Abstract
Polypeptide protease inhibitors are often found to inhibit targets with which they did not coevolve, as in the case of high-affinity inhibition of bacterial subtilisin by the leech inhibitor eglin c. Two kinds of contacts exist in such complexes: (i) reactive site loop-active site contacts and (ii) interactions outside of these that form the broader enzyme-inhibitor interface. We hypothesized that the second class of “adventitious” contacts could be optimized to generate significant increases in affinity for a target enzyme or discrimination of an inhibitor for closely related target proteases. We began with a modified eglin c, Arg-42–Arg-45–eglin, in which the reactive site loop had been optimized for subtilisin-related processing proteases of the Kex2/furin family. We randomized 10 potential adventitious contact residues and screened for inhibition of soluble human furin. Substitutions at one of these sites, Y49, were also screened against yeast Kex2 and human PC7. These screens identified not only variants that exhibited increased affinity (up to 20-fold), but also species that exhibited enhanced selectivity, that is, increased discrimination between the target enzymes (up to 41-fold for furin versus PC7 and 20-fold for PC7 versus furin). One variant, Asp-49–Arg-42–Arg-45–eglin, exhibited a Ki of 310 pM for furin and blocked furin-dependent processing of von Willebrand factor in COS-1 cells when added to the culture medium of the cells. The exploitation of adventitious contact sites may provide a versatile technique for developing potent, selective inhibitors for newly discovered proteases and could in principle be applied to optimize numerous protein–protein interactions.
Located in the trans-Golgi network and other late compartments of the eukaryotic secretory pathway, processing proteases of the Kex2/furin family convert proprotein substrates to their mature, active forms by cleaving after dibasic or multibasic sequences (1, 2). Each of these seven members (PC1/3, PC2, furin, Pace 4, PC4, PC5/6, and PC7/LPC) exhibits a unique pattern of expression and subcellular localization. In yeast, Kex2 cleaves a variety of secreted proteins including the precursors of the α-mating pheromone and killer toxin (2). Among the mammalian enzymes, furin stands out both because of the wide array of important endogenous substrates that it is thought to process and its likely involvement in the cleavage of key molecules essential for bacterial and viral pathogenesis. Such molecules include anthrax protective antigen, Pseudomonas exotoxin A, and the envelope glycoproteins of respiratory syncitial virus, avian influenza virus, measles virus, and Ebola virus, among numerous others (3–5). Although disruption of the gene encoding furin in mouse results in embryonic lethality (6), furindeficient cell lines are viable (7, 8). There is reason to believe, therefore, that short-term inhibition of furin might provide a mechanism to counter a wide spectrum of acute bacterial and viral pathogens (5). Indeed, a recent study demonstrates that a simple furin inhibitor, hexa-D-Arg, protects the mouse against Pseudomonas exotoxin A when introduced by i.p. injection (9).
As an approach to developing high-affinity, selective furin inhibitors, we chose the elastase inhibitor, eglin c, of the medicinal leech Hirudo medicinalis, as a starting point because it was known to inhibit subtilisin, the founding member of the superfamily of serine proteases to which Kex2/furin enzymes belong (10, 11). Analyzing the effects of substitutions at the P1, P2, P4, and P6 positions¶ of the substrate-like reactive site loop of eglin c, we found that introduction of Arg at P1 and P4 resulted in a form of eglin c, R1R4-eglin, that inhibited furin with a Ki of 2.5 nM and Kex2 with a Ki of 0.91 nM (11). These altered forms of eglin c exhibited slow, tight binding inhibition, formed stable complexes with both Kex2 and furin, and were not cleaved by either enzyme (11). The affinities of furin and Kex2 for R4R1-eglin were not improved on by substitutions at P2 or P6. Indeed, either Lys at P2 or Arg at P6 resulted in cleavage of the inhibitor by furin (11). Moreover, substitutions within the reactive site loop alone are not likely to provide highly selective inhibition of individual processing enzymes. Like furin, the other mammalian proprotein processing proteases also recognize basic residues at P1, P2, and P4, with a marked preference, in most cases, for Arg at P1 (10). Residues predicted to lie in the complementary S1,S2, and S4 subsites of these enzymes are, likewise, conserved (13).
Having thus exhausted the possibilities for enhanced affinity and selectivity offered by mutagenesis of the reactive site loop, we reasoned that contacts between enzyme and inhibitor beyond the enzyme/substrate-like interactions between the reactive site loop and enzyme might be exploited. Contacts between polypeptide-protease inhibitors and target proteases often involve an extensive interface that extends beyond the reactive site loop (14). In the case of a noncognate enzyme-inhibitor pair (i.e., a nonphysiological pairing not governed by coevolutionary constraints), adventitious contacts in this interface are unlikely to be optimal (Fig. 1A). Optimization of such contacts could provide both enhanced affinity for a noncognate enzyme (Fig. 1B) and discrimination between related noncognate enzymes (Fig. 1C). Based on this reasoning, we developed an approach to optimize the interaction between R4R1-eglin and Kex2/furin family proteases. Examining crystal structures of complexes of eglin c with degradative subtilisins (15), we identified 10 eglin c residues flanking the reactive site loop that were positioned within 7 Å of subtilisin surface residues (Fig. 2, Table 1). Although structural data for furin and Kex2 are not available, it is clear from sequence alignments that surface residues of Kex2/furin enzymes and subtilisin differ markedly in these regions (13). Codons for these 10 eglin c residues were randomized, creating a set of small libraries, which were screened, after expression and purification in a 96-well format, for inhibition of human furin and, in some cases, human PC7 and yeast Kex2. The best furin inhibitor obtained in this study, Asp-49–R4R1-eglin, inhibited processing of von Willebrand factor (vWF) in COS-1 cells when added to culture medium of COS-1 cells coexpressing furin and vWF. This approach has the potential to be generalized to the engineering of any noncognate protein–protein interaction.
