Abstract
CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone, which is thought to facilitate translation at low temperature by destabilizing mRNA structures. Here we demonstrate that CspA, as well as homologous RNA chaperones CspE and CspC, are transcription antiterminators. In vitro, the addition of physiological concentrations of recombinant CspA, CspE, or CspC decreased transcription termination at several intrinsic terminators and also decreased transcription pausing. In vivo, overexpression of cloned CspC and CspE at 37°C was sufficient to induce transcription of the metY-rpsO operon genes nusA, infB, rbfA, and pnp located downstream of multiple transcription terminators. Similar induction of downstream metY-rpsO operon genes was observed at cold shock, a condition to which the cell responds by massive overproduction of CspA. The products of nusA, infB, rbfA, and pnp—NusA, IF2, RbfA, and PNP—are known to be induced at cold shock. We propose that the cold-shock induction of nusA, infB, rbfA, and pnp occurs through transcription antitermination, which is mediated by CspA and other cold shock-induced Csp proteins.
Keywords: CspA proteins, cold shock, transcription antitermination
When exponentially growing Escherichia coli cells are shifted to a low temperature of 15°C, the cell growth stops, and the synthesis of most proteins is repressed. However, some proteins are synthesized at increased rate. The synthesis of one such protein, the 70-aa-long CspA, is spectacularly induced and comprises more than 10% of the total protein synthesis in cold-shocked cells. Eventually, CspA synthesis decreases, and cells gradually resume growth at low temperature. Over the years, more than 50 homologues of CspA have been identified in bacteria (1). CspA is also highly homologous to a domain called CSD (cold-shock domain) of eukaryotic Y-box proteins, which are involved in regulation of gene expression at the level of transcription or translation (2). E. coli contains nine members of the CspA family from CspA to CspI. In E. coli, CspA, CspB, CspG, and CspI are cold-shock inducible; CspC and CspE are constitutively produced at 37°C; CspD is induced at the early stationary phase (see ref. 1 for a review).
The exact physiological function of CspA and its homologues has not been defined yet, and at least some Csp proteins may be functionally redundant (3–5). CspA binds cooperatively to RNA or single-stranded DNA without apparent sequence specificity (6) with a Kd of around 2.7 × 10−5 M. On cold shock, the intracellular concentration of CspA reaches ≈100 μM, and it should be able to bind to mRNAs under physiological conditions (6, 11). CspA binding to RNA destabilizes RNA secondary structures. Thus, it was proposed that high levels of CspA could facilitate translation at low temperatures by destabilizing secondary structures in mRNA (6, 8–10).
Recent data implicate CspE in transcription regulation. Interactions of CspE with nascent RNA chains during transcription elongation were detected by RNA–protein crosslinking. Moreover, purified CspE interfered with Q-mediated transcription antitermination (11). CspE as well as CspA were also proposed to increase transcription pause recognition at the region following the “cold box” sequence, a negative cis element located in the 5′ untranslated region of the cspA mRNA (5, 12). In the present paper, we further examine the role of Csp proteins in transcription termination. We show that CspA, CspE, and CspC function as transcription antiterminators at ρ-independent terminators in vitro and in vivo, and we suggest that this occurs by preventing the formation of secondary structures in the nascent RNA. Further, we demonstrate that the in vivo activation of a cold-shock operon occurs at the level of antitermination and is likely mediated by Csp proteins.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli strain JM83 was used as a host for plasmid construction and for RNA isolation.
The pINCspA, pINCspC, and pINCspE plasmids are derived from pINIII (13) and contain cspA, cspC, and cspE, respectively, under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lpp and lac promoters
To construct pWBmy, a DNA fragment encompassing the region from + 112 to + 308 of the metY gene and containing two intrinsic terminators, t1 and t2 (14), was obtained by PCR by using the JM83 chromosomal DNA as a template with the following primers: the forward primer, 5′-CCGGATCCTACTGTGAAGACTTCG-3′, the sequence underlined represents an engineered BamHI site; the reverse primer, 5′-CCGGATCCTGCAGTTGCTCTAATGTGGAC-3′, the sequence underlined represents a PstI site. The PCR product was digested with BamHI and PstI and cloned into appropriately treated pRL407 (15), generating pWBmy
Proteins.
RNA polymerase (RNAP) reconstitution was performed as described (16). RNAP was further purified on a Mono-Q column attached to FPLC. Fractions containing RNAP holoenzymes were pooled, concentrated to ≈1 mg/ml, and stored in −20°C in the presence of 50% glycerol. CspA, CspB, and CspC were purified as described (17). CspE was purified as described in ref. 5.
