbors the promoter region of yeast msrA (11). This region (spanning positions 16978–17176 on chromosome V of S. cerevisiae) contains several TATA sequences and a consensus sequence for Skn7p, a reactive oxygen species-activated transcription factor (12). However, Skn7p does not seem to regulate msrA transcription because MsrA overexpression was not impaired in an Skn7p-null mutant yeast strain (J.M., unpublished data). According to database analysis (www.ncbi.nlm.nih.gov/GenBank/GenbankSearch.html), no other cis-regulatory elements for known transcription factors seem to be present. To gain further insight into msrA gene regulation during stress response, we grew yeast lacking the msrA gene until stationary phase; then we (i) isolated nuclear protein(s) that formed a specific binding complex with the msrA promoter region as described (11), (ii) identified the targeted protein by MS, and (iii) verified in vivo the functional role of a putative msrA regulator protein by using genetically manipulated yeast strains.
Materials and Methods
Yeast Strains and Media. The yeast strains used in this study were as follows: msrA-null mutant [(H9ΔmsrA::URA3) + YEp351] and its parent yeast strain (H9 + YEp351), and TEF3-null mutant (TKY 661), TEF4-null mutant (TKY 662), and their parent strain (TKY 660). All yeast strains were grown in appropriate SD media (Qbiogene, Carlsbad, CA).
Nuclear Protein Extract. A suspension of the msrA-null mutant yeast strain was centrifuged for 5 min at 5,000 rpm (Sorvall SS34) and washed with 1 M sorbitol containing 50 mM potassium phosphate (pH 7.8), 10 mM MgCl2, 20 mM DTT, and 0.5 mM PMSF. The pellet was resuspended in 1 M sorbitol containing 25 mM potassium phosphate (pH 7.8), 10 mM MgCl2, 2 mM DTT, 25 mM sodium succinate, and 0.5 mM PMSF and was incubated at 30°C with Zymolase 20T (final concentration 4 mg/ml Zy-molase 20T; ICN) until protoplast formation had occurred. The protoplasts were homogenized in the above-described buffer containing 0.2% Triton X-100 (glass/Teflon Potter–Elvehjem homogenizer) and centrifuged at 3,500 rpm for 5 min. The resuspended cells (in the above buffer containing 0.5% Triton X-100) were broken with glass beads and centrifuged for 15 min in a Beckman Coulter Microfuge at maximum speed. The resulting supernatant served as starting material for the following protein purification.
Protein Purification. Nuclear protein extract was fractionated with ammonium sulfate (30%, 60%, and 100% of saturation). The precipitates were redissolved in buffer A (10 mM Hepes buffer, pH 7.6/20 mM DTT/0.5 mM PMSF/0.05% Nonidet P-40/20% glycerol) and dialyzed against buffer A containing 0.15 M KCl. The resulting clear protein solution was passed over a HiTrap heparin-affinity column (2.0 mg of protein per milliliter; Amersham Biosciences) that was equilibrated with buffer A containing 0.15 M KCl. The column was washed with 10 column volumes of the same buffer and eluted stepwise with 5 column volumes of buffer A containing 0.30, 0.60, or 1.0 M KCl. The fractions were dialyzed against buffer A and concentrated to 100 μl in Centricon tubes (Amicon, Millipore). Each fraction was tested for DNA-binding activity by electrophoretic mobility-shift assay (EMSA) as described below. Fractions with DNA-binding activity were dialyzed against 50 mM Tris·HCl buffer, pH 6.6, containing 20 mM DTT, 0.5 mM PMSF, 0.05% Nonidet P-40, and 20% glycerol and were loaded on a CM Sepharose Fast Flow column (2.0 mg of protein per ml of CM Sepharose Fast Flow; Amersham Biosciences) previously equilibrated with the same buffer. After washing with 10-column volumes of the above buffer, stepwise elution was carried out with 5-column volumes of the above buffer containing 50, 100, 200, 300, or 500 mM NaCl. The eluates were concentrated to 100 μl on Centricon tubes and dialyzed against buffer A, and each fraction was tested for DNA-binding activity by EMSA as described below. The fractions that contained DNA-binding activity were dialyzed against 10 mM Tris·HCl buffer, pH 6.6, and were subjected to Tris/glycine/SDS/4–20% PAGE (NOVEX, San Diego).