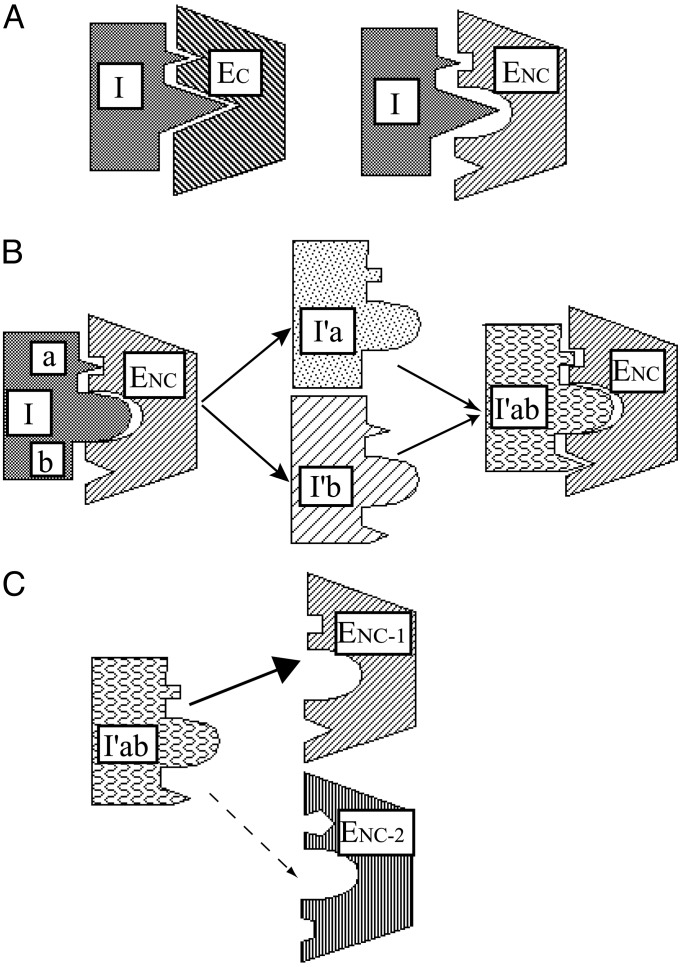
Fig. 1.
Optimization of adventitious inhibitor-enzyme contacts for affinity and selectivity. (A) In cognate inhibitor-enzyme pair (Left), both reactive-site loop contacts (larger triangle) and flanking, adventitious contacts are likely to be optimized through evolutionary selection. In noncognate inhibitor-enzyme pair (Right) neither type of interaction is optimal. (B) As a first step in optimizing interaction of inhibitor with noncognate enzyme, reactive site loop is first optimized (Left). In a second step (Center), randomization of inhibitor residues at multiple potential adventitious contacts leads to optimization of individual sites. These alterations may be combined (Right). (C) An inhibitor in which adventitious contacts have been optimized for one of two related enzymes that have similar substrate specificity can exhibit selective inhibition. I, inhibitor; EC, cognate enzyme; ENC, noncognate enzyme; I′, inhibitor with altered reactive site loop; a and b, flanking adventitious contact sites; I′a, I′b, and I′ab, I′ with substitutions at adventitious contact residues; ENC-1 and ENC-2, noncognate enzymes with distinct substrate specificities.
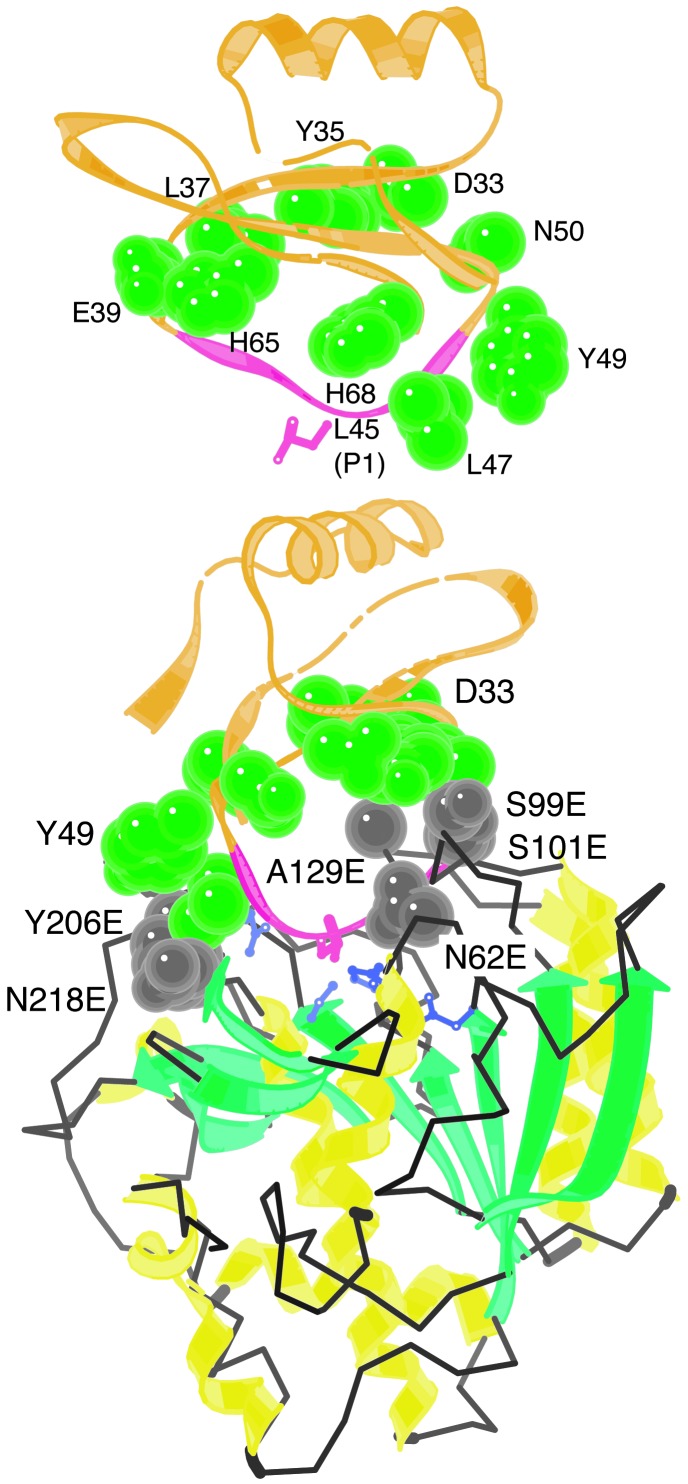
Fig. 2.