In Vitro Transcription.
DNA templates were prepared by PCR from plasmids carrying various ρ-independent terminators downstream from the T7 A1 promoter with the use of the following primers: 5′-GACCAGACCTAAAGACCAGAC-3′; and 5′-GCAGTTCCCTACTCTCGCATGC-3′. The trpL template was obtained from plasmid pRL407 as described previously (17). The trp template was obtained from plasmid pRL522 as described previously (18). The t1 and t2 of the metY-rpsO operon was obtained from the pWBmy plasmid described above. The tR2 template was obtained by PCR amplification of the 324-bp DNA fragment containing T7 A1 promoter as described previously (18).
To study the effect of Csp proteins on transcription, elongation complexes stalled at position + 20 (EC20) were prepared in 50 μl of transcription buffer containing 25 μl of Ni2+-nitrilotriacetic acid agarose, 20 nM of T7 A1 promoter–DNA fragments, 40 nM His-tagged RNAP, and 0.5 mM ApU. The standard transcription buffer contained 40 mM KCl, 40 mM Tris⋅HCl, pH 7.9, and 10 mM MgCl2. Reaction mixtures were incubated for 5 min at 37°C to form the open complexes and then transferred to room temperature, and 50 μM CTP and ATP, and 2.5 μM [α-32P]GTP (300 Ci/mmol) were added. After 10-min incubation, the agarose beads were washed five times with 1.5 ml of transcription buffer as described previously (19). Equal aliquots containing purified stalled EC20 were supplemented with Csp proteins and NTPs. To study termination efficiency, the final NTP concentration was 250 μM and reactions were incubated for 5 min at room temperature unless otherwise stated. Longer incubations did not result in changes in transcription termination levels, suggesting that reactions were complete. To study transcription pausing, the final NTP concentration was 5 μM. Reactions were allowed to proceed for various time intervals at room temperature and were terminated by the addition of formamide-containing loading buffer. Products were analyzed by urea-PAGE electrophoresis (7 M urea/10% polyacrylamide), followed by autoradiography and phosphorimager analysis.
Northern Blot Analysis.
Total RNA was isolated from cells grown at 37°C (OD600 of 0.6) and the cold-shocked cells (cells were initially grown at 37°C to OD600 of 0.6, shifted to 15°C, and incubated for additional 30 min) by the hot phenol method (20). For RNA isolation from cells overproducing Csp proteins at 37°C, cells transformed with pINCspA, pINCspC, or pINCspE were grown to OD600 of 0.3–0.4 and induced with 1 mM IPTG. After 60 min, half of the culture was removed and subjected to RNA isolation by the hot phenol method. The other half was processed for SDS/PAGE.
For Northern blot analysis, 20 μg of total RNA was loaded per lane. Hybridization probes were generated by asymmetric PCR amplification (21) of the antisense strands of metY, nusA, rbfA, and lpp in the presence of [α 32P]dCTP, by using chromosomal DNA as template. Blotting and hybridization were performed by using standard procedures (22). The blots were washed with 2 × SSC/0.1% SDS for 1 h at 60°C. To reprobe, the same blot was washed with 0.5 × SSC/0.1% SDS for 2 h at 80°C and rehybridized.
Ribonuclease Assay.
For preparation of the RNA substrate, the region corresponding to the 5′-UTR (from + 1 to + 142) of the cspA mRNA, which was previously used to show RNA chaperone activity of CspA (6), was placed under the control of T7 RNAP promoter, and in vitro transcription of the PCR-amplified template was carried out in the presence of [α 32P] CTP as described (6). 32P-labeled RNA substrates (≈1 × 104 cpm) were incubated for 15 min on ice in 15 μl of the buffer containing 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, 50 mM KCl, 7.4% glycerol in the presence or in the absence of ≈27 μM of Csp proteins. RNase T1 (Boehringer Mannheim) was added to the reaction mixture, and the reactions were kept at room temperature for 6 min and loaded onto a 8% acrylamide gel.
Results
Csp Proteins Act as Transcription Antiterminators at Intrinsic Transcription Terminators in Vitro.
