Abstract
The iscU gene in bacteria is located in a gene cluster encoding proteins implicated in iron–sulfur cluster assembly and an hsc70-type (heat shock cognate) molecular chaperone system, iscSUA-hscBA. To investigate possible interactions between these systems, we have overproduced and purified the IscU protein from Escherichia coli and have studied its interactions with the hscA and hscB gene products Hsc66 and Hsc20. IscU and its iron–sulfur complex (IscU–Fe/S) stimulated the basal steady-state ATPase activity of Hsc66 weakly in the absence of Hsc20 but, in the presence of Hsc20, increased the ATPase activity up to 480-fold. Hsc20 also decreased the apparent Km for IscU stimulation of Hsc66 ATPase activity, and surface plasmon resonance studies revealed that Hsc20 enhances binding of IscU to Hsc66. Surface plasmon resonance and isothermal titration calorimetry further showed that IscU and Hsc20 form a complex, and Hsc20 may thereby aid in the targeting of IscU to Hsc66. These results establish a direct and specific role for the Hsc66/Hsc20 chaperone system in functioning with isc gene components for the assembly of iron–sulfur cluster proteins.
Proteins containing iron–sulfur clusters possess important redox, catalytic, and regulatory functions, but the mechanism by which their clusters are formed or repaired is not known (see refs. 1–3). Much of the limited information on this subject has been provided by studies on the nif operon of nitrogen-fixing bacteria, which encodes proteins necessary for the assembly of the Fe/S centers present in the nitrogenase system. Recently, homologs of nif genes have been found in both nitrogen-fixing and non-nitrogen-fixing bacteria, and these coding regions have been designated isc genes for their putative roles in iron sulfur cluster formation (4). These genes include iscS, homologous to nifS, which encodes a cysteine desulfurase shown to provide sulfide for Fe/S cluster formation (5, 6); iscU, homologous to nifU, which encodes a protein implicated in Fe/S binding (7); and iscA, homologous to nif ORF 6, whose function is not known (8).
In bacteria, the isc genes are found in a highly conserved gene cluster, iscSUA-hscBA-fdx, which also encodes an hsp70-type molecular chaperone designated Hsc66 (hscA; refs. 9–11), a J type cochaperone designated Hsc20 (hscB; refs. 10 and 11), and a [2Fe-2S] ferredoxin (fdx; ref. 12). The exact cellular roles of the chaperones and ferredoxin are not known, but the conserved association of their genes with the isc genes suggests a possible function in the assembly and/or repair of iron–sulfur cluster proteins. Recent studies in which overexpression of the iscSUA-hscBA-fdx gene region in Escherichia coli yields increased production of recombinant iron sulfur proteins (13, 14) support such a function. It also seems likely that similar proteins are involved in iron–sulfur cluster assembly in eukaryotes. Earlier studies showed that bacteria and yeast are able to assemble the iron–sulfur center of a human [2Fe-2S] protein, suggesting a conserved mechanism of cluster assembly (15, 16), and recent studies with Saccharomyces cerevisiae have implicated homologs of IscS (17–19), IscU (20, 21), IscA (22), Hsc20 (17), Hsc66 (17), and Fdx (23) in iron–sulfur protein biogenesis.
To understand better the molecular mechanism of Fe/S cluster assembly, we have begun studies to investigate interactions between the isc proteins and the Hsc66/Hsc20 chaperone system. In the studies presented herein, we describe the initial characterization of the iscU gene product from E. coli and show that IscU functions together with Hsc20 to regulate the activity of Hsc66. The results provide direct evidence for a link between a class of proteins that mobilize iron and sulfur for cluster assembly/repair and a molecular chaperone system.
Experimental Procedures
Materials.
E. coli W3110 was from American Type Culture Collection (ATCC no. 27325); DH5αF′IQ cells were from GIBCO/BRL; and BL-21 (DE3)pLysS cells were from Novagen. Enzymes for DNA manipulation were obtained from Roche Molecular Biochemicals, New England Biolabs, or United States Biochemical. Synthetic oligonucleotides were obtained from Genosys (The Woodlands, TX). Bacterial growth medium components were from Difco, and other reagents were from Sigma.
Expression and Purification Methods.
The vector used for overexpressing IscU, pTrcIscU, was constructed by cloning the iscU gene into pTrc99a (Amersham Pharmacia). The coding region of iscU was amplified from genomic DNA isolated from E. coli K-12 strain by PCR with Taq DNA polymerase and primers designed to introduce a NcoI restriction site at Met-1 and a HindIII site following the termination codon. The NcoI/HindIII-digested PCR product was ligated into pTrc99a, and sequencing showed the iscU gene to be identical to that reported by the E. coli genome project (24).