Reduction and Alkylation. Coomassie blue-stained gel slices were excised and subjected to in-gel trypsin digestion. The digests were analyzed on an electrospray hybrid quadrupole time-of-flight mass spectrometer (Q-Tof 2, Micromass, Manchester, U.K.) integrated with Waters CapLC before MS. Liquid chromatography/tandem MS (LC-MS/MS) was carried out on a Zorbax ZC-10-C18SBW column (Microtech, Sunnyvale, CA). MS/MS data were processed with MASSLYNX V.3.5 (Micromass), and the generated peak list files were submitted for database search by using MASCOT (Matrix Science, London; peptide and MS/MS tolerance was ±0.2 Da).
EMSA. EMSA was performed essentially as described by Carey and Smale (13). The DNA segments containing different yeast msrA promoter sequences and their specific PCR primers are shown in Table 1. 32P-labeled probes were synthesized by PCR using Taq DNA polymerase (Promega), [α-32P]dCTP (0.1 μCi per 50-μl final incubation volume; 1 Ci = 37 GBq; ICN), 0.02 mM dCTP, 0.2 mM dATP, 0.2 mM dGTP, 0.2 mM dTTP, and synthetic oligonucleotide primers (BioSynthesis, Lewisville, TX; see Table 1; PCR cycle: 5 min at 94°C followed by 33 cycles of 30 sec at 94°C, 60 sec at 50°C, and 60 sec at 72°C). The PCR products were purified with the Wizard PCR Preps DNA Purification System (Promega). Short DNA fragments were obtained by annealing (at 72°C for 15 min) the 39-mer oligonucleotides and their complementary reverse oligonucleotides (Table 1) and end-labeling with Redivue [γ-32P]ATP (6,000 Ci/mmol; Amersham Biosciences) by using the T4 polynucleotide kinase system (Promega). The EMSA reaction mixture contained 1× gel shift buffer (Promega), yeast nuclear protein solution (20 μg of protein per lane) previously dialyzed against buffer A, and 32P-labeled DNA (≈100,000 cpm). The mixtures were incubated at 24°C for 20 min. To identify calcium phospholipid-binding protein (CPBP) in the DNA-binding complex, a postincubation procedure with either rabbit polyclonal anti-His6-CPBP antibody or His6-monoclonal antibody (CLON-TECH) was carried out. The reaction was stopped with 10× gel-shift sample buffer (Promega), and samples were loaded on a 4% polyacrylamide nondenaturing gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) and electrophoresed for 3 h at 125 mV. The dried gel was either exposed to x-ray film (BioMax MS, Kodak) with intensifying screens at -76°C or evaluated by phosphor imaging (PhosphorImager, Storm 860, Molecular Dynamics).
Table 1. Segments of yeast msrA promoter used as probes.
| Segment | Forward primer | Reverse primer |
|---|---|---|
| Whole msrA promoter (16977-17186) | ATG AGT TAA CGG GCA CAG CGT CAA AAA ATC CGC (16977-17019) | TAA ATC AAG TTG CAA TGT CGT CGC (17163-17186) |
| 5′ end segment (16978-17055) | ATG AGT TAA CGG GCA CAG CGT CAA AAA ATC CGC (16977-17019) | GC CGA AAA CGA GGG AGT TGC GAT GAG TAA A (17026-17055) |
| First TATA box* (17061-17186) | GGG CAT ATA TAA ACA AGA GTA GAC AGA GAC AC (17061-17092) | TAA ATC AAG TTG CAA TGT CGT CGC (17163-17186) |
| Second and third TATA box (17074-17148) | CAA GAG TAG ACA GAG ACA CAT TTT TCA (17074-17100) | GGA AAG ACA GTT ATA TTC TAA CGT C (17123-17148) |
| 3′ end segment† (17148-17186) | CAA AAA CTT AAC AAG TAA ATC AAG TTG CAA TGT CGT CGC (17148-17186) | GCG ACG ACA TTG CAA CTT GAT TTA CTT GTT AAG TTT TTG‡ (17148-17186) |
The numbers in parentheses indicate the positions spanned
The forward primer for a point mutation in the first TATA box (spanning positions 17061-17092) is GGG CAT ATG TAA ACA AGA GTA GAC AGA GAC AC
The 3′ end segment is 39 bp
This sequence is a complementary reverse primer
Expression and Purification of His6-Tagged CPBP. A PCR-amplified product (one cycle of 5 min at 94°C followed by 33 cycles of 30 sec at 94°C, 60 sec at 50°C, and 60 sec at 72°C) containing the CPBP coding region with engineered SphI and PstI sites was ligated into a pQE-30 vector with an N-terminal His tag by using T4 DNA ligase (Roche Molecular Biochemicals). Competent cells (MAX Efficiency DH5α′IQ, GIBCO/BRL) were transformed with the ligation mixture and grown at 37°C in LB Agar Amp IPTG/X-Gal (Fermentas, Hanover, MD). CPBP-positive clones were isolated and grown in 1 liter of LB/Amp Medium (Fermentas) until they reached 0.5 OD units (OD600). Protein expression was induced with 1 μM isopropyl 1-thio-β-d-galactopyranoside (final concentration). The cells were disrupted in a French press, and His6-tagged CPBP was purified with Ni-NTA agarose affinity resin (Qiagen, Valencia, CA). Polyclonal antibodies directed against His6-tagged CPBP were raised in rabbits.