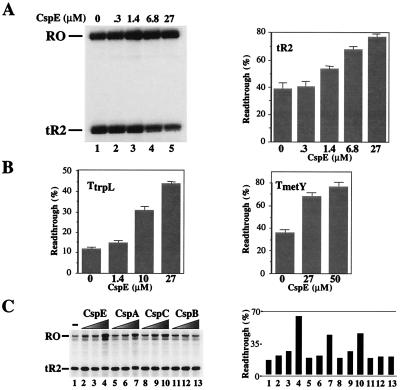
CspE, one of the nine members of the cold shock family of proteins present in E. coli, was recently shown to interact with nascent RNA chains during in vitro transcription in partially fractionated E. coli extracts (11). To better define the role of CspE in transcription, we studied the effect of highly pure recombinant CspE on transcription by E. coli RNAP. As a template, we used a DNA fragment containing the T7 A1 promoter that was followed by a ρ-independent terminator, tR2, at position + 105 relative to the transcription start point (18). Immobilized transcription complexes artificially stalled at position + 20 were prepared, and transcription was resumed by the addition of NTPs in the presence or in the absence of CspE. The addition of CspE decreased transcription termination at tR2 (Fig. 1A). The readthrough efficiency (RE) defined as the fraction of the run-off transcripts of the total transcripts produced was less than 40% in the absence of CspE and increased to almost 80% in the presence of 27 μM of CspE (lane 1 and lane 5, respectively). The effect of CspE on transcription termination on other intrinsic terminators, such as TtrpL (15) and TmetYt1 (14), was also tested (Fig. 1B). As can be seen, CspE also increased RE at these terminators. It should be noted that the maximum amount of CspE used in this experiment was roughly equal to its cellular concentration (approximately 30 μM; ref. 5).
Figure 1.
Csp proteins act as transcription antiterminators in vitro. (A) CspE induces transcription antitermination on the tR2 terminator. Transcription elongation complexes artificially stalled at position 20 (EC20) were prepared by using a DNA template containing the T7 A1 promoter fused to tR2 terminator and αHisRNAP immobilized on Ni2+-nitrilotriacetic acid agarose. The indicated concentrations of recombinant CspE were added to washed EC20, and transcription elongation was resumed by the addition of NTP. Reaction products were separated by denaturing PAGE and visualized by autoradiography. RO and tR2 indicate the runoff and the tR2-terminated transcripts, respectively. The gel shown (Left) was quantified by using phosphorimagery, and the readthrough efficiency, RE, was calculated (RE = {[RO transcripts]/[total transcripts (tR2 + RO)]} × 100%). The results are schematically presented (Right). (B) CspE induces transcription antitermination at the trpL and metY terminators. The in vitro transcription and product analysis were carried out under the same condition as described in A, except the template used contained TtrpL (Left) or TmetYt1 (Right) fused to the T7 A1 promoter. (C) Effects of CspA, CspB, CspC, and CspE on transcription termination at tR2. Csp proteins (CspE, CspA, CspB, and CspC) were used for in vitro transcription assays, as described in A: lane 1, no Csp protein; 1.4 μM Csp for lanes 2, 5, 8, and 11; 10 μM Csp for lanes 3, 6, 9, and 12; and 20 μM Csp for lanes 4, 7, 10, and 13. The RE values were quantified and are schematically presented (Right).
Purified recombinant CspE homologues, CspA, CspB, and CspC, were also tested for their antitermination activity in vitro (Fig. 1C). Both CspA and CspC increased RE at tR2. In contrast, CspB showed no effect on transcription termination in this system.
From these results, we conclude that some Csp proteins can act as transcription antiterminators in vitro. In the following sections, we present the effects of the most active Csp protein, CspE, on in vitro transcription. However, similar effects were observed with purified CspA and CspC proteins (data not shown).
The Mechanism of Csp-Induced Antitermination.
Antitermination through protein–protein interactions between RNAP and protein factors has been described in several systems (23–25). To test whether Csp proteins interact with RNAP core or holoenzyme, we used HPLC size-exclusion chromatography, native PAGE, and Ni2+-nitrilotriacetic acid agarose coimmobilization assays. Under none of the conditions used were we able to detect interaction between CspE and RNAP (data not shown). Therefore, the observed effect of Csp proteins on transcription termination is likely caused by the RNA-binding activity of Csp proteins or their ability to interact with transcription elongation complexes.
The C-terminal domain of RNAP α subunit, αCTD, is a well-characterized docking site for transcription regulators (25–27). RNAPα235, which lacks αCTD, elongates and terminates transcription normally, but does not interact with transcription termination factor NusA (25). To determine whether αCTD is necessary for Csp-mediated transcription antitermination, we investigated the effect of Csp addition on in vitro transcription by RNAPα235 by using the same conditions as used in Fig. 1A. RNAPα235 responded to the addition of CspE by decreasing transcription termination, as did the wild-type RNAP, indicating the lack of functional interaction between αCTD and CspE (data not shown).