BL-21 cells transformed with pTrcIscU were grown in Terrific Broth at 37°C, induced with 0.25 mM isopropyl β-d-thiogalactoside (IPTG) at A600 ≈ 1, and grown for ≈16 h to allow expression. Cells were harvested by centrifugation, frozen, thawed, and lysed by French press in TED8 buffer (50 mM Tris⋅HCl, pH 8.0/0.5 mM EDTA/1 mM DTT) containing 0.1 mM phenylmethylsulfonyl fluoride. The soluble supernatant fluid after centrifugation at 31,000 × g was used for purification, and all subsequent steps were carried out in TED8 buffer at 4°C. The IscU protein was monitored by SDS/PAGE and purified by ion-exchange chromatography with DEAE-cellulose (Whatman DE-52), DEAE-Biogel (Bio-Rad), and elution gradients of 0–0.15 M NaCl, followed by reverse-phase chromatography with phenyl-Sepharose (Amersham Pharmacia) and an elution gradient of 30–15% (vol/vol) saturated ammonium sulfate. Purified fractions of IscU protein were combined, dialyzed against 50 mM Hepes, pH 7.3/0.5 mM EDTA/1 mM DTT, concentrated by ultrafiltration, and stored at −70°C.
Methods for the preparation of Hsc66 and Hsc20 (11) and of DnaK and DnaJ (25) have been described.
Formation of the Iron–Sulfur Complex of IscU.
After dialysis to remove EDTA, IscU was combined with 2.5 equivalents each of ferric citrate and sodium sulfide under anaerobic conditions (Plas-Labs Controlled Atmosphere Chamber, Lansing, MI) in 50 mM Tris⋅HCl (pH 7.3) containing 5 mM DTT. The mixture was applied to a DEAE-cellulose column (DE-52, Whatman) to remove excess iron–sulfide precipitate, and the red-brown Fe/S complex of IscU was eluted from the column by washing with 50 mM Tris⋅HCl, pH 7.3/5 mM DTT/500 mM NaCl. Aliquots of the IscU–Fe/S complex were removed from the anaerobic chamber in sealed ampoules. ATPase and surface plasmon resonance (SPR) assays were carried out under semianaerobic conditions (see below), and the IscU–Fe/S complex was stable for >30 min under conditions of the ATPase and >5 min under conditions of the SPR assay as assessed by a lack of change in the visible-UV absorption spectrum. Under anaerobic conditions, the complex also remained stable in the presence of a 100-fold molar excess of EDTA (5 mM) for >30 min, indicating that iron is lost only very slowly from the cluster. Under conditions required for isothermal titration calorimetry (see below), the IscU–Fe/s complex decomposed too rapidly to obtain reliable measurements. A more extensive characterization of the complex is in preparation (K.G.H. and L.E.V., unpublished observations).
ATPase Assays.
ATPase activities were determined at 23°C as before (11) by measuring phosphate released with a coupled enzyme assay (26) with the EnzCheck phosphate assay kit (Molecular Probes). The basal ATPase activity of Hsc66 was 0.08 mol ATP hydrolyzed per mol Hsc66 per min; purified IscU and Hsc20 alone did not exhibit detectable ATPase activity (<0.0001 min−1). Curves shown in figures represent a least-squares fit of the data to a hyperbolic saturation function.
SPR Analysis.
SPR studies were carried out at 25°C with a Biacore X instrument (Piscataway, NJ). Hsc66 in the presence of 1 mM ATP or Hsc20 was randomly crosslinked to the surface of the sensor chip by amine coupling as recommended by the manufacturer. Experiments were conducted in HKM buffer (50 mM Hepes, pH 7.3/150 mM potassium chloride/10 mM magnesium chloride) containing 5 mM DTT as the running buffer at a flow rate of 20 μl/min. Hsc20 or IscU that had been dialyzed against running buffer was used for injections; maximal signals were measured 1–2 min after injections. Binding of IscU seemed to be specific, because no interaction was observed by sensor chips prepared without Hsc66 or Hsc20. Experiments were repeated to verify that changes in the sensor chip did not occur during the course of the measurements. Curves shown in figures represent least-squares fits of the data to a hyperbolic saturation function, and error bars are shown where these fell outside of the symbols used. The amount of immobilized Hsc66 or Hsc20 that was capable of binding IscU was estimated by comparison of the response signals obtained after immobilization of Hsc66 or Hsc20 on the sensor chip, and those resulting from IscU binding were corrected for the relative masses of the proteins.
