Abstract
Many fish, amphibians, reptiles, birds, and some mammals use UV vision for such basic activities as foraging, mate selection, and communication. UV vision is mediated by UV pigments in the short wavelength-sensitive type 1 (SWS1) group that absorb light maximally (λmax) at ≈360 nm. Reconstructed SWS1 pigments of most vertebrate ancestors have λmax values of ≈360 nm, whereas the ancestral avian pigment has a λmax value of 393 nm. In the nonavian lineage, UV vision in many modern species is inherited directly from the vertebrate ancestor, whereas violet vision in others has evolved by different amino acid replacements at ≈10 specific sites. In the avian lineage, the origin of the violet pigment and the subsequent restoration of UV pigments in some species are caused by amino acid replacements F49V/F86S/L116V/S118A and S90C, respectively. The use of UV vision is associated strongly with UV-dependent behaviors of organisms. When UV light is not available or is unimportant to organisms, the SWS1 gene can become nonfunctional, as exemplified by coelacanth and dolphin.
Ultraviolet (UV) vision in insects has been a topic of interest for many years. By the early 1930s it was already known that the behavior of a variety of insects was strongly affected by their UV vision (1). A survey of UV vision in vertebrates started much more recently, but we now know that many fish, amphibian, reptilian, avian, and some mammalian species use UV vision (2). Many birds can identify UV-reflected nectar and berries (3). UV-reflecting plumages in birds and scales in fishes are used for recognition of others (4, 5). Similarly, throat dewlaps in some reptiles and scent marks in rodents are used for communications more effectively under UV light than under visible light (6, 7). UV vision is also used in determining the sex ratio (8) in addition to sexual selection (9, 10).
UV vision is mediated by visual pigments that absorb light maximally (λmax)at ≈360 nm. These UV pigments and violet (or blue) pigments with λmax values of 390–440 nm belong to a short wavelength-sensitive type 1 (SWS1) pigment group (11). Visual pigments consist of a transmembrane (TM) protein, opsin, and the chromophore, usually an 11-cis-retinal. The variable λmax values are generated by the interactions between the chromophore and various types of opsins, which is referred to as the spectral tuning of visual pigments (12). Experimental analyses of the mechanisms of the spectral tuning of the SWS1 pigments started only recently (13). So far, based on a limited number of species, eight amino acid sites that determine the λmax values of the SWS1 pigments have been identified (14–19).
Comparative sequence analyses of SWS1 pigments suggest that most contemporary pigments have evolved from the UV pigment in the vertebrate ancestor (16, 17). This hypothesis has not been tested and, consequently, the evolutionary processes of the SWS1 pigments are not well understood. To explore these issues, we reconstructed ancestral pigments considering 10 representative SWS1 pigments from a wide range of vertebrates. The results show that most vertebrate ancestors indeed used UV pigments, whereas the avian ancestor made the UV pigment violet-sensitive, but some of its descendants have restored UV-sensitivity. Here, we also present a new amino acid site that is involved in the spectral tuning of the SWS1 pigments.
Materials and Methods
Background Information. Many vertebrates have five paralogous groups of visual pigments: rhodopsin (RH1), RH1-like (RH2), SWS1, SWS type 2 (SWS2), and long wavelength- and middle wavelength-sensitive (LWS/MWS) pigments with the phylogenetic relationship of ((((RH1, RH2), SWS2), SWS1), LWS/ MWS) (11). Within each group, organisms sampled generally have the relationship of (bony fishes, (amphibians, ((reptiles, birds), mammals))) (11). This is consistent with the organismal tree based on molecular and paleontological data (e.g., see refs. 20 and 21).