Overexpression of MsrA in TEF3 Mutant (TKY 661), TEF4 Mutant (TKY 662), and Their Parental Strain (TKY 660) and Immunoblotting. The vector YEp351 alone (control for the overexpression strain) and YEp351 encompassing the coding region of msrA (American Type Culture Collection no. 37672) and upstream region (217 bp; ref. 12) were used in the transformation of TKY 660, TKY 661, and TKY 662 yeast strains with the Alkali-Cation Yeast Transformation Kit (Qbiogene). Colonies from each agar plate were grown in ampicillin SD medium (Qbiogene) with or without l-leucine as required until the stationary phase. The cells were washed with 1 M sorbitol, resuspended in 50 mM Tris·HCl buffer, pH 7.4, and disrupted by glass beads. Proteins (30 μg per lane) were analyzed by Western blotting (Invitrogen). Immunoblotting was carried out with rabbit anti-MsrA polyclonal antibodies and goat anti-rabbit IgG (H and L chains)-horseradish peroxidase conjugate (Bio-Rad). The bands were detected with SuperSignal West Pico Chemiluminescent substrate (Pierce).
Determination of MsrA Activity. The ability of MsrA to reduce protein-bound methionine sulfoxide was assayed by using 4-dimethylaminoazobenzene-4′-sulfonyl (dabsyl)-methionine sulfoxide as previously described by Moskovitz et al. (11)
RNA Isolation and Northern Blot Analysis. Total RNA was extracted with the RNAwiz kit (Ambion) according to the manufacturer's instructions. The total RNA content was determined spectrophotometrically. The A260 to A280 ratios ranged between 1.907 and 2.065. Gel electrophoresis (20 μg of total RNA per lane), capillary transfer to nylon membranes (BrightStar-Plus, Ambion), and probing and washing of the blots were carried out with the glyoxal-based system for Northern blots (Ambion) according to the manufacturer's instructions. Membranes were prehybridized for 1 h at 42°C and hybridized at 42°C overnight. The radiolabeled msrA coding region was used as probe. The membranes were analyzed by phosphor imaging (Storm 860, PhosphorImager).
Results
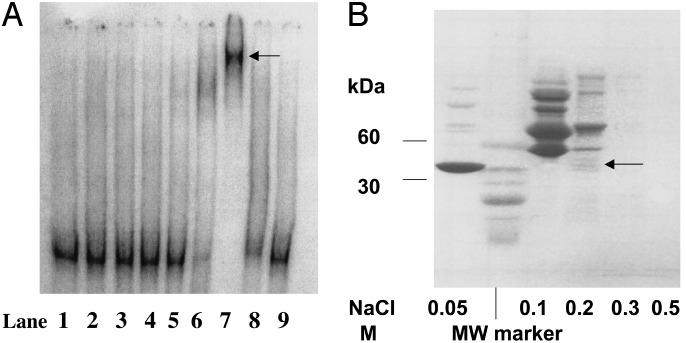
Isolation and Identification of Yeast Nuclear Protein msrA Promoter DNA-Binding Complex. In the studies aimed to isolate a DNA-binding protein from nuclear protein extracts of msrA-null mutant yeast strain grown until stationary phase, we consistently obtained an abundant binding complex with the 32P-labeled msrA promoter DNA (data not shown). The 60% saturated ammonium sulfate fraction of nuclear extract contained protein(s) that formed a binding complex with the 32P-labeled msrA promoter (data not shown). In a consecutive chromatography step with a HiTrap heparin-affinity column, the DNA-binding activity was eluted with 0.3 M KCl (data not shown). The concentration of KCl (0.3 M) was kept in the range suggested for transcription factor elution (13). This fraction was chromatographed on a CM Sepharose Fast Flow column by stepwise elution with buffers containing NaCl. Abundant DNA-binding activity was detected in the 0.2 M NaCl fraction (Fig. 1 A, lane 7, arrow). The different fractions eluted from the CM Sepharose column were resolved on an SDS/Tris/glycine/4–20% polyacrylamide gel (Fig. 1B). Various protein bands in the 0.2 M NaCl fraction were cut out and processed for MS. Liquid chromatography MS/MS analysis of the tryptic digests identified, besides CPBP, three other proteins (data with sequence of peptides obtained by MS/MS not shown). We have chosen to study CPBP for two reasons: first, its band intensity (Fig. 1B, arrow) correlated well with the observed promoter-binding activity; second, CPBP originally was cloned as a suppressor of the cold-sensitive drs2 yeast mutant (14) and thus might be capable of regulating msrA gene expression. In addition, CPBP was shown to have a 64.5% homology to yeast elongation factor (EF) 1γ (14). During the sequencing of tryptic digests of different protein bands (0.2 M NaCl fraction), EF-1γ was not detected by liquid chromatography-MS/MS (E.S.B. and I.H., unpublished data).