Isothermal Titration Calorimetry.
A Microcal (Amherst, MA) Omega titration calorimeter was used to investigate the binding of IscU and Hsc20 by using procedures described previously (27).
Analytical Methods.
SDS/PAGE was carried out according to the method of Laemmli (28), and N-terminal amino acid sequencing used Edman degradation (determined by A. Henshen-Edman, Univ. of California, Irvine). Antiserum to purified IscU was prepared from a single rabbit by Bethyl Laboratories (Montgomery, TX), and serum was used without further purification. Western immunoblotting was carried out (29) by using enhanced chemiluminescence detection (Amersham Pharmacia). A 1:750 dilution of rabbit anti-IscU serum was followed by a 1:10,000 dilution of an anti-rabbit horseradish peroxidase conjugate. Absorption spectra were recorded with a Cary 1 spectrophotometer (Varian). The UV extinction coefficient of IscU, ɛ280 = 11,200 (M⋅cm)−1, was calculated based on average absorptivities of one tryptophan and four tyrosine residues (30–32); concentrations of IscU–Fe/S were determined by the Coomassie blue G250 method (Bio-Rad; ref. 33) with apo-IscU as a standard.
Results
Overexpression and Purification of IscU.
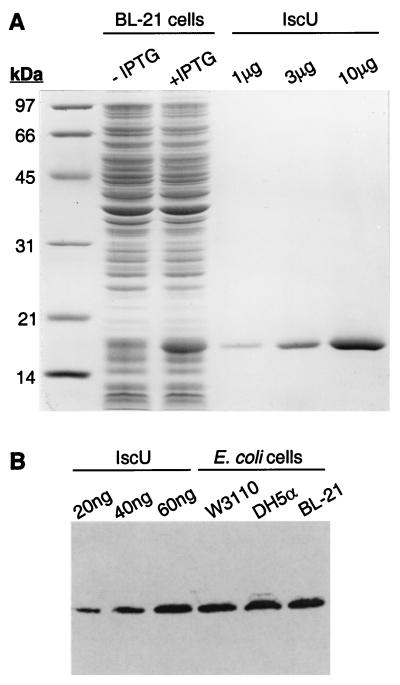
The iscU gene was subcloned into the IPTG-controlled expression plasmid pTrc99 to yield pTrcIscU, and Fig. 1A shows an SDS/PAGE analysis of E. coli BL-21 cells harboring the plasmid. IscU is predicted to have a mass of 13.8 kDa, and a prominent band of ≈17 kDa is present in IPTG-induced cells. Recombinant IscU was isolated from overproducing cells by anion exchange and hydrophobic chromatography with an overall yield of ≈50 mg/liter of culture, and a single major band was observed on SDS/PAGE analysis of the purified protein (Fig. 1A). N-Terminal sequence analysis revealed a single sequence, Ala-Ser-Glu-Lys-Val-, indicating removal of Met-1 as expected for proteins in which alanine is the penultimate residue (34, 35).
Figure 1.
Analysis of IscU expression and purification. (A) SDS/PAGE of overproduction and purification. Lane 1, molecular weight standards; lane 2, BL-21 cells harboring pTrcIscU (25 μg of protein); lane 3, BL-21 cells harboring pTrcIscU plasmid grown for ≈16 h after IPTG induction (25 μg of protein); lanes 4–6, purified IscU. (B) Immunoblot analysis of constitutive IscU expression in W3110, DH5α, and BL-21 cells grown to stationary phase in Terrific Broth. Lanes 1–3, purified IscU; lanes 4–6, E. coli whole cells (15 μg of protein each).
Antiserum to purified IscU was raised in rabbits and used to estimate the cellular level of the IscU protein. As shown in the Western immunoblot in Fig. 1B, IscU is readily detected in cultures of the prototrophic K-12 strain, W3110, the laboratory K-12 derived strain, DH5α, and the B strain, BL-21. Comparing the band intensities observed for these cells with purified IscU, we estimate that for cultures grown overnight to stationary phase at 37°C in rich medium, IscU comprises ≈0.4% of the total cell protein, corresponding to a cellular concentration of ≈40 μM IscU; similar results were obtained with cells grown to log phase (data not shown). The cellular level of IscU is slightly greater than that estimated for Hsc66 and Hsc20 (20 and 10 μM, respectively; ref. 11).