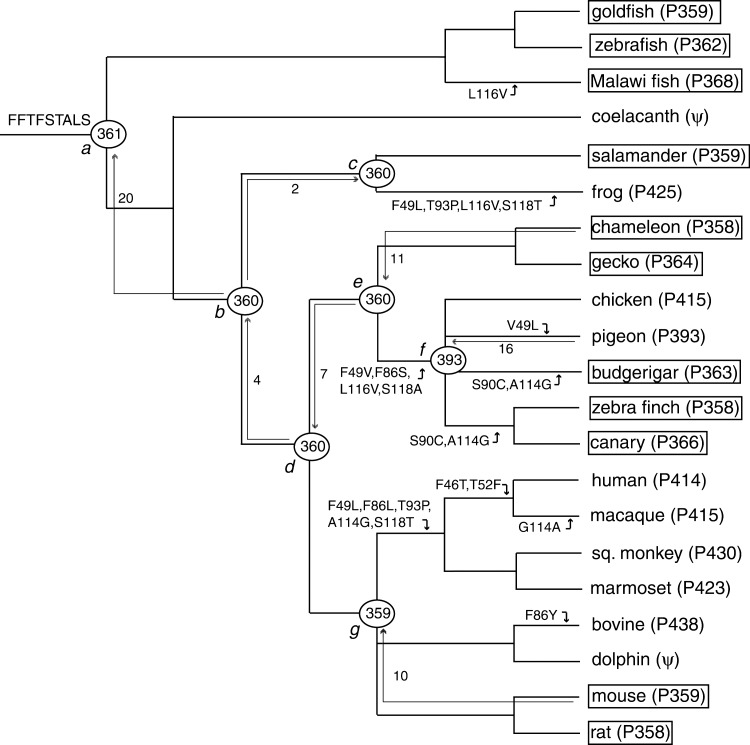
So far, both amino acid sequence and λmax are characterized for 21 SWS1 pigments. They are cloned from goldfish (Carassius auratus), zebrafish (Danio rerio), Malawi fish (Metriaclima zebra; GenBank accession no. AF191220), coelacanth (Latemeria chalamnae), frog (Xenopus laevis), salamander (Ambystoma trigrinum; AF038948), chicken (Gallus gallus), pigeon (Columba livia), budgerigar (Melopsittacus undulatus), zebra finch (Taeniopygia guttata), canary (Serinus canaria), chameleon (Anolis carolinensis), gecko (Gekko gekko; AY024356), human (Homo sapiens), macaque (Macaca fascicularis), squirrel monkey (Sciurus carolinensis), marmoset (Callithrix jacchus), bovine (Bos taurus; U92557), dolphin (Tursiops truncatus), mouse (Mus musculus), and rat (Rattus norvegicus) (for the date source, see ref. 11). Among these, the SWS1 genes in the coelacanth (22) and dolphin (23) are pseudogenes. The evolutionary tree based on molecular and paleontological data (20, 21) is shown in Fig. 1, where the phylogenetic positions of the avian and mammalian pigments are not specified.
Fig. 1.
An evolutionary tree of 21 vertebrate SWS1 pigments. FFTFSTALS refers to the amino acids at the critical sites 46, 49, 52, 86, 90, 93, 114, 116, and 118 for the ancestral pigment. The UV pigments are boxed. The numbers after P and those at the nodes a–g refer to λmax values. The numbers next to nodal arrows indicate the total numbers of amino acid changes introduced in constructing ancestral pigments.
Construction of the Chimeric and Ancestral Pigments. To infer the amino acid sequences of ancestral pigments at nodes a–g in Fig. 1, we have selected 10 representative pigments: goldfish (P359), frog (P425), salamander (P359), chicken (P415), pigeon (P393), zebra finch (P358), chameleon (P358), human (P414), bovine (P436), and mouse (P359). In the inference, we considered essentially the same phylogenetic relationships of the 10 pigments given in Fig. 1. One exception, however, is the phylogeny of the three avian pigments: (chicken (P414), (pigeon (P393), zebra finch (P358))). This tree topology is based not only on the amino acid sequences of SWS1 pigments (14) but also on DNA–DNA hybridization data (24). When this tree topology was used, the seven ancestral amino acid sequences were inferred by using a likelihood-based Bayesian method (25, 26).
Point mutations were generated by using QuikChange site-directed mutagenesis kit (Stratagene). Various chimeric pigments were also constructed by recombining different SWS1 cDNAs using restriction sites SphI, NdeI, and MfeI (Fig. 2). NdeI and MfeI sites were introduced and removed after recombination by site-directed mutagenesis. To rule out spurious mutations, all WT and mutated opsins were sequenced by using the Sequitherm Excel II Long-read kits (Epicentre Technologies, Madison, WI) with dye-labeled M13 forward and reverse primers. Sequencing reactions were run on a LI-COR 4200LD automated DNA sequencer (LI-COR, Lincoln, NE).
Fig. 2.