Fig. 1.
(A) Representative EMSA of DNA-binding activity-containing fractions eluted from CM Sepharose. DNA-binding activity-containing fraction eluted from a HiTrap heparin column (0.3 M KCl) was loaded on CM Sepharose (1 mg of protein per ml) and stepwise eluted with buffer A containing NaCl. Lane 1, free 32P-labeled DNA probe; lane 2, effluent of CM Sepharose column; lanes 3 and 4, washes with buffer A; lane 5, 0.05 M NaCl; lane 6, 0.1 M NaCl; lane 7, 0.2 M NaCl; lane 8, 0.3 M NaCl; lane 9, 0.5 M NaCl. (B) Representative Tris/Glycine/SDS/4–20% PAGE of the CM Sepharose fractions that were dialyzed against 10 mM Tris·HCl buffer, pH 6.6 (10 μg of protein per well). The gel was stained with Coomassie blue. The protein bands in the lane designated as 0.2 M NaCl were cut out, and tryptic digests were prepared as described in Materials and Methods.
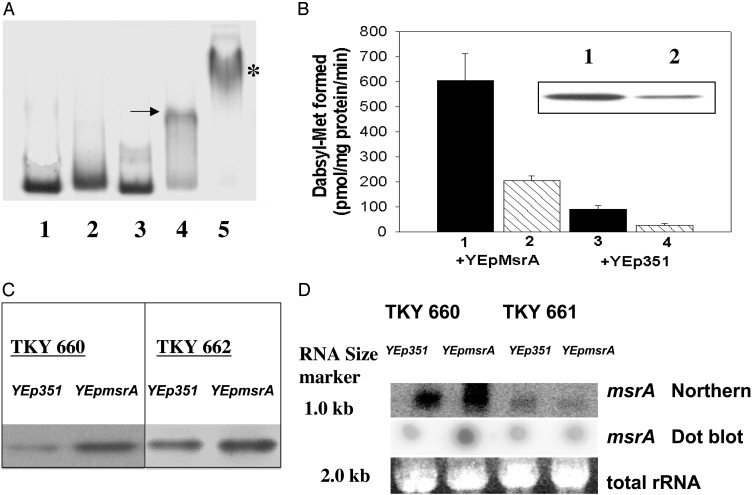
CPBP Interacts by Cooperative Binding with msrA Promoter DNA and Enhances MsrA Activity, msrA mRNA Levels, and MsrA Protein Expression. To determine whether CPBP was a functional component of the observed DNA-binding complex, we studied whether the formation of the DNA-binding complex may be enhanced by cooperative interaction of CPBP with other, yet unknown nuclear proteins. His6-tagged CPBP by itself formed a complex with the msrA promoter when high concentrations were used (8 μg of protein per well; data not shown). The representative EMSA shown in Fig. 2 A indicates that subthreshold protein concentrations of yeast nuclear extract (60% ammonium sulfate fraction; 10 μg of protein per well; lane 2) or His6-tagged CPBP (1.0 μg of protein per well; lane 3) failed to produce detectable DNA-binding complexes. However, mixing both protein solutions (same subthreshold concentrations) produced a prominent DNA–protein binding complex (lane 4, arrow). This complex was supershifted by postincubation with rabbit polyclonal anti-His6-CPBP antibodies (lane 5, asterisk). A supershifted band also was obtained with mouse His6-monoclonal antibody (data not shown), suggesting that CPBP is a component of the DNA-binding complex.
Fig. 2.