Stimulation of Hsc66 ATPase Activity.
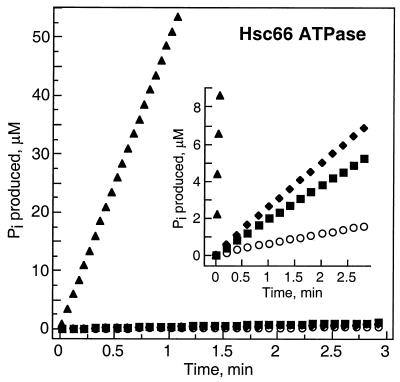
Initial studies on possible interactions between IscU and the Hsc66/Hsc20 chaperone system were carried out by determining the effect of IscU on the ATPase activity of Hsc66. Fig. 2 shows that addition of 50 μM IscU increases the steady-state ATPase activity of Hsc66 ≈5-fold over basal activity. The observed increase can be compared with the ≈4-fold increase that is observed with a similar concentration of the cochaperone Hsc20. Addition of both Hsc20 and IscU to Hsc66 resulted in a dramatic stimulation, increasing the ATPase activity of Hsc66 ≈100-fold over the level observed in the presence of Hsc20 or IscU alone and >400-fold over the basal intrinsic activity.
Figure 2.
Effects of IscU and Hsc20 on the ATPase activity of Hsc66. Time course of ATP hydrolysis at 23°C by 0.5 μM Hsc66 alone (○) or in the presence of 50 μM Hsc20 (■), 50 μM IscU (♦), or 50 μM IscU + 50 μM Hsc20 (▴). (Inset) An experiment with 5 μM Hsc66 and an expanded ordinate scale to show more clearly the activities in the presence of individual components.
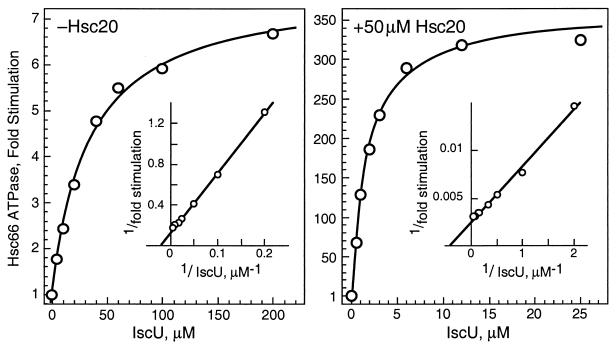
The synergistic effect of Hsc20 on the IscU stimulation of Hsc66 ATPase activity was investigated further by comparing the concentration dependence of IscU stimulation in the absence and presence of Hsc20 (Fig. 3). In the absence of Hsc20, IscU elicits a maximal stimulation of ≈8-fold, and half-maximal stimulation is observed at ≈34 μM IscU, a value in the range of the estimated cellular concentration of IscU. These values are similar to those observed for Hsc20 effects on activity (maximal stimulation ≈5-fold, with half-maximal activation occurring near the cellular concentration ≈10 μM; refs. 11 and 25). In the presence of a near-saturating level of Hsc20, a maximal stimulation by IscU of ≈360-fold over basal activity is observed. In addition, the concentration of IscU required for half-maximal stimulation decreases to ≈2 μM IscU. Thus, Hsc20 both enhances the stimulatory activity of IscU on Hsc66 and lowers the concentration of IscU required to activate the ATPase activity of Hsc66. In similar experiments, IscU had no significant effect on the concentration dependence of Hsc20 for stimulation of Hsc66 ATPase activity (data not shown). Thus, although Hsc20 enhances the interaction of IscU with Hsc66, IscU does not affect the affinity of Hsc20 for Hsc66.
Figure 3.
Effect of IscU on the ATPase activity of Hsc66 in the absence and presence of Hsc20. Results are reported as the increase in basal ATPase rates at 23°C. (Insets) Double reciprocal plots of the data. The curves shown represent best fits to the data for maximal stimulation of 7.7-fold and half-maximal stimulation at 33.7 μM IscU in the absence of Hsc20 and for maximal stimulation of 362-fold and half-maximal stimulation at 1.8 μM IscU in the presence of Hsc20.
Effect of the IscU Fe/S Cluster on Activity.