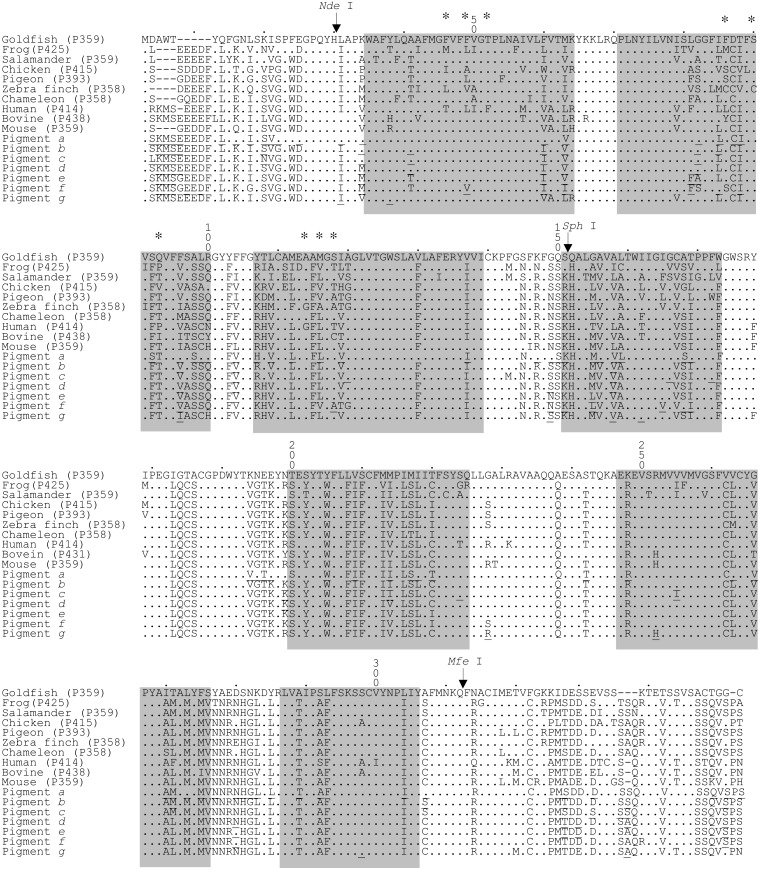
Aligned amino acid sequences of the contemporary and ancestral SWS1 pigments in vertebrates. The numbers after P refer to λmax values. Dots indicate the same amino acids as those of the goldfish (P359) pigment. Gaps in sequences required for optimal alignment are indicated by dashes. The amino acid site numbers are those of bovine rhodopsin. The positions of nine critical amino acid sites, 46, 49, 52, 86, 90, 93, 114, 116, and 118, are marked by asterisks. The ancestral amino acids that have a posterior probability of <90% are underlined. Seven shaded segments indicate TM I–VII helices (30). NdeI, SphI, and MfeI denote the positions of three restriction sites used in this study.
In Vitro Assays of Visual Pigments. The SWS1 cDNAs in an expression vector, pMT5, were expressed in COS1 cells by transient transfection (27). The visual pigments were regenerated by incubating these opsins with 11-cis-retinal (Storm Eye Institute, Medical University of South Carolina, Charleston) in the dark. The resulting visual pigments were then purified by immunoaffinity chromatography using monoclonal antibody 1D4 Sepharose 4B (Cell Culture Center, Minneapolis) in buffer consisting of 50 mM N-(2-hydroxyethyl) piperazine-N′-2-ethanesulfonic acid (pH 6.6), 140 mM NaCl, 3 mM MgCl2, 20% (wt/vol) glycerol, and 0.1% dodecyl maltoside. The absorption spectra of visual pigments were recorded at 20°C by using a Hitachi (Tokyo) U-3000 dual beam spectrophotometer. Recorded spectra were analyzed by using sigmaplot software (Jandel, San Rafael, CA).
In addition, visual pigments were not only exposed to a 366 nm UV light illuminator and a 60-W room lamp with 440-nm cutoff filter, but also denatured by sulfuric acid (H2SO4) at pH 1.8 in the dark. The λmax values of all ancestral and mutant pigments under light exposure and in sulfuric acid moved to new peaks at ≈380 and 440 nm, respectively, showing that the observed λmax values are caused by opsins covalently linked to 11-cis-retinal via a Schiff base bond (27).