(A) Representative EMSA for the cooperative binding interaction among 32P-labeled msrA promoter DNA, His6-tagged CPBP, and other, not-yet-defined nuclear protein(s). Lane 1, free 32P-labeled msrA promoter DNA probe; lane 2, nuclear protein (60% ammonium sulfate fraction; 10 μg of protein per well); lane 3, His6-tagged CPBP (1.0 μg of protein per well); lane 4, mixture of both proteins (the arrow indicates cooperative binding complex); lane 5, mixture of both proteins reacted with rabbit polyclonal anti-His6-CPBP antibodies. The asterisk indicates supershift of binding complex. (B) MsrA catalytic activities in TKY 660 and TKY 661 yeast strains transformed with either YEp351 or YEp351msrA plasmid. Filled bars represent TKY 660 and hatched bars represent TKY 661. The results are expressed in pmol of dabsylmethionine per mg of protein formed per min as mean + SD (n = 3). (Inset) Western blot showing the protein bands that cross-reacted with rabbit polyclonal anti-MsrA antibodies. Lane 1, TKY 660 yeast strain overexpressing YEp351msrA; lane 2, TKY 661 yeast strain overexpressing YEp351msrA;20 μg of protein per well. (C) Western blot showing the protein bands (40 μg of protein per well) that cross-reacted with rabbit polyclonal anti-MsrA antibodies. TKY 660 and TKY 662 yeast strains were transformed with either YEp351 or YEp351msrA. (D) Total RNA was extracted, blotted, and hybridized with a 32P-labeled random prime msrA probe as described in Materials and Methods. For Northern blots, 30 μg of total RNA per well was loaded, and bands were stained with ethidium bromide and further processed as described in Materials and Methods. In dot blots, 30 μg of total RNA per well was pipetted on nylon filters and hybridized with 32P-labeled random prime msrA. Total RNA shows ethidium bromide-stained bands of indicated molecular size for comparing equal loading.
To determine whether CPBP may regulate MsrA activity, the conversion of dabsylmethionine sulfoxide to dabsylmethionine was compared in wild-type and TEF3-null mutant yeast strains containing either YEp351 or YEp351msrA (plasmid harboring the upstream activating sequence and msrA coding region). The formation of dabsylmethionine in the MsrA-overexpressing wild-type yeast strain (TKY 660; Fig. 2B, column 1) was 6-fold higher than in the strain harboring the plasmid alone (Fig. 2B, column 3). Interestingly, the TEF3-null mutant yeast strain (TKY 661; Fig. 2B, column 4) had only a third of the enzymatic activity of the TKY 660 wild-type strain (Fig. 2B, column 3). In comparison, MsrA overexpression in the TEF3-null mutant (Fig. 2B, column 2) increased the formation of dabsylmethionine by 2-fold over the wild-type yeast strain (Fig. 2B, column 3). These results were consistent with changes in MsrA protein content. The immunoblots depicted in the Fig. 2B Inset show that the MsrA protein level in wild-type yeast strain (TKY 660) harboring YEp351msrA (lane 1) was markedly higher than in the TEF3 mutant strain (TKY 661) harboring the YEp351msrA (lane 2). Because CPBP and EF-1γ share a 64.5% identity (14) we also studied whether EF-1γ might play a role in the regulation of MsrA expression. Fig. 2C shows that deletion of the TEF4 gene (TKY 662) failed to impair MsrA overexpression and was comparable with that in the wild-type yeast strain (TKY 660) transformed with YEp351msrA.
In addition, as shown by the representative Northern and dot blots (Fig. 2D), the msrA mRNA level in the YEp351msrA-transformed wild-type strain (TKY 660) was increased over that in the wild type transformed with YEp351 alone. In contrast, disruption of the TEF3 gene impaired the increase of the msrA mRNA level in the YEp351msrA-transformed mutant yeast strain (TKY 661). The absence of an msrA mRNA increase in the TEF3-null mutant suggests that CPBP may either enhance msrA gene expression or stabilize msrA mRNA.