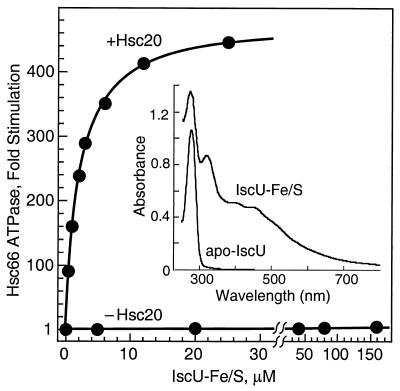
As isolated, recombinant IscU does not contain bound metal ions, but it is able to form an Fe/S cluster when reconstituted with Fe3+ and sulfide under anaerobic conditions. Dean and coworkers (7) have shown recently that the N-terminal domain of NifU, which is homologous to IscU, also forms a transient [2Fe-2S] cluster. To determine whether Fe/S cluster formation affects the ability of IscU to interact with Hsc66, we prepared the IscU–Fe/S complex and measured its effect on the ATPase activity of Hsc66 (Fig. 4). The visible/near-UV absorption spectrum of the Fe/S complex of IscU is similar to that described for the NifU-1 domain of NifU (7), consistent with a similar [2Fe-2S] cluster.† In the absence of Hsc20, the IscU–Fe/S complex elicits a low level of stimulation (≈7-fold maximal increase) similar to that produced by apo-IscU. The concentration required for half maximal stimulation (≈160 μM), however, is much greater than that for apo-IscU, suggesting that IscU–Fe/S binds to Hsc66 with lower affinity. In the presence of Hsc20, on the other hand, IscU–Fe/S is very active in stimulating Hsc66. Half-maximal stimulation occurs at ≈2 μM IscU–Fe/S, a value similar to that observed for apo-IscU, and maximal stimulation (≈480-fold) is similar to or slightly greater than that for apo-IscU.
Figure 4.
Effect of IscU–Fe/S on the ATPase activity of Hsc66. Results are reported as the increase in basal ATPase rates at 23°C. The curve shown for results in the presence of 50 μM Hsc20 represents a best fit to the data with maximal stimulation of 481-fold and half-maximal stimulation at 2.1 μM IscU–FeS; in the absence of Hsc20, maximal stimulation of 7.4-fold with half-maximal stimulation at 162 μM IscU. (Inset) The visible and near UV region absorption spectra for the metal-free form of IscU (apo-IscU, 81 μM) and IscU reconstituted with ferric iron and sulfide (IscU–Fe/S, 80 μM; see Experimental Procedures).
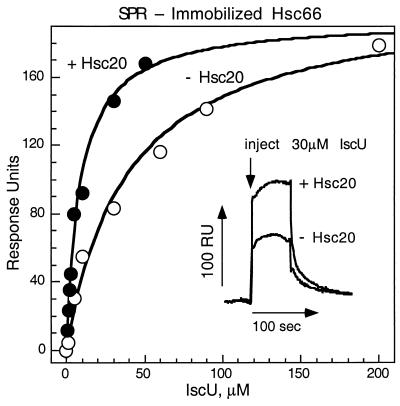
Binding of IscU to Hsc66.
SPR analysis was used to investigate the interaction of IscU with Hsc66 further. Fig. 5 shows the results of titrations in which Hsc66 was randomly crosslinked to the sensor chip and then exposed to different concentrations of IscU in the presence or absence of Hsc20. In the absence of Hsc20, IscU binds to Hsc66 with an apparent half-maximal affinity that is close to the concentration of IscU required for half-maximal stimulation of Hsc66 activity. In the presence of Hsc20, IscU exhibits enhanced affinity with half-maximal binding occurring at a lower concentration. The maximal signal observed as a result of IscU binding was similar in the absence and presence of Hsc20, consistent with binding to the same fraction (≈27%) of immobilized Hsc66.
Figure 5.
SPR analysis of the binding of IscU to Hsc66. IscU was injected into solutions passing over a sensor chip containing immobilized Hsc66 in the presence of 1 mM ATP (3,550 relative response units, RU) at 25°C, and maximal resonance signals observed are plotted as a function of the concentration of IscU. The equilibration solution contained HKM buffer, 1 mM ATP, and 5 mM DTT without (○) or with (●) 40 μM Hsc20 before injection of IscU. Experiments were carried out in duplicate, and error bars shown correspond to individual measurements when these fell outside the symbol used. The curves shown represent best fits to the data with RUmax = 206 and Kd = 8.9 μM in the presence of Hsc20 and RUmax = 193 and Kd = 39 μM in the absence of Hsc20. (Inset) Sensorgrams recorded after injections of 30 μM IscU in the absence or presence of 40 μM Hsc20.