Results and Discussion
Amino Acids in the N and C Termini and λmax. All amino acids that are involved in the spectral tuning of various visual pigments have been localized in TM I–VII (11). To evaluate the effects of amino acid differences in the N and C termini on the λmax shift of SWS1 pigments, we replaced the two segments of the mouse pigment with those of the goldfish, chameleon, and human pigments separately. Here the N and C termini are amino acids between sites 1 and 30 and between sites 313 and 348, respectively, where the amino acid site numbers are those of the bovine rhodopsin (GenBank accession no. M21606). These two termini are separated from the “internal segment” by NdeI and MfeI sites (Fig. 2). The three chimeric pigments have λmax values of 359–360 ± 1 nm and are identical to the λmax of the mouse pigment (13). These analyses demonstrate that different amino acids at the two termini are not involved in the spectral tuning of SWS1 pigments.
Ancestral Pigments. In inferring ancestral pigments, we consider the tree topology of (goldfish (P359), ((frog (P425), salamander (P359)), (((chicken (P415), (pigeon (P393), zebra finch (P358))), chameleon (P358)), (human (P414), bovine (P436), mouse (P359))))), which is based on molecular as well as paleontological data (see Materials and Methods). The amino acid sequences of the pigments at nodes a–g (pigments a–g) were inferred by using paml (26) with the JTT model of amino acid replacements (28) (Fig. 2). Most of these inferred amino acids have posterior probabilities ≥0.9, but those of many amino acids in the N and C termini are <0.9 (Fig. 2). We have also inferred the ancestral amino acid sequences by using the Dayhoff model of amino acid replacements (29). The two sets of inferred amino acid sequences of pigments a–g differ at 11, 2, 3, 2, 3, 5, and 1 sites, respectively. Amino acids at most of these sites have a posterior probability of <0.9, where amino acids with the two highest probabilities in one inference are often reversed in another. Thus, the uncertain amino acid inference based on a certain model of amino acid replacements and different amino acids inferred by the two models are closely interrelated.
To reconstruct these ancestral pigments, we first cloned the cDNAs of the chameleon, pigeon and mouse pigments into pMT5 separately. Before reconstructing the ancestral pigments, however, the N and C terminal segments of the pigeon and mouse pigments were replaced by those of the chameleon pigment. Although the amino acid differences at these segments should not affect the λmax values of the SWS1 pigments, this operation will eliminate any possible differential effects of the interactions between the TM and terminal segments on the λmax shift. The seven ancestral pigments were then engineered by introducing a total of 70 aa changes into the “internal segment” of the modified mouse and pigeon pigments and chameleon pigment (Fig. 1). In vitro assays show that the engineered pigments a–e and g are UV-sensitive, whereas pigment f is violet-sensitive (Fig. 1). Standard errors associated with the λmax values in Fig. 1 are all within 1 nm. It should be cautioned that these results are based on the amino acids with the highest posterior probabilities, some of which are still <0.9. Furthermore, they also depend on the phylogenetic relationship of (chicken (P414), (pigeon (P393), zebra finch (P358))). Before we draw any conclusions, therefore, these points must be checked.
To evaluate the effects of the ambiguous amino acids on the λmax, we first replaced the amino acids 152–348 of pigment a by those of the chicken and bovine pigments separately. Compared with pigment a, each pigment has 31 different amino acids at this region. These two chimeric pigments have λmax values of 362 ± 1 nm (data not shown), showing that different amino acids at sites 152–348 do not cause any λmax shift in the SWS1 pigments (see ref. 17). At amino acid sites 31–151, ancestral pigments a–g contain 20, 5, 6, 7, 7, 8, and 3 amino acids with posterior probabilities of <0.9, respectively. For pigment a, we replaced the 20 amino acids with those with the second highest probabilities in six sets: 1, 1, 9, 1, 6, and 2 changes in TM I, segment C–I, TM II, segment E–I, TM III, and segment C-II, respectively, where C and E denote cytoplasmic and extracellular loops (30). None of the λmax values of these mutant pigments differs from that of pigment a (Table 1). For pigment f, when we introduced eight mutations in four sets, most of which have no effect on the λmax shift, except that A118S (amino acid changes from A to S at site 118) in TM III increases the λmax by 5 nm (Table 1). In fact, with the exceptions of an amino acid site 116 in pigment a, and 49 and 118 in pigment f, none of the ambiguous amino acids in the seven ancestral pigments are located at functionally important sites (see below). Thus, the effect of the ambiguous amino acids on the λmax is not an important factor.