Eglin c residues with the potential to make adventitious contacts with Kex2/furin family enzymes. (Upper) Structure of eglin c alone, from crystal structure of a complex of eglin c with subtilisin Carlsberg. (Lower) Crystal structure of the complex (coordinates from 1cse.pdb, ref. 15). Structure was drawn with kinemage (28). Ten eglin c residues (exclusive of P4-P′1) whose side chains come within 7 Å of the subtilisin surface and that were mutagenized in this study are shown in green. Interacting enzyme residues are shown in gray and are indicated by residue numbers followed by E. P1 Leu-45 of eglin c is shown in pink and catalytic His (His-64) is shown in purple.
Table 1. Substitutions at potential adventitious contact sites that enhanced inhibition of furin, relative to R4R1-eglin or R4K1-eglin.
| Position of substitution* | Major effect† | Minor effect |
|---|---|---|
| R4R1 construct | ||
| Glu-39‡ | nd | Pro |
| Gly-40‡ | nd | Ala, Arg, Pro |
| Tyr-49 | Asp | Ala |
| R4K1 construct | ||
| Asp-33 | nd | Val |
nd, none detected
No substitutions were found at Tyr-35, Leu-47, His-65, or His-68 (R4R1 context) or at Leu-37 or Asn-30 (R4K1 context) that enhanced affinity
Substitutions that exhibited a major effect (>5-fold) or a minor effect (<5-fold) on furin affinity are shown. These results are based on precise inhibition assays using purified inhibitors as described in Materials and Methods
Glu-39 and Gly-40 substitutions generated temporary inhibitors for furin as described in the text
Materials and Methods
Expression and Purification of Processing Enzymes. Secreted, soluble Kex2, human furin, and human PC7 were purified and active site-titrated as described (11, 16–18). For cell culture assays, pSR-furin plasmid DNA (18), encoding full-length furin, was transfected into COS-1 cells (19).
Mutant Library Construction. Synthetic genes encoding either R4R1-eglin or R4K1-eglin cloned into Escherichia coli expression vector pET27b(+) were as described (11). Codons encoding eglin residues 33, 37, and 50 were mutagenized in the vector encoding R4K1-eglin; codons encoding eglin residues 35, 39, 40, 47, 49, 65, and 68 were mutagenized in the vector encoding R4R1-eglin. Eglin codons 35, 39, and 40 were randomized by oligonucleotide cassette mutagenesis using unique AatII and BglII sites (11). Codons encoding residues 33, 37, 47, 49, 50, 65, and 68 were mutagenized by using PCR-based Quick Change site-directed mutagenesis with Pfu Turbo (Stratagene). In the first experiments, all 10 codons, 33, 35, 37, 39, 40, 47, 49, 50, 65, and 68, were partially randomized: the first two nucleotides were fully randomized and the third nucleotide was restricted to C or T. This strategy eliminated the possibility of introducing stop codons and permitted each library to be screened in one 96-well plate. Although codons for Gln, Glu, Lys, Met, or Trp were excluded, substitution of similar residues (Asn, Asp, Arg, Ile, or Leu and Phe) would occur. In a second series of experiments, libraries were constructed in which all three nucleotides of the codons encoding residues 49, 65, and 68 were fully randomized. Mutagenized vectors were transformed into E. coli DH5α (20) to produce libraries of ≥500 independent transformants. For screening libraries and larger-scale expression of individual eglin species, purified library DNA was transformed into BL21-(DE3) (11).
Expression and Screening of Libraries. Individual BL21-(DE3) transformants were grown in 200 μl of LB containing kanamycin (30 μg/ml) by shaking at 37°C in 96-well, 0.45-μm GHP membrane filter plates (Gelman). When OD600 values reached 0.4, isopropyl β-D-thiogalactoside (IPTG) was added to a final concentration of 1 mM, and eglin variant expression was induced for 1 h in the filter plate [before IPTG addition, 60 μl of culture was transferred to a 96-well master plate (Falcon) in which wells were adjusted to 10% glycerol for storage at -80°C]. Cells were harvested in filter plates by centrifugation at 1,000 rpm, washed with 0.125 M NaCl, and stored overnight at -80°C. Eglin c variants were quantitatively extracted by osmotic shock (21) by resuspension in 50 mM Tris·HCl, pH 8.0, as described (11). Extracts were filtered from cells by centrifugation into a 96-well reservoir plate. Well-to-well variation of protein production was tested by expressing R4R1-eglin in 96-well format. Randomly selected wells were analyzed by SDS/PAGE. Fluctuation of protein yield was within ±10%.
Filtrate (5–10 μl) containing eglin variants was incubated with furin (≈5 nM) in furin assay buffer (20 mM NaMES, pH 7.0 containing 1 mM CaCl2, 0.1% Triton X-100) for 20 min at room temperature in an opaque 96-well plate (Costar). Boc-Arg-Val-Arg-Arg-MCA (2 μM) was added to a final volume of 100 μl, and residual furin activity was recorded for least 20 min by using an fmax fluorescence microtiter plate reader (Molecular Devices) with a 360-nm excitation filter and 480-nm emission filter. In a 96-well plate 88 variants were screened. Two wells were used for monitoring cell growth and six wells for expression of controls (R4R1-eglin or R4K1-eglin). R4K1-eglin, having a lower affinity (38 nM, ref. 11) for furin than R4R1-eglin, was used to monitor subtle improvements in affinity.