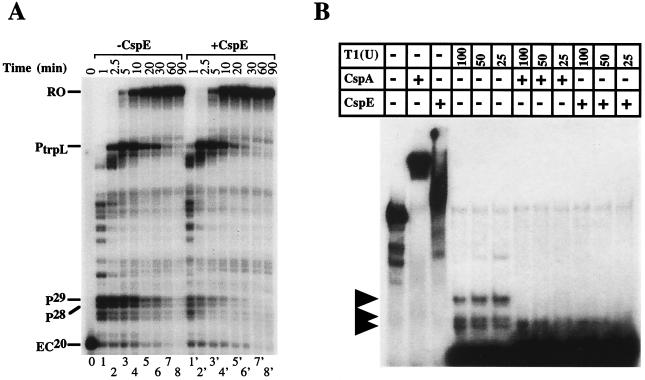
In E. coli, intrinsic transcription terminators consist of two essential elements: a stem–loop structure and a stretch of U residues (28, 29). Csp proteins may affect termination at intrinsic terminators by (i) preferentially interacting with the stem–loop RNA structure, or (ii) by preferentially interacting the U-rich stretch (a combination of the two is also possible). As an initial test of these two possibilities, the effect of CspE on transcription elongation at a hairpin pause site was examined. Such pause sites contain the stem–loop structure but lack the U stretch. Immobilized transcription was performed by using the T7 A1 promoter-driven template containing PtrpL, a hairpin pause site (15). Transcription complexes stalled at position + 20 were prepared, and transcription was resumed by the addition of low concentrations of NTP in the presence or absence of CspE. Reaction aliquots were withdrawn at various time points, and the products were analyzed by denaturing PAGE (Fig. 2A). In the absence of CspE, RNAP paused at PtrpL and at an additional previously uncharacterized strong pause site, which is more proximal to the promoter. This additional pause site was mapped to positions + 28 and + 29 relative to transcription start site (P28 and P29 in the Fig. 2A). In the presence of 27 μM CspE, the pausing efficiency at P28/P29 decreased significantly. The effect of CspE on P28/P29 pausing was quantified by using phosporimagery. In the absence of CspE, more than 80% of transcription complexes were paused at P28/P29 1 min after the addition of NTP (Fig. 2A, lane 2). In contrast, in the presence of CspE less than 30% of complexes were paused after 1 min of elongation (lane 2′). CspE also decreased the pause half-life from ≈10 min to less than 5 min. As a result, in the presence of CspE RNAP reached PtrpL and the end of the template sooner (compare lanes 3 and 3′). Pausing at PtrpL also appeared to decrease in the presence of CspE, although this effect was difficult to quantify because of the upstream P28/P29 signal. CspE and the other Csp proteins presumably act by binding nascent RNA; one mechanism of reducing pausing would be to prevent RNA hairpin formation. In fact, one of our laboratories had demonstrated earlier that the CspA protein can act as an “RNA chaperone,” and that CspA binding to double-stranded RNA destabilizes RNA secondary structure and increases RNA susceptibility to RNase attack (6). Because no comparable experiments were performed with CspE or CspC, we repeated the RNase T1 susceptibility experiment of Jiang et al. (6) by using purified CspC/E (Fig. 2B). Radioactively labeled in vitro transcribed RNA derived from the 5′-UTR of the cspA promoter was used as a substrate. Addition of either CspA or CspE to RNA substrate in the absence of RNase treatment resulted in supershift (compare lane 1 with lanes 2 and 3), indicating that both Csp proteins were interacting with RNA in our assay conditions. As is explained elsewhere, the 142-base-long RNA substrate forms extensive secondary structure (6). Therefore, treating this RNA with RNase T1 results in the appearance of several resistant fragments (ref. 6, lanes 4–6). As expected, the RNase-resistant fragments disappeared when CspA was added to the reaction (lanes 7–9). Importantly, addition of CspE (lanes 10–12) and CspC (data not shown) also lead to disappearance of resistant RNA fragments. We conclude that CspC and CspE facilitate the ribonuclease activity of RNase T1 by destabilizing the secondary structures of RNA substrate and thus function as RNA chaperones in a manner similar to CspA (6). We propose that the RNA-chaperoning activity of CspA, CspC, and CspE is the cause of the antitermination activity of these proteins.