Table 1. Effects of ambiguous amino acids in TM I-III on the λmax shift.
| Pigment | Segment | Amino acid changes* | λmax, nm |
|---|---|---|---|
| a | WT | 361 ± 1 | |
| I | F4260I34 | 360 ± 1 | |
| C-I | V6663I26 | 361 ± 1 | |
| II | G6882A31/L7385I21/C3987V22/I8188T9/S7392F14/F5896V27/S8698A12/L5799S21/Q63100R17 | 362 ± 1 | |
| E-I | V47104I35 | 361 ± 1 | |
| III | H57107R31/V82109L8/L84112M13/L50116M49/V68119I28/I66137V33 | 361 ± 1 | |
| C-II | K64147R36/G49149S45 | 361 ± 1 | |
| f | WT | 393 ± 1 | |
| I | V5049L35/I7160V29/V5963I41 | 392 ± 1 | |
| II | F8181V13/I5185M20/I7588V25 | 392 ± 1 | |
| III | A71118S18 | 398 ± 1 | |
| C-II | N87149S13 | 392 ± 1 |
Subscripts show posterior probabilities of inferred amino acids
To test the effect of different tree topologies on the λmax, let us now assume that the three avian pigments are equally distantly related (Fig. 1). Then, for pigment a, of 20 amino acid changes, the orders of amino acids with the two highest posterior probabilities are reversed at three sites. For pigment f, the highest posterior probability of each inferred amino acid increases significantly. For example, the probability for A118 increases from 0.71 (Table 1) to 0.97. Because the inference of A118 is now highly reliable, a slight increase in the λmax caused by A118S may be disregarded. Importantly, the new tree topology of the three avian pigments does not introduce any new amino acids. Because amino acids with the two highest probabilities at ambiguous sites do not change the λmax (Table 1), the difference in the effects of the two avian tree topologies on the λmax is also negligible.
The order of ambiguous amino acids with the two highest posterior probabilities in one inference is often reversed in the other and, therefore, our mutagenesis results also imply that different amino acids inferred by the different models of amino acid replacements do not change the λmax significantly. All of these results show that most ancestral pigments were UV-sensitive, whereas the ancestral avian pigment was violet-sensitive (Fig. 1).
Spectral Tuning of the SWS1 Pigments. One curious aspect of the evolution of SWS1 pigments is that the avian ancestor achieved violet vision, but some of its descendants changed it back to UV vision (Fig. 1). It is of considerable interest to find some biological significance associated with these changes (see below). At the same time, these evolutionary changes provide an excellent opportunity to test our current understanding of the spectral tuning of the SWS1 pigments: amino acid sites at 46, 49, 52, 86, 90, 93, 114, and 118 mediate the spectral tuning of the SWS1 pigments (14–19).
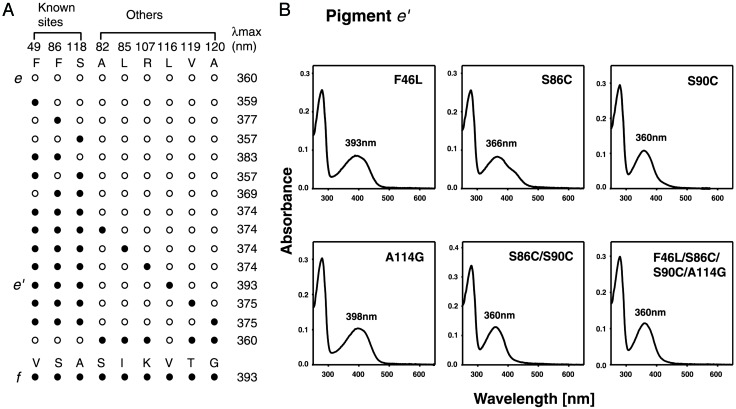
Of the eight critical sites, amino acids at 49, 86, and 118 differ between pigments e and f. When F49V/F86S/S118A (amino acid changes F49V, F86S, and S118A) are introduced into pigment e, the mutant pigment has a λmax of 374 ± 1 nm (Fig. 3A). Thus, it is clear that the eight amino acid sites are not sufficient to explain the transition from pigment e to pigment f. It turns out that pigments e and f have six different amino acids in addition to those at 49, 86, and 118 at the “internal segment.” When the amino acids at these six sites in pigment e with F49V/F86S/ S118A were replaced by those of pigment f individually, all but one quadruple mutant have λmax values of 374–375 ± 1 nm (Fig. 3A). The pigment e with F49V/F86S/L116V/S118A (referred to as pigment e′) has a λmax of 393 ± 1 nm (Fig. 3A), which is identical to that of pigment f. When the four reverse mutations are introduced into pigment f, the mutant pigment has a λmax of 360 ± 1 nm (Fig. 3A). The acquisition of violet sensitivity by the ancestral avian pigment can now be fully explained by F49V/ F86S/V116L/S118A. Thus, we have discovered a new amino acid site that is involved in the spectral tuning of the SWS1 pigments.