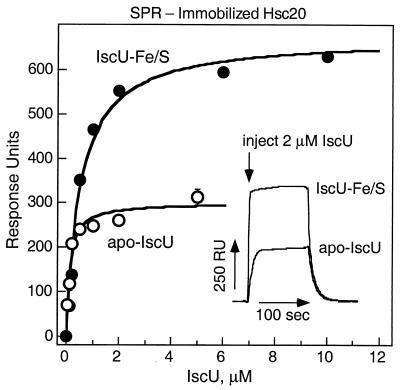
Binding of IscU to Hsc20.
Because the ATPase and SPR studies revealed that Hsc20 enhances the affinity of IscU for Hsc66, we investigated the possibility of direct interactions between Hsc20 and IscU with SPR. Fig. 6 shows the results of titrations in which Hsc20 was randomly crosslinked to the sensor chip and exposed to different concentrations of IscU or IscU–Fe/S. Both forms bind to immobilized Hsc20 with high affinity. The maximal signal observed for binding of IscU–Fe/S, however, was approximately twice that observed for IscU. For IscU, the maximal relative response signal observed corresponds to binding to ≈38% of the immobilized Hsc20, if binding is assumed to yield a 1:1 complex. For IscU–Fe/S, the signal strength suggests binding either to ≈86% of the immobilized Hsc20 as a 1:1 complex or to ≈43% as a 2:1 IscU–Fe/S:Hsc20 complex. Assuming that IscU and IscU–Fe/S bind to the same site on Hsc20, these results suggest that IscU–Fe/S binds to Hsc20 as an oligomer twice the size of the form of IscU that binds to Hsc20.‡
Figure 6.
SPR analysis of the binding of IscU to Hsc20. IscU (○) or IscU–Fe/S (●) was injected into solutions passing over sensor chips containing immobilized Hsc20 (1,200 RU). Curves shown represent best fits to the data with RUmax = 300 and Kd = 0.10 μM for IscU and RUmax = 673 and Kd = 0.54 μM for IscU–Fe/S. Sensorgrams recorded for individual injections from one titration are shown (Inset). A titration of apo-IscU was carried out initially, followed by two sequential titrations of IscU–Fe/S and a duplicate titration of apo-IscU. No decay of the sensor chip was detected, and error bars shown correspond to individual measurements when these fell outside the symbol used.
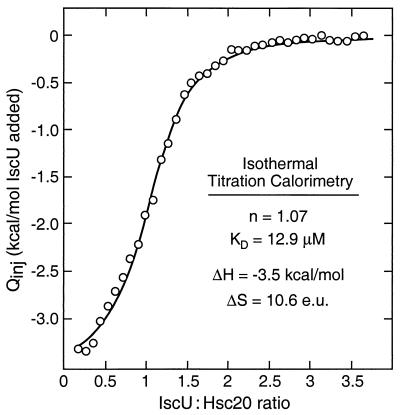
Because of possible complications in the SPR binding studies arising from surface and/or immobilization effects, we used isothermal titration calorimetry to quantitate the interaction between IscU and Hsc20 in solution more accurately. Fig. 7 shows the results of an experiment in which successive additions of IscU were made to a solution containing Hsc20. The data are plotted as the integrated heats of binding vs. the molar ratio of IscU to Hsc20, and the best fit curve shown corresponds to binding of 1.07 mol IscU/mol Hsc20 with Kd = 12.9 μM. It was not possible to carry out isothermal titration calorimetry studies on the interaction of the IscU–Fe/S complex with Hsc20 because of the limited stability of the IscU–Fe/S complex under the partially aerobic conditions of the experiment. The higher affinity observed for IscU in the SPR studies in Fig. 6 may have resulted from nonspecific effects at the sensor surface and/or cooperative effects of multimeric binding sites as a result of immobilization.§ Both methods, however, support the formation of an Hsc20–IscU complex, and this complex may account for the enhanced affinity of IscU for Hsc66 in the presence of Hsc20 that was observed in the ATPase studies (Figs. 2–4).
Figure 7.
Calorimetric analysis of the binding of IscU to Hsc20. A series of 42 equivalent 7-μl aliquots of 3.2 mM IscU were injected into a cell containing 1.348 ml of 0.19 mM Hsc20 at 25°C in HKM buffer containing 5 mM DTT. Integrated heats caused by binding, Qinj (corrected for the heats of dilution and divided by the moles of IscU injected), are plotted vs. the molar ratio of IscU and Hsc20 in the titration cell. The solid line represents a best-fit curve assuming 1.07 binding sites. Kd = 12.9 μM; ΔH = −3.53 kcal/mol; ΔS = 10.6 entropy units (e.u.).