Figure 2.
Effect of CspE on transcription pausing, and RNA chaperoning activity of Csp proteins. (A) In vitro transcription experiment was performed by using DNA fragment consisting of the T7 A1 terminator fused to the trpL pause site (PtrpL) as template. To observe pausing, low concentration of NTP (5 μM) was used for chasing the primary EC (EC20). Reactions were terminated at the times indicated. (B) Stimulation of RNase activity by CspE and CspA. Ribonuclease assays were carried out as described in Materials and Methods. The RNA substrate used is the same as used previously for the ribonuclease experiment for CspA (6). RNase T1 was added as indicated (Top) in units. Lane 1, RNA substrate alone; lane 2, RNA substrate plus CspA (27 μM); lane 3, RNA substrate plus CspE (27 μM); lanes 4–6, 100, 50, 25 unit of RNase T1, respectively; lanes 7–9, the same as lanes 4–6, respectively, except that CspA (27 μM) was added before adding RNase T1; lanes 10–12, the same as lanes 4–6, respectively, except that CspE (27 μM) was added before adding RNase T1. The reaction mixtures were incubated on ice for 10 min and loaded onto the 8% acrylamide gel.
Csp Proteins Antiterminate Transcription of a Cold-Shock Operon in Vivo and in Vitro.
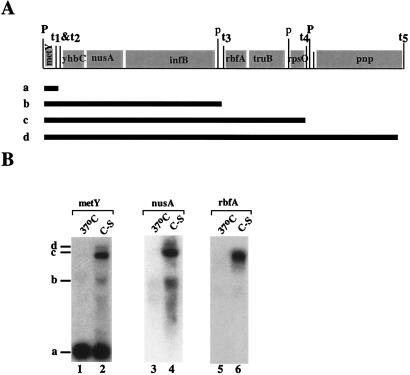
A set of genes encoding four cold-shock proteins, NusA, IF2, RbfA, and PNP, is located in two adjacent operons called the metY and rpsO operons (refs. 30–35; see also Fig. 3A). In the present paper, we consider metY-rpsO as a single operon, because metY is cotranscribed with pnp (Fig. 3A; ref. 33). The operon contains four cold-shock genes, nusA, infB, rbfA, and pnp, and four intercistronic intrinsic terminators, from t1 to t4. Except for the promoter downstream of rpsO, promoters located in the middle of this operon are very weak. The shortest metY transcript is the most abundant because of the transcriptional polarity (30, 33, 35).
Figure 3.
The downstream genes of the metY-rpsO operon are induced by cold shock. (A) The gene structure of the metY and rpsO operons (33), which contains metY (tRNAf2), yhbC (an unidentified ORF), nusA (NusA), infB(IF2), rbfA (RbfA, 34), truB (P35, E. coli Psi synthase), rpsO (S15), and pnp (PNPase). The positions of four intercistronic intrinsic terminators from t1 to t4 and promoters are indicated. Capital “P”s represent promoters that are relatively strong. Lowercase “p”s represent weak promoters. The hypothetical transcripts, which are initiated from the metY promoter but terminated at different terminators sequentially, are shown and indicated with letters, “a–d”. (B) RNA was isolated from cells grown at 37°C and cold-shocked cells (C-S). An equal amount of RNA (20 μg) was loaded in each lane. Northern blot analysis was carried out by using the metY gene as a probe (Left). After the membrane was washed, it was reprobed with the rbfA gene (Center) and the nusA gene (Right). The transcripts are indicated with the letters “a–d,” whose identities are shown in A.
It was previously suggested that on temperature downshift, the relative overexpression of NusA, IF2, RbfA, and PNPase might occur because of the readthrough from metY (8, 33). To test this idea, we compared transcripts produced from the metY-rpsO operon in cells grown at 37°C and in cold-shocked cells (Fig. 3B). RNA was purified from cells and probed with radioactively labeled DNA fragment that contained the entire metY gene. In the 37°C sample, the major transcript observed corresponded to tRNAf2Met, which is encoded by promoter-proximal metY (“a” in Fig. 3B). Cold shock resulted in a dramatic change in the spectrum and length of metY-rpsO transcripts (compare lanes 1 and 2). Based on the estimates of their length, transcripts labeled “b” and “d” correspond to RNAs from metY to infB and from metY to pnp, respectively. It should be noted that the metY probe used in lane 1 and 2 of Fig. 3B extended into the area downstream of the metY gene by more than 200 nucleotides and thus could hybridize to longer transcripts from the processed primary message. That the metY probe detected only promoter-proximal metY transcript at 37°C indicates that most of metY-rpsO transcripts terminate at t1 and/or t2.