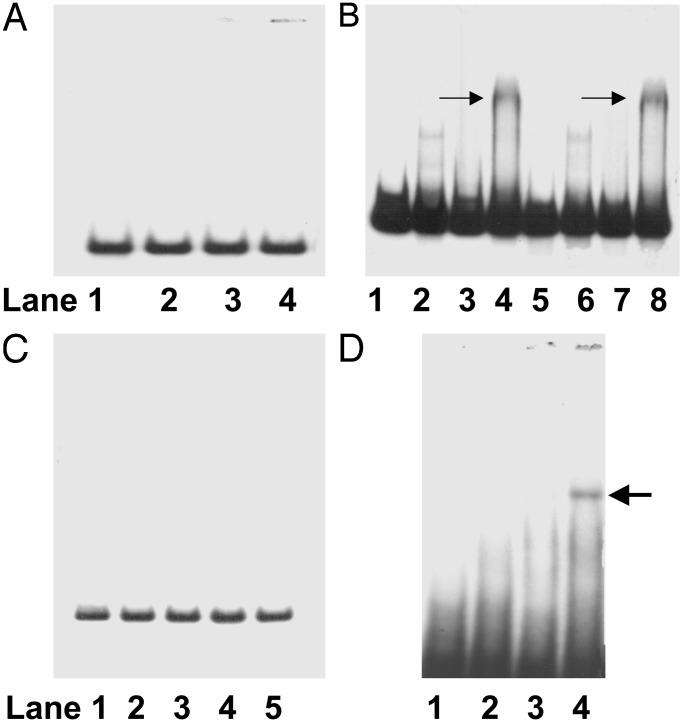
Mapping of the CPBP Binding Domain on the msrA Promoter. To characterize the CPBP binding domain on the msrA promoter, EMSA was performed with various DNA fragments (Table 1) by using the cooperative binding model described in Fig. 2 A. As shown in Fig. 3A, no DNA-binding protein complex was obtained with the 5′ end DNA segment. In contrast, the DNA segment with a point mutation in the first TATA sequence and the corresponding nonmodified DNA segment formed cooperative DNA-binding complexes of similar intensity (Fig. 3B, lanes 4 and 8, respectively, arrow). However, a DNA fragment (spanning from 17074 to 17148) that contained the other two TATA sequences but excluded the 3′ end DNA segment of the promoter resulted in a total loss of DNA-binding activity (Fig. 3C). Therefore, we tested the 3′ end DNA segment (region upstream from the start codon; see Table 1) and observed a cooperative binding interaction with His6-tagged CPBP and yeast nuclear extract. As shown in Fig. 3D, the mixture of His6-tagged CPBP (1.0 μg of protein per well) and yeast nuclear protein (60% ammonium sulfate fraction, 10 μg of protein per well) enhanced the formation of a cooperative DNA-binding complex (lane 4, arrow).
Fig. 3.
Representative EMSA for cooperative binding interaction of His6-tagged CPBP (1 μg of protein per well) and nuclear protein (60% ammonium sulfate fraction, 10 μg of protein per well). (A) 5′ end DNA segment (lane 1, free 32P-labeled probe; lane 2, 60% ammonium sulfate fraction; lane 3, His6-tagged CPBP; lane 4, mixture of lanes 2 and 3). (B) DNA segment with the first TATA sequence (lane 1, free 32P-labeled probe; lane 2, 60% ammonium sulfate fraction; lane 3, His6-tagged CPBP; lane 4, mixture of lanes 2 and 3) and corresponding DNA segment with point mutation in the first TATA sequence (lane 5, free 32P-labeled probe; lane 6, 60% ammonium sulfate fraction; lane 7, His6-tagged CPBP; lane 8, mixture of lanes 2 and 3). (C) DNA with the second and third TATA sequences (lane 1, free 32P-labeled probe; lane 2, 60% ammonium sulfate fraction; lane 3, His6-tagged CPBP; lane 4, mixture of lanes 2 and 3; lane 5, mixture of lanes 2 and 3 and postincubation with rabbit polyclonal anti-His6-CPBP antibody). (D) 3′ end DNA segment (lane 1, free 32P-labeled probe; lane 2, 60% ammonium sulfate fraction, 10 μg of protein per well; lane 3, His6-tagged CPBP, 1 μg of protein per well; lane 4, mixture of lanes 2 and 3). Arrows indicate DNA-binding complex.
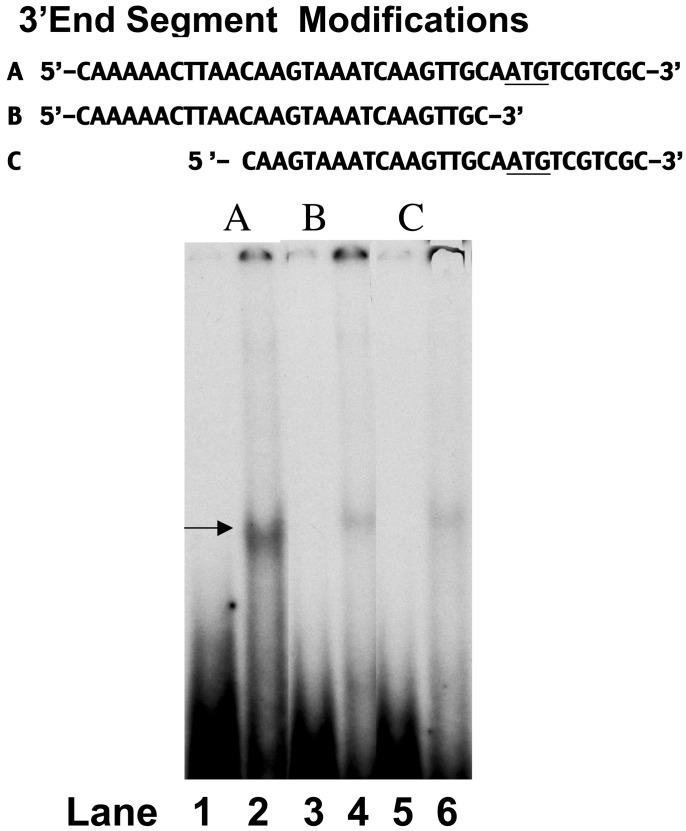
In addition, we examined whether the whole 39-bp DNA segment was essential for the cooperative interaction with His6-tagged CPBP and nuclear protein extract. A representative EMSA, depicted in Fig. 4, indicated that yeast nuclear protein (60% ammonium sulfate fraction; 20 μg of protein per well) formed a DNA-binding complex with the 39-bp DNA segment (lane 2, arrow). However, omission of 10 bases at either the downstream or upstream end of the 3′ end DNA segment (Fig. 4 Upper) greatly diminished the formation of a DNA protein-binding complex (Fig. 4, lanes 4 and 6, respectively).