Characterization of Candidate Variants. Vectors encoding eglin variants that exhibited inhibition greater than the control were recovered from master plates and sequenced. Promising variants were expressed on a 50-ml scale and purified to homogeneity, and their identities were verified by matrix-assisted laser desorption ionization MS, as described (11). Inhibition of Kex2 and furin by purified eglin variants was assayed, and Ki (1/Ka) values were determined as described (11). PC7 inhibition was assayed in 20 mM BisTris·HCl (pH 7.0), 1 mM CaCl2 using 10 μM pyroGlu-Arg-Thr-Lys-Arg-MCA (Bachem) as substrate (17), and Ki (1/Ka) values were determined as described (11).
Furin-Dependent Processing of vWF in COS-1 Cells. Simian virus 40-derived vectors, pMT2-vWF (19) and pSR-furin (18), were cotransfected into COS-1 cells as described (22). To monitor the effect of Asp-49–R4R1-eglin on furin-dependent pro-vWF processing, Asp-49–R4R1-eglin or WT (Leu-45) eglin (5 μM) was added to the medium of 10-cm plates containing COS-1 cells (≈5 × 106) 4 h before a pulse-labeling of cells by addition of [35S]Met/Cys mixtures (200 μCi) for 30 min. After pulse labeling, a chase with unlabeled Met and Cys (200 mM) was performed for 6 h. Cell lysates were prepared by using Nonidet P-40 lysis buffer (22) and precleared by incubating with 25 μl of protein A-Sepharose (Amersham Pharmacia) for 2 h then centrifuging. Precleared extracts were incubated at 4°C with 1 μl of anti-vWF polyclonal antibody (Dako) overnight then with 50 μl of protein A-Sepharose for 2 h. Immunoprecipitates were collected by centrifugation and washed (22). Samples were analyzed by SDS/PAGE (6%), treated with ENHANCE (NEN), dried, and autoradiographed (22).
Results
Initial Screening of Eglin C Libraries. Ten eglin c codons were subjected to partial randomization (third position limited to T or C) as described in Materials and Methods, and the resulting libraries were screened for furin inhibition (Fig. 3). Variants that appeared to give enhanced inhibition relative to the control (R4R1-eglin or R4K1-eglin) in the initial rapid screen were sequenced at the DNA level and purified to homogeneity for more precise determinations of affinity. Only substitutions at 4 of the 10 positions showed reproducible enhancements of furin inhibition, substitutions for Asp-33, Glu-39, Gly-40, and Tyr-49 (Table 1). Purified inhibitors that yielded reproducible enhancements of affinity for furin were tested for their effects on Kex2 and PC7 also. In line with previous findings that Arg substitution at P6 (Gly-40) resulted in a temporary furin inhibitor (11), Ala, Arg, and Pro substitutions at position 40 that were obtained in the screen all were cleaved by furin (data not shown). Likewise, substitution of Pro for Glu-39 (P7) also resulted in a temporary inhibitor. Other than these P6 and P7 mutants, all others examined in this work formed 1:1 stable complexes (data not shown). Substitution of Val for Asp-33 resulted in 2-fold increased affinity for both furin and Kex2 relative to R4K1-eglin (see below). Substitution of Asp for Tyr-49 (P′4) exhibited a substantial enhancement in affinity for both Kex2 and furin.
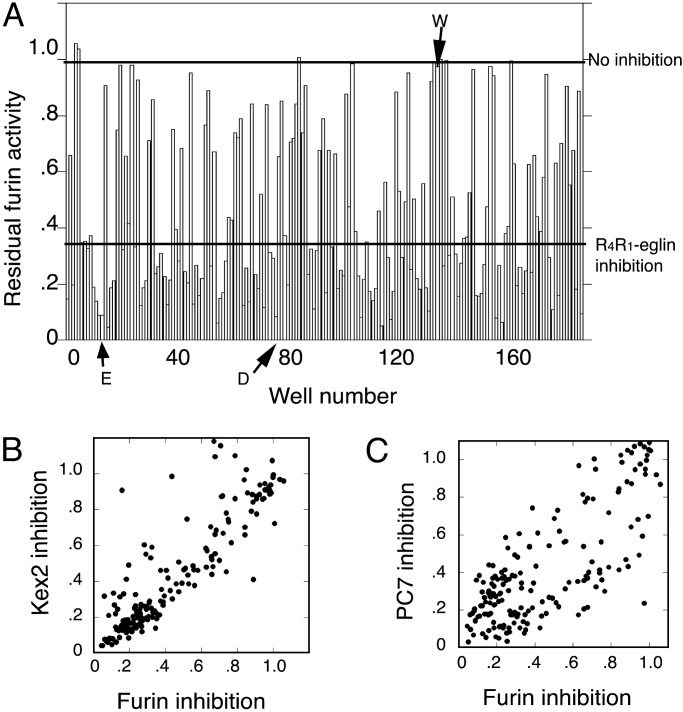
Fig. 3.
Rapid inhibition assay for novel eglin variants generated by randomization at position 49 in R4R1-eglin. (A) Furin inhibition by newly generated variants in two 96-well plates. The lower bar shows the level of inhibition by Tyr-49–R4R1-eglin. The upper bar indicates no inhibition. Glu (E), Asp (D), and Trp (W) substitutions are indicated by arrows. (B) Correlation plot of extent of inhibition of furin versus Kex2. (C) Correlation plot of extent of inhibition of furin versus PC7.