To confirm the assignments of multiple metY-rpsO transcripts in cold-shocked cells, the membrane was reprobed with the rbfA gene (located between t3 and t4) or with a fragment of the nusA gene. The results are presented in Fig. 3B, lanes 3 and 4, and 5 and 6, respectively. In the 37°C sample, no transcript was detected by the rbfA probe, whereas in the cold-shocked cells, a long transcript corresponding to transcript “c” was detected. In the 37°C sample, the nusA probe did not detect transcript “a” but hybridized with band “b,” which results from transcription termination at t3. In the cold-shocked sample, the longer transcripts “c” and “d” became detectable by the nusA probe. These data strongly suggest that at 37°C, most of metY-rpsO transcription is terminated after metY, whereas on cold shock, expression of distal genes occurs through a transcription antitermination mechanism. Alternatively, the observed changes in the pattern of transcripts can be caused by altered RNA processing. We strongly favor the first possibility. The following experiments support this view.
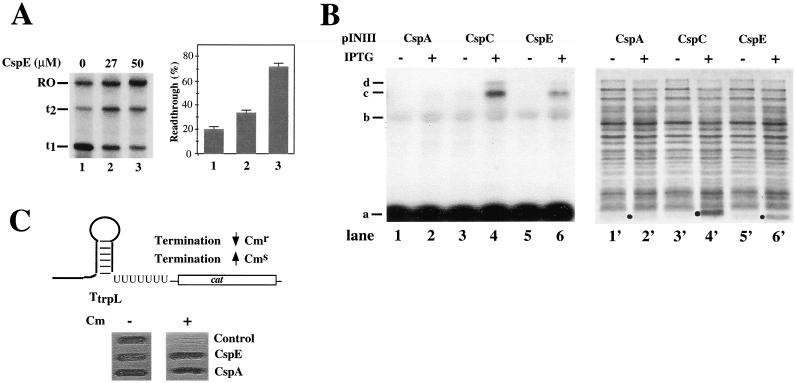
Csp proteins acted as antiterminators of metY-rpsO terminators in vitro. Two consecutive intrinsic metY-rpsO terminators, t1 and t2, which are located downstream of the metY and are thus primarily responsible for observed polarity at 37°C, were fused to the T7 A1 promoter, and the antitermination effect of Csp proteins was examined by using immobilized transcription system. The result obtained with purified CspE is presented in Fig. 4A and shows that CspE increased RE at both t1 and t2. In the absence of CspE (lane 1), 80% of the nascent RNA chains were terminated at t1, 6% were terminated at t2, and only 14% reached the end of the template (labeled RO on Fig. 4). At physiological concentration of 27 μM of CspE, these values were 36, 27, and 37%, respectively (lane 2). At high CspA concentration (50 μM, lane 3), most RNAP molecules reached the end of template (72%); 13% terminated at t1 and 15%, at t2.
Figure 4.
Csp proteins cause transcription antitermination at the metY-rpsO operon in vitro and in vivo. (A) A DNA fragment containing two consecutive intrinsic terminators, t1 and t2, found downstream of metY and genetically fused to the T7 A1 promoter, was used as a template to examine the effect of CspE on the termination at these terminators. The experiment was performed and data analyzed as described in the Fig. 1 legend. The combined RE values at t1 and t2 were calculated (RE = {[RO transcripts]/[total transcripts (t1+ t2+ RO)]} × 100%) and are graphically presented (Right). (B) E. coli cells were transformed with the pINCspA, pINCspC, or pINCspE plasmids, grown at 37°C to OD600 of 0.3–0.4, and csp expression was induced by the addition of 1 mM IPTG. After 60 min incubation at 37°C, half of the culture was subjected to the isolation of total RNA and Northern blot analysis by using the metY gene as a probe (lanes 1–6), whereas the other half was analyzed by SDS/PAGE on a 17.5% gel (lanes 1′-6′). The overexpressed proteins are indicated by black dots. (C) RL211 E. coli cells containing a cat gene cassette positioned downstream of the trpL terminator (36) were transformed with pINCspA, pINCspC, or control pINIII, and spotted on plates containing 30 μg/ml chloramphenicol and1 mM IPTG. The results of overnight cell growth are presented.