Fig. 4.
Modifications of the 3′ end DNA segment (shown in Upper; the start codon of msrA gene is underlined). Lane 1, free 32P-labeled DNA (A); lane 2, binding complex of A and nuclear protein (60% ammonium sulfate fraction, 20 μg of protein per well); lane 3, free 32P-labeled DNA segment (B); lane 4, binding complex of B with nuclear protein (60% ammonium sulfate fraction, 20 μg of protein per well); lane 5, free 32P-labeled DNA segment (C); lane 6, binding complex of C with nuclear protein (60% ammonium sulfate fraction, 20 μg of protein per well). Arrow indicates DNA-binding complex.
Discussion
In this study we detected a DNA-binding protein that is functionally involved in the regulation of yeast msrA gene. MS analysis of the tryptic digests of protein bands isolated through screening of msrA promoter-binding activity resulted in the identification of CPBP (molecular mass of 47 kDa). In S. cerevisiae, CPBP is encoded by the TEF3 gene and has a 64% sequence homology with EF-1γ, which is encoded by the TEF4 gene (14). We failed to detect EF-1γ during CPBP purification, which is in line with a previous report (15). Although the EF-1 complex plays an important role in protein translation (16), disruption of the TEF3 and TEF4 genes failed to be lethal (14), leading to the conclusion that CPBP and its homologue, EF-1γ, may be redundant in translational regulation but may have other, not-yet-described functions (14).
By using a yeast strain with a disrupted TEF3 gene, we provided evidence that in the absence of CPBP the expression of msrA mRNA and MsrA protein levels were decreased and the ability of MsrA to reduce methionine sulfoxide to methionine was impaired greatly. In contrast, disruption of the TEF4 gene failed to prevent MsrA overexpression, indicating that EF-1γ may not play a role in msrA gene regulation (Fig. 2C). Another difference between the biological functions of EF-1γ and CPBP emerged from previous studies of the cold-sensitive drs2 mutant that has a defect in the 40S ribosomal protein assembly (17). CPBP suppressed the drs2 mutant-specific cold sensitivity, whereas EF-1γ was devoid of such an effect (17). Taken together, these two independent results indicate that, in yeast, CPBP may act as a response regulator during environmental stress conditions. Comparison of the expression stimulation during H2O2 exposure or growth to stationary phase (Saccharomyces Genome Database, www.yeastgenome.org) revealed a similarity in the stress response between the msrA and TEF3 genes, although they differ in their stimulation efficiency (2-fold higher for the msrA gene). The nature or relevance of this difference presently is not understood. A recent review on yeast transcription factors including Msn2p/Msn4p, Skn7p, and Yap1p pointed out that a disruption of their encoding genes was not lethal but reduced cell viability in oxidative or environmental stress conditions (18). It seems that CPBP, by regulating MsrA expression, may play a similar role targeted at correcting environmental stress-induced cell damage.
Studies on the cooperative binding interaction of CPBP on the msrA promoter yielded additional support for a regulatory function of CPBP. CPBP binding was mapped to the 3′ end of the untranslated DNA sequence upstream from the start codon of the msrA gene. By clipping off 10 bases on either end of this 39-bp DNA segment, we observed that the whole segment might be required for binding efficiency. This finding suggests that the binding complex may not require a consensus sequence but rather may be an unstructured intervention with the 3′ end of the untranslated activating sequence. Such a mode of DNA interaction tentatively suggests a supportive role in the positioning of RNA polymerases on the promoter and/or a role in transcript elongation.