Full Randomization of Position 49 (P′4). After the discovery of the significant effect of the substitution at position 49, which corresponds to P′4, a library was constructed in which Tyr-49 codon was fully randomized. From this library, proteins were expressed in two 96-well plates and subjected to the rapid inhibition assay for Kex2, furin, and PC7. The inhibition pattern for furin is shown in Fig. 3A. Correlation plots of inhibition of Kex2 and PC7 versus inhibition of furin (Fig. 3 B and C) demonstrated that many variants inhibited all three enzymes with equal potency, but that others inhibited one or two enzymes selectively. Kex2 and furin exhibited similar inhibition patterns (Fig. 3B). The pattern of inhibition of PC7 was more distinctive (Fig. 3C). Table 2 shows substitutions for Tyr-49 that enhanced affinity for each enzyme. Kex2 and furin exhibited a preference for acidic residues (Asp and Glu), whereas these substitutions had little or no effect on PC7. Molecules with Cys at 49 purified as mixtures of monomeric and disulfide-linked dimeric forms (data not shown), and precise inhibition assays were not pursued.
Table 2. Substitutions for Tyr-49 that were found to inhibit Kex2, furin, and PC7.
| Enzyme | Substitution* |
|---|---|
| Kex2 | Val, Asp, Glu, Cys |
| Furin | Gly, Ser, Asp, Glu, Cys, Cys+† |
| PC7 | Ala, Val, Met, Trp, Cys+ |
Within the context of R4R1-eglin
Cys+ contained, in addition to Cys substitution at 49, a spontaneous substitution at Leu for Pro at position 38
Substitutions for Tyr-49 Have Dramatic Effects on Affinity and Selectivity for Kex2, Furin, and PC7. To obtain accurate Ki values for Tyr-49 substitutions, selected variants were purified to homogeneity and precise inhibition assays were performed. Asp-49–R4R1-eglin and Glu-49–R4R1-eglin exhibited substantially higher affinity for furin and Kex2 than R4R1-eglin did (Table 3). Both gave ≈6-fold higher selectivity for furin than for PC7. In contrast, substitution of Trp at position 49 increased affinity for PC7 but had no effect on Kex2 and decreased affinity for furin, resulting in substantially increased selectivity for PC7 versus furin (20-fold). Of the inhibitors with substitutions at position 49, Asp-49–R4R1-eglin exhibited the highest affinity for furin (Ki = 330 pM), Glu-49–R4R1-eglin the highest for Kex2 (Ki = 36 pM), and Trp-49–R4R1-eglin the highest for PC7 (KI = 470 pM).
Table 3. Ki values (M) for R4R1-eglin containing the indicated substitution at position 49.
| Residue at position 49* | Kex2 | Furin | PC7 |
|---|---|---|---|
| Tyr | 9.1 × 10-10† | 2.5 × 10-9† | 1.3 × 10-9 |
| Ala | 1.0 × 10-9 | 1.8 × 10-9 | 2.9 × 10-9 |
| Asp | 5.6 × 10-11 | 3.3 × 10-10 | 1.8 × 10-9 |
| Glu | 3.6 × 10-11 | 5.3 × 10-10 | 2.9 × 10-9 |
| Ser | 2.1 × 10-10 | 4.8 × 10-9 | 2.5 × 10-9 |
| Trp | 1.2 × 10-9 | 1.0 × 10-8 | 4.7 × 10-10 |
| Met | 1.0 × 10-9 | 1.6 × 10-9 | 1.4 × 10-9 |
All values were determined by using purified inhibitors as described in Materials and Methods. Experimental errors were ±20%
In the context of R4R1-eglin
Ki values for Tyr-49-R4R1-eglin with Kex2 and furin are from ref. 11
Inhibition of Kex2, Furin, and PC7 by Val-33–Asp-49–R4R1-Eglin. The Asp-33–Val and Tyr-49–Asp substitutions were combined to generate Val-33–Asp-49–R4R1-eglin. Val-33–Asp-49–R4R1-eglin inhibited furin with a Ki of 270 pM and Kex2 with a Ki of 42 pM (Table 4), indicating, somewhat disappointingly, that the combination of the two mutations did not have an additive effect on affinity for these enzymes. In the case of PC7, however, this combination of mutations had a negative, synergistic effect. Whereas the single Asp-33–Val and Tyr-49–Asp substitutions had almost no effect on affinity for PC7, the combination resulted in a 6- to 8-fold decrease in affinity (Table 4). Thus, the synergistic, negative effect of this combination of substitutions yields an inhibitor that shows enhanced discrimination (41-fold) between furin and PC7.
Table 4. Effect of combining substitutions at positions 33 and 49 in R4R1-eglin.
| Substitutions* | Kex2 | Furin | PC7 |
|---|---|---|---|
| Asp-33-Tyr-49 | 9.1 × 10-10† | 2.5 × 10-9 | 1.3 × 10-9 |
| Asp-33-Asp-49 | 5.6 × 10-11 | 3.3 × 10-10 | 1.8 × 10-9 |
| Val-33-Asp-49 | 4.2 × 10-11 | 2.7 × 10-10 | 1.1 × 10-8 |
All values were determined by using purified inhibitors as described in Materials and Methods. Experimental errors were ±20%
In the context of R4R1-eglin
Inhibition constants (Ki, M) determined by purified variants for Kex2, furin, and PC7
P6 Substitutions. As noted earlier, substitutions of Ala, Arg, and Pro for Gly-40 (P6) in R4R1-eglin were identified as residues that slightly enhanced affinity for furin but also resulted in temporary inhibition. However, P6 substitutions affected PC7 differently. In the context of Met at P4 and Arg at P1, the Gly-40–Arg-substituted form of eglin exhibited a 13-fold enhanced affinity for PC7 (Table 5). The Gly-40–Arg substitution in R4R1-eglin resulted in only a 2-fold increase in affinity for PC7, but unlike the case of furin and like the case of Kex2, inhibition was persistent rather than temporary, indicating that whereas furin cleaved Gly-40–Arg–R4R1-eglin, both Kex2 and PC7 formed stable 1:1 complexes with this inhibitor (Table 5).