Fig. 3.
Reconstruction of the evolution of avian SWS1 pigments. (A) Schematic representation of the point mutations of pigment e. An open circle indicates an unchanged amino acid, whereas a filled circle indicates the change from the amino acid of the pigment e to that of the corresponding site of the pigment f. Among the nine amino acid differences between pigments e and f, previously identified functionally important sites, 46, 86, and 118 are labeled as known sites. (B) Mutations are introduced into the mutant pigment e′, and the absorption spectra measured by the in vitro assay are shown. Mutations introduced are marked in the upper right corner, and the λmax of each pigment is specified near the peak of the spectra.
It has been claimed that the SWS1 pigments of zebra finch, canary, and budgerigar achieved their UV sensitivities by S90C (14, 15). Indeed, when S90C is introduced into pigment e′, the mutant pigment becomes UV-sensitive (Fig. 3B). However, in addition to S90C, F46L/S86C/A114G probably occurred in the common ancestor of the zebra finch and canary pigments. When they are introduced singly into pigment e′, F46L, S86C, and A114G shift the λmax by 0, -27, and 5 nm, respectively (Fig. 3B). When S86C/S90C and F46L/S86C/S90C/A114G are introduced into pigment e′, the mutant pigments have λmax values of 360 ± 1 nm (Fig. 3B), identical to the effect of S90C alone (14). Thus, both S86C and S90C decrease the λmax by ≈30 nm either separately or jointly. The budgerigar, zebra finch, and canary pigments, and ancestral pigment f have amino acids Ala, Cys, Cys, and Ser at site 86, and Cys, Cys, Cys, and Ser at site 90, respectively. Thus, it is most likely that S90C occurred in the common ancestor of the three avian pigments, whereas S86C occurred in that of the zebra finch and canary pigments, strongly suggesting that S90C preceded S86C. Thus, despite its ability, S86C probably was not involved in the λmax shift.
Having new information that the λmax values of SWS1 pigments are determined mainly by nine amino acid sites 46, 49, 52, 86, 90, 93, 114, 116, and 118, we can identify a total of 43 amino acid replacements at these sites during vertebrate evolution. At these sites, with the exception of the three amino acids, all amino acids in pigments a–g have a posterior probability of >0.97. Amino acid changes that are known to cause some λmax shifts in some mammalian and avian SWS1 pigments are shown in Fig. 1. Clearly, the contemporary UV pigments in fish, salamander, chameleon, gecko, mouse, and rat are mostly free of these amino acid replacements, showing that these pigments have maintained their UV-sensitivities during vertebrate evolution. F49L/T93P/L116V/S118T and L116V seem to have increased the λmax values of the frog and Malawi fish pigments, respectively (Fig. 1). On the other hand, V49L in the pigeon pigment and G114A in the macaque pigment do not seem to change the λmax. The actual effects of these and other amino acid changes at the nine critical sites on the λmax shift remain to be evaluated.
Ecological and Physiological Requirements for UV Vision. A wide range of species has maintained UV vision during vertebrate evolution and used it for such basic behavioral traits as foraging, mate choice, and communication. This finding suggests that organisms with UV vision have a selective advantage over those without it. However, we have also seen that many other species exchanged UV vision by violet vision. Therefore, the selective advantage of organisms having UV vision may occur under special circumstances. Because UV vision works under UV light, it is reasonable for organisms to switch UV vision to violet vision when UV light is not available to them. In the extreme cases, as we have seen in the coelacanth and dolphin (Fig. 1), the SWS1 gene can become nonfunctional when UV and violet light are not available or are unimportant to them.