To demonstrate that Csp proteins also act as transcription antiterminators in vivo, we wished to overexpress, in a controlled manner, a Csp protein at 37°C, and to monitor cold-shock operon transcripts at these conditions. Plasmids overproducing CspA, CspE, and CspC were introduced in E. coli cells. The cells were grown at 37°C, plasmid-borne csp genes were induced, cells were harvested, and the metY-rpsO transcripts were analyzed by Northern blotting (Fig. 4B, lanes 1–6). In parallel, the amount of overproduced Csp proteins was examined by SDS/PAGE (Fig. 4B, lanes 1′-6′). The results presented in Fig. 4B demonstrate that no significant overproduction of CspA was achieved (compare lanes 1′ and 2′). In contrast, CspC and CspE were significantly overproduced (lanes 4′ and 6′, respectively). Strikingly, cells overproducing CspC/E contained longer metY-rpsO transcripts (lanes 4 and 6), and the pattern of transcripts in CspC/E-overproducing cells was very similar to that in cold-shocked cells. We conclude that CspC/E overproduction is sufficient to stimulate antitermination of the cold-shock metY-rpsO operon in vivo.
To demonstrate the in vivo antitermination effect of CspA, we used the E. coli RL211 strain devised by Landick et al. (36). RL211 contains the cat gene preceded by ρ-independent trpL terminator and is sensitive to chloramphenicol. However, when transcription termination at trpL is reduced, RL211 becomes chloramphenicol resistant. RL211 cells were transformed with plasmids overproducing either cspE or cspA, and cells were plated on medium containing chloramphenicol. As shown in Fig. 4C, cells harboring control plasmid (pINIII) did not grow on the plate containing chloramphenicol, whereas cells harboring either cspE or cspA expression plasmid became resistant to chloramphenicol. These results indicate that CspA/E overproduction is sufficient to stimulate antitermination at the trpL terminator in vivo.
Discussion
In this paper, we demonstrate that overproduction of CspE/C proteins at 37°C causes increased expression of the promoter-distal nusA, infB, rbfA, and pnp cold-shock-inducible genes of the metY-rpsO operon. Dramatically, on temperature downshift, a condition to which the cell responds by gross overproduction of CspA and other Csp proteins, a similar derepression of nusA, infB, rbfA, and pnp is observed. In addition, we report here that Csp protein act as transcription antiterminators in a purified in vitro system. In principle, the cold-shock induction of the metY-rpsO operon can be caused by CspA-mediated transcription antiterimination or CspA-mediated changes in mRNA stability. Although we cannot rigorously exclude alterations in RNA processing, we believe the following results make this possibility unlikely. First, as already mentioned, the addition of physiological concentrations of recombinant Csp proteins to pure transcription system increases readthrough efficiency at several intrinsic terminators, including metY t1 and t2 in vitro. Second, Csp overexpression allows the growth of the sensitive RL211 tester strain on chloramphenicol. RL211 has been used extensively by the Landick group to obtain numerous RNAP mutants, which were affected in termination efficiency in vitro (36, 37). Therefore, we propose that CspA and its 37°C degree homologues CspE and CspC act as transcription antiterminators in vivo, and that Csp-induced transcription antitermination is responsible for increased expression of promoter-distal genes of the metY-rpsO cold-shock operon.
It is likely that Csp proteins act by binding RNA. One potential target is RNA secondary structure, which is known to be inhibiting to various cellular functions such as translation and RNA processing (29, 32). Hairpin formation of nascent RNAs causes destabilization of transcription elongation complexes leading to either pausing or intrinsic termination, depending on the surrounding DNA sequences (29, 39). Because RNA hairpins become more stable at low temperature (38, 40), one possibility is that Csp proteins facilitate transcription elongation by destabilizing these hairpin structures. However, they might also act in other ways through RNA binding, e.g., by inhibiting backtracking at pause sites. We were unable to detect Csp protein interaction with RNAP. However, Hanna and Liu (11) recently demonstrated specific interaction of CspE with the nascent RNA. Thus, Csp proteins might be able to differentiate between the nascent RNA chains and the rest of the cellular RNA, and specifically target transcription elongation complexes. Interestingly, Hanna and Liu (11) did not observe CspE-induced antitermination. This could be a result of the lower concentration of CspE in their reactions.
E. coli genome contains nine genes coding for CspA-like proteins. The exact function of these proteins is not known, and at least some of them can substitute for each other. It is attractive to speculate that different Csp proteins antiterminate transcription of different sets of genes, thus allowing the cell to adequately respond to different environmental challenges. Our in vivo results are consistent with this notion, because low levels of CspA expression allowed readthrough past the trpL terminator but apparently had no effect on termination at t1 and t2 of metY-rpsO. It has been shown recently by one of our laboratories that different Csp proteins have different nonoverlapping optimal RNA-binding sequences (41). Experiments designed to uncover terminator-specific effects of different Csp proteins are currently in progress.