Table 5. Stable, potent inhibition of PC7 by Arg-40-Arg-42-Arg-45-eglin.
| Kex2 | Furin | PC7 | |
|---|---|---|---|
| Met-42-Arg-45-eglin | 3.4 × 10-10* | 3.6 × 10-6* | 3.2 × 10-6 |
| Arg-42-Arg-45-eglin | 9.1 × 10-10* | 2.5 × 10-9* | 1.3 × 10-9 |
| Arg-40-Met-42-Arg-45-eglin | 5.3 × 10-10 | 2.7 × 10-6 | 2.4 × 10-7 |
| Arg-40-Arg-42-Arg-45-eglin | 2.9 × 10-11* | 2.8 × 10-9*† | 6.7 × 10-10 |
All values were determined by using purified inhibitors as described in Materials and Methods. Experimental errors were ±20%
Published values from ref. 11 are provided for comparison
Temporary inhibition
Inhibition of Cellular Processing of Pro-vWF by Asp-49–R4R1-Eglin. We tested the ability of Asp-49–R4R1-eglin to inhibit intracellular furin activity by examining the effect of adding inhibitor to culture medium on the furin-dependent processing of vWF in COS-1 cells (19). Furin and vWF were coexpressed in COS-1 cells and pulse-labeled for 30 min with [35S]Met/Cys and chased for 6 h with unlabeled Met and Cys as described in Materials and Methods, and vWF cross-reacting species were immunoprecipitated from cell lysates, separated by SDS/PAGE, and autoradiographed. Asp-49–R4R1-eglin or WT eglin (Leu-45) (5 μM) was added to the medium 4 h before pulse labeling. COS-cells were transfected with a vector encoding Δpro-vWF, which lacks the vWF pro-domain but retains the signal peptide, to provide a size control for mature vWF (Fig. 4B, lane 3). As previously observed (23), transformation of vWF alone resulted in appearance of only pro-vWF (Fig. 4B, lane 2). Cotransfection with furin (Fig. 4B, lane 6) resulted in appearance of mature vWF although persistence of pro-vWF species, presumably in early secretory compartments, was observed. Addition of WT eglin c had no effect on production of mature vWF (Fig. 4B, lane 5) but addition of Asp-49–R4R1-eglin completely blocked furindependent processing (Fig. 4B, lane 4). Analysis of species immunoprecipitated from conditioned media gave the same results (data not shown).
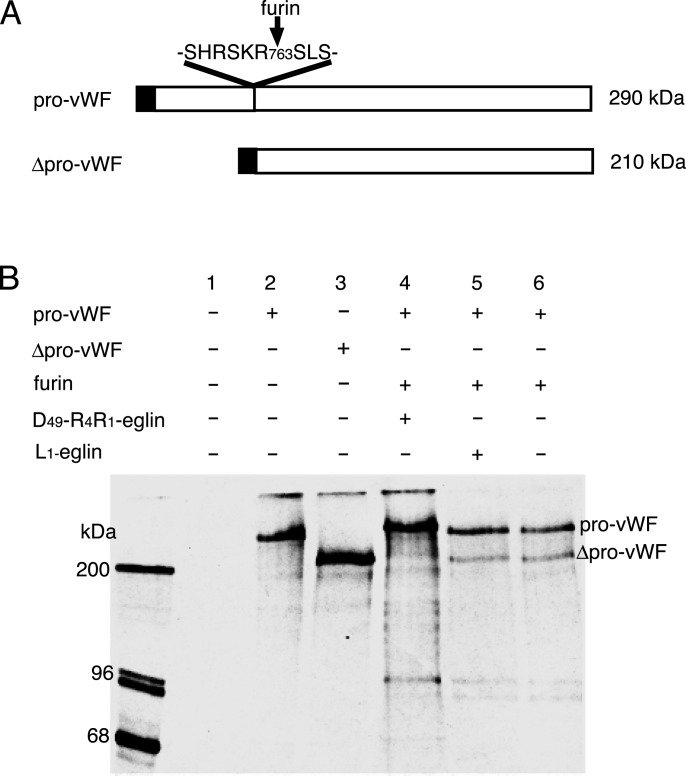
Fig. 4.
Furin-dependent processing of pro-vWF in COS-1 cells in the presence and absence of Asp-49–R4R1-eglin. (A) Schematic representations of pro-vWF and mature vWF. (B) After pulse labeling and a 6-h chase, cell extracts were prepared with Nonidet P-40 lysis buffer and immunoprecipitated with anti-vWF antibody. Immunoprecipitates were subjected to SDS/PAGE and analyzed by autoradiography. Lane 1, mock transfection; lane 2, pro-vWF; lane 3, Δpro-vWF, (lacks the pro-domain and serves as size control for processed vWF); lanes 4–6, furin and vWF cotransfection. At 4 h before pulse labeling, Asp-49–R4R1-eglin was added to the medium (lane 4), and WT eglin (Leu at P1) was added to the medium (lane 5).
Discussion
This article presents an approach to engineering protein–protein interactions to optimize, simultaneously, both affinity and selectivity. Enhancements in affinity and selectivity are achieved by optimization of adventitious interactions, that is, interactions involving residues outside the site(s) of primary recognition that are not subject to evolutionary constraints. We have applied this approach to creating high-affinity inhibitors selective for specific members of the Kex2/furin family of proprotein-processing enzymes. Creating selective, potent inhibitors for the seven mammalian enzymes in this family presents a considerable challenge because of the similarity of these enzymes in substrate recognition (10). Developing selective inhibitors for each member of the furin family logically demanded exploitation of the differences between the enzymes that can affect inhibitor interactions, most of which will lie outside the active site, at so-called “exosites” (14). The interface between a protein protease inhibitor, such as eglin c, and a target protease, such as Kex2 or furin, extends well beyond the reactive site loop of the inhibitor and the active site of the enzyme and encompasses exosites that become potential sites for adventitious interactions. Because eglin c did not evolve to inhibit Kex2, furin, or other processing enzymes, it seemed likely that these interactions were not optimal, but could be optimized by randomization of individual eglin c residues followed by screening for inhibitors of increased affinity. It also followed that because any two mammalian enzymes, such as furin and PC7, would differ extensively outside of the active site, substitutions optimal for one enzyme would often not be optimal for the other, and indeed might be interfering. We have applied this strategy to the optimization of R4R1-eglin for three targets, yeast Kex2 protease, mammalian furin, and PC7. In the course of the work, substitutions that increased affinity and selectivity for each of the enzymes tested, Kex2, furin, and PC7, were identified. Substitutions at two positions, 33 and 49, had significant effects on all three enzymes.