Given an abundance of UV light in their environment, why have so many organisms switched from UV vision to violet vision? Two major reasons can be considered for this change. First, UV light, even at ≈360 nm, can damage retinal tissues (31). Indeed, the yellow pigments in the lenses or corneas in many species, including human, are devised to obviate most UV light from reaching the retina (2). This change in the eye structure must be responsible for the transition from UV vision to violet vision. Second, by achieving violet vision, organisms can improve visual resolution and subtle contrast detection (2). On the other hand, in the avian lineage, its ancestor lost UV vision, but some of its descendants restored it (Fig. 1). This restoration of UV vision seems to have been caused by avian migration. For migratory birds, the pineal gland senses changes in day length and releases hormones that cause the restless behavior that precedes each period of migration (32). Indeed, in a “relatively closely related” American chameleon (Anolis carolinensis), some opsin genes, including the UV gene, are expressed in the pineal gland (33). In addition, UV pigments in the migratory birds seem to be essential in the orientation based on the “sun compass” (34).
Rodents distinguish themselves from other mammals by using UV vision (2). It has been observed that voles (Microtus agrestis) mark their runaways with urine and feces, which can be detected much more easily under UV light than under visible light (7). Importantly, a high proportion of the light available to animals around dawn and dusk is of short wavelength (35). Consequently, nocturnal rodents that are also active at these times of the day can use UV pigments for a range of tasks requiring vision. Under similar dim light conditions, UV pigments are also important, but in another simple but highly structured photoreceptor organ, called the third (or parietal) eye (36). The UV pigments are the major visual pigments expressed in the third eye of the American chameleon (33), strongly suggesting that UV detection through this special organ is important to lizards in addition to UV vision.
It should also be noted that many fish have UV vision, but this does not necessarily mean that they use UV vision their entire lives. On the contrary, UV vision in many of them may decline during development. For example, young brown trout (Salmo trutta) has UV vision, but the adults do not use it (37). This change in gene expression is closely related to the change in their habitats: young fish live in shallow water and feed on plankton, where UV light is essential, whereas adults live in deeper water and do not receive much UV light.
Taken together, the use of UV pigments by organisms is strongly associated with their light environments and behaviors. Compared with organisms with violet vision, those with UV vision have an advantage of recognizing certain UV-reflecting objects much more quickly, but they lack precision in viewing their surroundings and are also subjected to a higher chance of developing retinal damages caused by UV light. Whether organisms use UV vision or violet vision must depend mostly on a relative importance of these and other conflicting characteristics associated with UV vision to them. Expression of UV opsins in the pineal gland and third eye in lizards strongly suggests that the importance of UV pigments is not limited to vision. To appreciate the evolution of UV pigments in nature, it is necessary to study the roles of UV and violet pigments of many species in different photic environments. The functional differentiation of these SWS1 pigments must be related not only to ecological and behavioral changes of organisms, but to physiological changes of the eye and other photosensitive organs during evolution.
Acknowledgments
We thank M. Nei, P. Dunham, and R. Yokoyama for their valuable comments on the manuscript. This work was supported by the National Institutes of Health.
Abbreviations: SWS1, short wavelength-sensitive type 1; TM, transmembrane.
See commentary on page 8045.
References
- 1.Menzel, R. & Backhaus, W. (1991) in The Perception on Colour, ed. Gouras, P. (CRC, Boca Raton, FL), pp. 262-293.
- 2.Jacobs, G. H. (1992) Am. Zool. 342 544-554. [Google Scholar]
- 3.Burkhardt, D. (1982) Naturwissenschaften 69 153-157. [DOI] [PubMed] [Google Scholar]
- 4.Burkhardt, D. (1989) J. Comp. Physiol. A 164 787-796. [Google Scholar]
- 5.Harosi, F. I. (1985) in The Visual System, eds. Fein, A. & Levine, J. S. (Liss, New York), pp. 41-55.