Interactions of IscU with DnaK and DnaJ.
Possible interactions of IscU with the DnaK/DnaJ chaperone system were also investigated. IscU had a much weaker effect on the basal ATPase activity of DnaK compared with its effects on the Hsc66 (1.35-fold vs. 5-fold stimulation with 50 μM IscU) and showed no enhancement of the DnaJ-stimulated DnaK ATPase activity. Furthermore, unlike Hsc20, DnaJ did not enhance the IscU stimulation of Hsc66 ATPase activity (data not shown). These results suggest that IscU interacts specifically with the Hsc66/Hsc20 chaperone system.
Discussion
Previous studies established that Hsc66 and Hsc20 comprise a separate hsp70-type chaperone system with substrate specificity and ATPase kinetics distinct from those of the DnaK/DnaJ/GrpE system (11, 25, 26). Structural differences between Hsc20 and DnaJ as well as differences in their stimulatory effects on Hsc66 and DnaK further suggested that the two systems might be subject to different regulatory control mechanisms. Although the exact cellular role of the Hsc66/Hsc20 system was not known, the close association in bacteria of the hscA and hscB genes with genes encoding proteins implicated in iron–sulfur cluster assembly suggested a role for Hsc66 and Hsc20 in Fe/S protein biogenesis.
The results presented herein provide direct evidence that Hsc66 and Hsc20 are involved in Fe/S cluster assembly. IscU seems to serve a role similar to that proposed for NifU as a template site for assembly of a [2Fe-2S] cluster (7), and both Hsc66 and Hsc20 interact with the IscU protein. Binding of IscU to Hsc66 was shown by stimulation of the intrinsic ATPase activity of Hsc66 and by SPR measurements, and both effects were observed at physiologically relevant concentrations of IscU. The mechanism of stimulation of the ATPase activity of Hsc66 by IscU is not known. It could arise from actions of IscU as an additional regulatory protein in addition to Hsc20. Another possibility is that the increase in Hsc66 ATPase activity could result from interaction of IscU with the peptide-binding domain of Hsc66, i.e., with IscU serving as a “substrate” for Hsc66. Hsc66 may assist in Fe/S cluster formation by maintaining IscU in a conformation suitable for cluster assembly; alternatively, Hsc66 may facilitate the transfer of the cluster from IscU–Fe/S to an acceptor apoprotein.
In addition to its interactions with Hsc66, IscU also directly interacts with Hsc20, and in the presence of Hsc20, the affinity of IscU for Hsc66 is increased >18-fold. The mechanism whereby Hsc20 enhances the affinity of IscU for Hsc66 is not known. The increased affinity may involve direct stabilization of the IscU–Hsc66 complex. Alternatively, interactions of Hsc20 with Hsc66 may cause conformational changes in Hsc66 that lead to more productive and higher affinity interactions with IscU. Russell et al. (36) have shown the “targeting” of peptide substrates to DnaK by DnaJ is the result of the ATPase stimulatory activity of DnaJ, which leads to formation of the high-affinity ADP conformational state and thereby contributes to DnaK substrate specificity. A similar type of kinetic mechanism may be important in Hsc20–IscU–Hsc66 interactions and may reflect the role of IscU as a substrate for Hsc66. In contrast to DnaJ, however, Hsc20 does not display intrinsic chaperone activity and lacks the C-terminal domains necessary for Hsp40 interactions with unfolded polypeptides (37–39). Instead, Hsc20 has a unique C-terminal domain that is predicted to exist as a coiled coil (40), and this region may be responsible for the specific interactions with IscU.¶
Hsc20 also has a synergistic effect on the IscU stimulation of the ATPase activity of Hsc66, increasing the rate >50-fold over that found at saturating levels of IscU alone. Binding of both IscU and Hsc20 would thus enhance the rate of conversion of Hsc66 from a state exhibiting rapid substrate exchange and low substrate affinity (the ATP-bound “tense” state) to one with greater substrate affinity and slower exchange rates (the ADP-bound “relaxed” state; ref. 27). In this manner, Hsc20 may serve to control the association of IscU with Hsc66.
It will be interesting to investigate the interaction of Hsc66, Hsc20, and IscU with other components of the iron–sulfur cluster assembly system. Several lines of evidence have linked the functions of yeast homologs of IscS, IscA, and Fdx to those of IscU, Hsc20, and Hsc66 as well as Fe/S biogenesis (17–23), and elucidation of the complete molecular mechanism of cluster assembly will require detailed studies with reconstituted systems containing each of the components.