Importance of Position 49 (P′4). Substitutions for Tyr-49, which is equivalent to P′4, improved affinity for all three enzymes and permitted discrimination between Kex2 and furin, on the one hand, and PC7, on the other. Substitution of Asp for Tyr-49 enhanced affinity 8-fold for furin and 20-fold for Kex2 but slightly decreased affinity with PC7. Likewise, substitution of Glu for Tyr-49 increased affinity for both furin (5-fold) and Kex2 (25-fold) but decreased affinity for PC7 (2.2-fold). In contrast, substitution of Trp for Tyr-49 enhanced affinity 3-fold for PC7 but decreased affinity for furin 3-fold and slightly decreased affinity for Kex2 (Table 3). These results suggest that P′4 may contribute both to substrate recognition and discrimination by the proprotein-processing proteases.
Recently, the P′4 specificity of granzyme B was determined by alanine scanning mutagenesis of the reactive site loop of proteinase inhibitor 9, a serpin (24). This finding suggests that substitutions may be made at P′4 in serpins without destroying the inhibitory conformation. Substitution of an acidic residue at P′4 in α1-PDX (25), a variant of α1-antitrypsin engineered as a furin inhhibitor by inclusion of Arg at P1 and P4, might further enhance affinity and selectivity for furin.
Combination of Asp-33–Val and Tyr-49–Asp Mutations. Although combination of the Asp-33–Val and Tyr-49–Asp substitutions in R4R1-eglin did not have an additive effect on affinity for any of the enzymes, it did lead to greater discrimination by the inhibitor between furin and PC7, principally by decreasing affinity for PC7 (Table 4). Whereas Val-33–Asp–R4R1-eglin preferred furin to PC7 by 6-fold and Tyr-49–Asp–R4R1-eglin preferred furin to PC7 by 6-fold, Val-33–Asp–Tyr-49–Asp–R4R1-eglin exhibited 41-fold discrimination, in favor of furin, between the two enzymes. Thus, combination of substitutions at sites of adventitious interaction can generate unexpected gains in selectivity and, similarly, may offer unanticipated increases in affinity as well.
Interactions That Modulate Inhibitor Turnover Provide a Second Approach to Achieve Selectivity. R6R4R1-eglin was stable inhibitor of PC7 but a temporary inhibitor of furin. The use of substitutions that modulate the interaction of an inhibitor with noncognate target enzymes provides another potential means of producing inhibitors that function selectively in vivo. Thus, R6R4R1-eglin would be expected to provide more sustained inhibition with PC7 in vivo because interaction with furin would result in cleavage. Although not yet tested experimentally, we predict that introduction of the Tyr-49–Trp substitution into R6R4R1-eglin should produce a molecule that provides increased selectivity for PC7 through a combination of factors: enhanced affinity for PC7, decreased affinity for furin, and enhancement of furin cleavage. Given that furin and PC7 are widely expressed in mammalian tissues, inhibitor discrimination between these two enzymes may be crucial for in vivo applications.
Cellular Inhibition by Tyr-49–Asp–R4R1-Eglin. Pro-vWF is thought to be processed by furin during transit of the secretory pathway, presumptively in the trans-Golgi network, although recent studies suggest that at least some newly synthesized proteins may be processed by furin in post-trans-Golgi network compartments (26). In either case, inhibition of processing of pro-vWF by Asp-49–R4R1-eglin reflects disruption of an intracellular processing reaction, a conclusion reinforced by the fact that the experiment in Fig. 4 reports on cell-associated vWF species. A very high concentration of Asp-49–R4R1-eglin, 5 μM, was used to block processing in this case. Asp-49–R4R1-eglin at 2.5 μM was also effective, but 1 μM only partially blocked processing (data not shown). A very high concentration (8 μM) of the R4R1 variant of α1-antitrypsin, α1-PDX, was found to be required to block processing of the cytomegalovirus gB glycoprotein (25). This was attributed to a requirement to inactivate all cell-associated furin as it cycled through the plasma membrane, although the reason that concentrations 1,000-fold above the Ki would be required to accomplish this is unclear. An alternative explanation is that the inhibitor enters the endomembrane system by an inefficient process such as pinocytosis. The rate of internalization by this pathway is likely to be enhanced by a high external concentration of inhibitor. In support of this interpretation, we find that processing of a substrate known to be cleaved by furin at the cell surface can be completely blocked by 100 nM and partially inhibited by 20 nM Tyr-49–Asp–R4R1-eglin (data not shown). Pinocytotic internalization rates might be increased by increasing membrane association by acylation of the inhibitor, which increased the effectiveness of furin-targeted peptidyl chloromethanes by ≈100-fold in cellular inhibition assays (27). Alternatively, attachment of ligands for receptors that cycle between the plasma membrane and endomembrane systems might also enhance inhibitor uptake.
As shown in this work, exploiting potential adventitious contacts between a protease and a protein-protease inhibitor provides a powerful approach to creating high-affinity, selective inhibitors customized for interruption of a specific cellular function. This same method could be applied to engineering other protein–protein interactions, particularly those in which contacts involve a primary contact sequence in the context of a larger contact surface. Examples of such contacts can be found in binding of protein kinases to both protein substrates and inhibitors.