- 6.Fleishman, L. J., Loew, E. R. & Leal, M. (1993) Nature 365 397. [Google Scholar]
- 7.Viitala, J., Korpimaki, E., Palokangas, P. & Koivula, M. (1995) Nature 373 425-427. [Google Scholar]
- 8.Sheldon, B. C., Anderson, S., Griffith, S. C., Ornborg, J. & Sendecka, J. (1999) Nature 402 874-877. [Google Scholar]
- 9.Bennett, A. T. D., Cuthill, I. C. & Partridge, J. C. (1996) Nature 380 433-435. [Google Scholar]
- 10.Hunt, S., Cuthill, I. C., Swaddle, J. P. & Bennett, A. T. D. (1997) Anim. Behav. 54 1383-1392. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama, S. (2000) Prog. Ret. Eye Res. 19 385-419. [DOI] [PubMed] [Google Scholar]
- 12.Kochendoerfer, G. G., Lin, S. W., Sakmar, T. M. & Mathies, R. A. (1999) Trends Biochem. Sci. 24 300-305. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama, S., Radlwimmer, F. B. & Kawamura, S. (1998) FEBS Lett. 423 155-158. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama, S., Radlwimmer, F. B. & Blow, N. (2000) Proc. Natl. Acad. Sci. USA 97 7366-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkie, S. E., Robinson, P. R., Cronin, T. W., Poopalasundaram, S., Bowmaker, J. K. & Hunt, D. M. (2000) Biochemistry 39 7895-7901. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama, S. & Shi, Y. (2000) FEBS Lett. 486 167-172. [DOI] [PubMed] [Google Scholar]
- 17.Shi, Y., Radlwimmer, F. B. & Yokoyama, S. (2001) Proc. Natl. Acad. Sci. USA 98 11731-11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasick, J. I., Applebury, M. L. & Oprian, D. D. (2002) Biochemistry 41 6860-6865. [DOI] [PubMed] [Google Scholar]
- 19.Cowing, J. A., Poopalasundaram, S., Wilkie, S. E., Robinson, P. R., Bowmaker, J. K. & Hunt, D. M. (2002) Biochem. J. 367 129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaughlin, P. J. & Dayhoff, M. O. (1972) in Atlas of Protein Sequence and Structure, ed. Dayhoff, M. O. (National Biomedical Research Foundation, Washington, DC), Vol. 5, pp. 47-66. [Google Scholar]
- 21.Carroll, R. L. (1988) Vertebrate Paleontology and Evolution (Freeman, New York).
- 22.Yokoyama, S., Zhang, H., Radlwimmer, F. B. & Blow, N. S. (1999) Proc. Natl. Acad. Sci. USA 96 6279-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasick, J. L., Cronin, T. W., Hunt, D. M. & Robinson, P. (1998) Biochemistry 38 432-438. [Google Scholar]
- 24.Sibley, C. G. & Ahlquist, J. E. (1990) Phylogeny and Classification of Birds (Yale Univ. Press, New Haven, CT).
- 25.Yang, Z., Kumar, S. & Nei, M. (1995) Genetics 141 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, Z. (1997) Comput. Appl. Biosci. 13 555-556. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama, S. (2000) Methods Enzymol. 315 312-325. [DOI] [PubMed] [Google Scholar]
- 28.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8 275-282. [DOI] [PubMed] [Google Scholar]
- 29.Dayhoff, M. O., Schwartz, R. M. & Orcutt, B. C. (1978) in Atlas of Protein Sequence and Structure, ed. Dayhoff, M. O. (National Biomedical Research Foundation, Washington, DC), Vol. 5, pp. 345-352. [Google Scholar]
- 30.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289 739-745. [DOI] [PubMed] [Google Scholar]
- 31.van Norren, D. & Schellenkens, P. (1990) Vision Res. 30 1517-1520. [DOI] [PubMed] [Google Scholar]
- 32.Alcock, J. (1997) Animal Behavior (Sinauer, Sunderland, MA).
- 33.Kawamura, S. & Yokoyama, S. (1997) Vision Res. 37 1867-1871. [DOI] [PubMed] [Google Scholar]
- 34.Bennett, A. T. D. & Cuthill, I. C. (1994) Vision Res. 34 1471-1478. [DOI] [PubMed] [Google Scholar]
- 35.Lythgoe, J. N. (1979) The Ecology of Vision (Clarendon, Oxford).
- 36.Solessio, E. & Engbretson, G. A. (1993) Nature 364 442-445. [DOI] [PubMed] [Google Scholar]
- 37.Bowmaker, J. K. & Kunz, Y. W. (1987) Vision Res. 27 2102-2108. [DOI] [PubMed] [Google Scholar]