Acknowledgments
We thank Dr. R. Landick and his colleagues for their generous gift of plasmids. We are grateful to S. Nechaev for providing materials and for his advice with RNAP-CspE-binding experiments. We thank Drs. S. Phadtare and K. Yamanaka for critical reading of the manuscript and for providing purified CspC protein. This work was supported by grants GM19043 (to M. I.) and GM59295 (to K. S.) from the National Institutes of Health and by the Burroughs Wellcome Fund Career Development Award to K.S.
Abbreviation
- IPTG
isopropyl-β-d-thiogalactopyranoside
- RNAP
RNA polymerase
References
- 1.Yamanaka K, Fang L, Inouye M. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe A P. Bioassays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 3.Graumann P, Wendrich T M, Weber M H W, Schroder K, Marahiel M A. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 4.Bae W, Jones P G, Inouye M. J Bacteriol. 1997;179:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae W, Phadtare S, Severinov K, Inouye M. Mol Microbiol. 1999;31:1429–1442. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, Hou Y, Inouye M. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein J, Pollott N S, Inouye M. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones P G, Inouye M. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 9.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 10.Graumann P, Marahiel M A. Mol Gen Genet. 1997;253:745–752. doi: 10.1007/s004380050379. [DOI] [PubMed] [Google Scholar]
- 11.Hanna M, Liu K. J Mol Biol. 1998;282:227–239. doi: 10.1006/jmbi.1998.2005. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Fang L, Inouye M. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masui Y, Coleman J, Inouye M. Experimental Manipulation of Gene Expression. New York: Academic; 1983. pp. 15–31. [Google Scholar]
- 14.Ishii S, Kuroki K, Imamoto F. Proc Natl Acad Sci USA. 1984;81:409–413. doi: 10.1073/pnas.81.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landick R, Stewart J, Lee D N. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 16.Tang H, Severinov K, Goldfarb A, Ebright R H. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee S, Jiang W, Emerson S D, Inouye M. J Biochem (Tokyo) 1993;114:663–669. doi: 10.1093/oxfordjournals.jbchem.a124234. [DOI] [PubMed] [Google Scholar]
- 18.Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- 19.Kashlev M, Martin E, Polyakov A, Severinov K, Nikiforov V, Goldfarb A. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Sarmientos P, Sylvester J E, Contente S, Cashel M. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- 21.Macabe P C. In: PCR Protocols, A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. 1990. pp. 76–83. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, N.Y.: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Roberts J W, Yarnell W, Bartlett E, Guo J, Marr M, Ko D C, Sun H, Roberts C W. Cold Spring Harbor Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 24.Greenblatt J, Mah T F, Legault P, Mogridge J, Li J, Kay L E. Cold Spring Harbor Symp Quant Biol. 1998;63:327–336. doi: 10.1101/sqb.1998.63.327. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Zhang Y, Severinov K, Das A, Hanna M. EMBO J. 1996;15:150–161. [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi K, Ishihama A. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 27.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uptain S M, Kane C M, Chamberlin M J. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 29.Platt T. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 541–574. [Google Scholar]
- 30.Nakamura Y, Mizusawa S. EMBO J. 1985;4:527–532. doi: 10.1002/j.1460-2075.1985.tb03660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takata R, Mukai T, Hori K. Nucleic Acids Res. 1985;13:7289–7296. doi: 10.1093/nar/13.20.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunberg-Manago M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, et al., editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1386–1409. [Google Scholar]
- 33.Sands J F, Regnier P, Cummings H S, Grunberg-Manago M, Hershey W B. Nucleic Acids Res. 1988;16:10803–10816. doi: 10.1093/nar/16.22.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dammel C S, Noller H F. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- 35.Regnier P, Portier C. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 36.Landick R, Stewart J, Lee D N. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 37.Weilbaecher R, Hebron C, Feng G, Landick R. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 38.de Smit M H. In: RNA Structure and Function. Simons R W, Grunberg-Manago R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]
- 39.Landick R. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 40.Fresco J R. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 1–35. [Google Scholar]
- 41.Phadtare S, Inouye M. Mol Microbiol. 1999;33:1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x. [DOI] [PubMed] [Google Scholar]