Acknowledgments
We thank Edward McEwen for help in COS-1 cell tissue culture and Jennifer Blanchette and Laura Rozan for careful reading of the manuscript. This work was supported in part by National Institutes of Health Grants GM39697 (to R.S.F.) and HL53777 (to R.J.K.), a Gerard Fellowship (University of Michigan) (to B.V.), a Fonds pour la Formation de Chercheurs et l'Aide à la Recherche–Fonds de la Recherche en Santé du Québec studentship (to M.F.), and Canadian Institutes of Health Research grants (to R.D.). R.D. is a Fonds de la Recherche en Santé du Québec Scholar.
Abbreviation: vWF, von Willebrand factor.
Footnotes
According to the nomenclature of Schechter and Berger (12), residues surrounding a cleavage site are designated, from N terminus to C terminus, P3-P2-P1 ↓P′1–P′2-P′3, with the scissile bond between P1 and P′1. Corresponding binding sites in the enzyme for substrate side chains are designated S1, S′1, etc.
References
- 1.Steiner, D. F. (2001) in The Enzymes XXII, eds. Dalbey, R. E. & Sigman, D. S. (Academic, San Diego) pp. 163-198.
- 2.Rockwell, N. C. & Fuller, R. S. (2001) in The Enzymes XXII, eds. Dalbey, R. E. & Sigman, D. S. (Academic, San Diego), pp. 260-289.
- 3.Miller, C. J., Elliott, J. L. & Collier, R. J. (1999) Biochemistry 38 10432-10441. [DOI] [PubMed] [Google Scholar]
- 4.Hatta, M., Gao, P., Halfmann, P. & Kawaoka, Y. (2001) Science 293 1840-1842. [DOI] [PubMed] [Google Scholar]
- 5.Thomas, G. (2002) Nat. Rev. Mol. Cell Biol. 3 753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roebroek, A. J. M., Ulmans, L., Pauli, I. G. L., Robertson, E. J., van Leuven, F., Van de Ven, W. J. M. & Constam, D. B. (1998) Development (Cambridge, U.K.) 125 4863-4876. [DOI] [PubMed] [Google Scholar]
- 7.Spence, M. J., Sucic, J. F., Foley, B. T. & Moehring, T. J. (1995) Somatic Cell Mol. Genet. 21 1-18. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi, S., Kasai, K., Hatsuzawa, K., Kitamura, N., Misumi, Y., Ikehara, Y., Murakami, K. & Nakayama, K. (1993) Biochem. Biophys. Res. Commun. 195 1019-1026. [DOI] [PubMed] [Google Scholar]
- 9.Sarac, M. S., Cameron, A. & Lindberg, I. (2002) Infect. Immun. 70 7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockwell, N. C., Krysan, D. J., Komiyama, T. & Fuller, R. S. (2002) Chem. Rev. 102 4525-4548. [DOI] [PubMed] [Google Scholar]
- 11.Komiyama, T. & Fuller, R. S. (2000) Biochemistry 39 15156-15156. [DOI] [PubMed] [Google Scholar]
- 12.Schechter, I. & Berger, A. (1967) Biochem. Biophys. Res. Commun. 27 157-162. [DOI] [PubMed] [Google Scholar]
- 13.Siezen, R. J. & Leunissen, J. A. M. (1997) Protein Sci. 6 501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bode, W. & Huber, R. (2000) Biochim. Biophys. Acta 1477 241-252. [DOI] [PubMed] [Google Scholar]
- 15.Bode, W., Papapamokos, E., Musil, D. & Seemuller, U. (1986) EMBO J. 5 813-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner, C. & Fuller, R. S. (1992) Proc. Natl. Acad. Sci. USA 89 922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fugere, M., Limperis, P. C., Beaulleu-Audy, V., Gagnon, F., Vavigne, P., Klaskov, K., Leduc, R. & Day, R. (2002) J. Biol. Chem. 277 7648-7656. [DOI] [PubMed] [Google Scholar]
- 18.Bravo, D. A., Gleason, J. B., Sanchez, R. I., Roth, R. A. & Fuller, R. S. (1994) J. Biol. Chem. 269 25830-25837. [PubMed] [Google Scholar]
- 19.Wise, R. J., Wong, P. A., Kiefer, M. C., Brake, A. J. & Kaufman, R. J. (1990) Proc. Natl. Acad. Sci. USA 87 9378-9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue, H., Nojima, H. & Okayama, H. (1990) Gene 96 23-38. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez-Laslop, N., Lee, H., Hu, R. & Neyfakh, A. (2001) J. Bacteriol. 183 2399-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorner, A. J. & Kaufman, R. J. (1990) Methods Enzymol. 185, 577-596. [DOI] [PubMed] [Google Scholar]
- 23.Rehemtulla, A. & Kaufman, R. J. (1992) Blood 79 2349-2355. [PubMed] [Google Scholar]
- 24.Sun, J., Whisstock, J. C., Harriott, P., Walker, B., Novak, A., Thompson, P. E., Smith, A. I. & Bird, P. I. (2001) J. Biol. Chem. 276 15177-15184. [DOI] [PubMed] [Google Scholar]
- 25.Jean, F., Stella, K., Thomas, L., Liu, G., Xiang, Y., Reason, A. & Thomas, G. (1998) Proc. Natl. Acad. Sci. USA 95 7293-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Band, A. M., Maatta, J., Kaariainen, L. & Kuismanen, E. (2001) FEBS Lett. 505 118-124. [DOI] [PubMed] [Google Scholar]
- 27.Garten, W., Stieneke, A., Shaw, E., Wikstrom, P. Q. & Klenk, H. D. (1989) Virology 172 25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson, D. C. & Richardson, J. S. (1992) Protein Sci. 1 3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]