The finding that CPBP may operate as a regulator of yeast msrA gene expression may provide a model to study mammalian msrA. Several reports on postmortem brain tissues of Alzheimer's disease patients revealed an increased protein oxidation (19) together with a decrease of MsrA protein expression and MsrA enzymatic activity (20), supporting the working hypothesis that defects at the transcriptional, translational, and/or post-translational levels may have flawed the antioxidant function of MsrA.
Acknowledgments
We thank Dr. T. G. Kinzy (Robert Wood Johnson Medical School, Piscataway, NJ) for providing the TKY 660 wild-type and TKY 661 TEF3 mutant yeast strains and Drs. E. Stadtman, T. Stadtman, and H. Fales for useful critiques of this manuscript.
Abbreviations: Msr, methionine sulfoxide reductase; EF, elongation factor; TEF, translation EF; EMSA, electrophoretic mobility-shift assay; MS/MS, tandem MS; CPBP, calcium phospholipid-binding protein; dabsyl, 4-dimethylaminoazobenzene-4′-sulfonyl.
References
- 1.Stadtman, E. R. (2002) Free Radical Biol. Med. 33 597-604. [DOI] [PubMed] [Google Scholar]
- 2.Orr, W. C. & Sohal, R. S. (1994) Science 263 1128-1130. [DOI] [PubMed] [Google Scholar]
- 3.Larsen, P. L. (1993) Proc. Natl. Acad. Sci. USA 90 8905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mockett, R. J., Orr, W. C., Rahmandar, J. J., Sohal, B. H. & Sohal, R. S. (2001) Exp. Gerontol. 36 441-463. [DOI] [PubMed] [Google Scholar]
- 5.Levine, R. L., Berlett, B. S., Moskovitz, J., Mosoni, L. & Stadtman, E. R. (1999) Mech. Aging Dev. 107 323-332. [DOI] [PubMed] [Google Scholar]
- 6.Moskovitz, J., Poston, J. M., Berlett, B. S., Nosworthy, N. J., Szczepanowski, R. & Stadtman, E. R. (2000) J. Biol. Chem. 275 14167-14172. [DOI] [PubMed] [Google Scholar]
- 7.Moskovitz, J., Singh, V. K., Requena, J., Wilkinson, B. J., Jayaswal, R. K. & Stadtman, E. R. (2002) Biochem. Biophys. Res. Commun. 290 62-65. [DOI] [PubMed] [Google Scholar]
- 8.Moskovitz, J., Rahman, M. A., Strassman, J., Yancey, S. O., Kushner, S. R., Brot, N. & Weissbach, H. (1995) J. Bacteriol. 177 502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moskovitz, J., Bar-Noy, S., Williams, W. M., Requena, J., Berlett, B. S. & Stadtman, E. R. (2001) Proc. Natl. Acad. Sci. USA 98 12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskovitz, J., Berlett, S. B., Poston, M. & Stadtman, E. R. (1997) Proc. Natl. Acad. Sci. USA 94 9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moskovitz, J., Flescher, E., Berlett, S. B., Azare, J. A., Poston, M. & Stadtman, E. R. (1998) Proc. Natl. Acad. Sci. USA 95 14071-14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan, B. A., Banks, G. R., Toone, W. M., Raitt, D., Kuge, S. & Johnson, L. L. (1997) EMBO J. 16 1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey, M. & Smale, S. T. (2000) Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 493-504.
- 14.Kinzy, T. G., Ripmaster, T. L. & Woolford, J. L., Jr. (1994) Nucleic Acids Res. 22 2703-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kambouris, N. G., Burke, D. J. & Creutz, C. E. (1993) Yeast 9 151-163. [DOI] [PubMed] [Google Scholar]
- 16.Riis, B., Rattan, S. I. S., Clark, B. F. C. & Merrick, W. C. (1990) Trends Biochem. Sci. 15 420-424. [DOI] [PubMed] [Google Scholar]
- 17.Ripmaster, T. L., Vaughn, G. P. & Woolford, J. L., Jr. (1993) Mol. Cell. Biol. 13 7901-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moye-Rowley, W. S. (2002) Antioxid. Redox. Signal. 4 123-140. [DOI] [PubMed] [Google Scholar]
- 19.Smith, M. A., Perry, G., Richey, P. L., Sayre, L. M., Andersen, V. E., Beal, M. F. & Kowan, N. (1996) Nature 382 120-121. [DOI] [PubMed] [Google Scholar]
- 20.Gabbita, S. P., Aksenov, M. Y., Lovell, M. A. & Markesbery, W. R. (1999) J. Neurochem. 73 1660-1666. [DOI] [PubMed] [Google Scholar]