Acknowledgments
We thank Dennis Ta for expert technical assistance. This work was supported by National Institutes of Health Grant GM54264 and Training Grant GM07311.
Abbreviations
- SPR
surface plasmon resonance
- IPTG
isopropyl β-d-thiogalactoside
- RU
response units
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130201997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130201997
The extinction coefficient that we observe for IscU–Fe/S (ɛ410 ≈6.2 mM−1⋅cm−1) is ≈2-fold greater than that reported for the NifU-1 fragment of NifU (7) and may reflect the limited reconstitution or instability of the NifU-1 cluster. The IscU–Fe/S complex seems to be more stable than NifU-Fe/S complexes under the conditions tested.
IscU seems to exist as a dimer in solution (unpublished observations) and may bind to immobilized Hsc20 in the dimeric state. Thus, the observed binding of IscU would correspond to ≈19% of the immobilized Hsc20, and binding of IscU–Fe/S would thus correspond to binding of a tetrameric form to ≈22% of the immobilized Hsc20.
The oligomeric state of IscU and IscU–Fe/S may lead to enhanced binding to immobilized Hsc20 via multiple binding sites. We have observed that the apparent affinity of IscU for Hsc20 depends on the amount of immobilized Hsc20, suggestive of multimeric attachment (data not shown).
SPR experiments have shown that IscU is able to bind to a mutant of Hsc20 (His32Cys) specifically crosslinked to the sensor chip via the J domain, consistent with a role for the C-terminal coil domain in binding interactions (unpublished work).
References
- 1.Beinert H, Holm R H, Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Bian S, Cowan J A. Coord Chem Rev. 1999;190:1049–1066. [Google Scholar]
- 3.Beinert H. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 4.Zheng L, Cash V L, Flint D H, Dean D R. J Biol Chem. 1998;21:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 7.Yumaniyama P, Agar J N, Cash V L, Johnson M K, Dean D R. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson M R, Cash V L, Weiss M C, Laird N F, Newton W E, Dean D R. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 9.Seaton B S, Vickery L E. Proc Natl Acad Sci USA. 1994;91:2066–2070. doi: 10.1073/pnas.91.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawula T H, Lelivelt M J. J Bacteriol. 1994;176:610–619. doi: 10.1128/jb.176.3.610-619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickery L E, Silberg J J, Ta D T. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ta D N, Vickery L E. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- 13.Nakamura M, Saeki K, Takahashi Y. J Biochem. 1999;126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi Y, Nakamura M. J Biochem. 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 15.Coghlan V M, Vickery L E. Proc Natl Acad Sci USA. 1989;86:835–839. doi: 10.1073/pnas.86.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seaton B L, Vickery L E. Arch Biochem Biophys. 1992;294:603–608. doi: 10.1016/0003-9861(92)90731-b. [DOI] [PubMed] [Google Scholar]
- 17.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Kogan M, Knight S A B, Pain D, Dancis A. J Biol Chem. 1999;274:33025–33034. doi: 10.1074/jbc.274.46.33025. [DOI] [PubMed] [Google Scholar]
- 19.Kispal G, Csere P, Prohl C, Lill R. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schilke B, Voisine C, Beinert H, Craig E. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garland S A, Hoff K G, Vickery L E, Culotta V C. J Mol Biol. 1999;294:897–907. doi: 10.1006/jmbi.1999.3294. [DOI] [PubMed] [Google Scholar]
- 22.Jensen L T, Culotta V C. Mol Cell Biol. 2000;20:3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange H, Kaut A, Kispal G, Lill R. Proc Natl Acad Sci USA. 2000;97:1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 25.Silberg J J, Hoff K G, Vickery L E. J Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb M R. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silberg J J, Vickery L E. J Biol Chem. 2000;275:7779–7786. doi: 10.1074/jbc.275.11.7779. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill S C, Von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 31.Mach H, Middaugh C R, Lewis R V. Anal Biochem. 1992;200:74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]
- 32.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Flinta C, Oersson B, Jornvall H, von Heijne F. Eur J Biochem. 1986;154:193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 35.Hirel P-H, Schmitter J-M, Dessen P, Fayet F, Blanquet S. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell R, Wali Karzai A, Mehl A F, McMacken R. Biochemistry. 1999;38:4165–4176. doi: 10.1021/bi9824036. [DOI] [PubMed] [Google Scholar]
- 37.Szabo A, Korszun R, Hartl R U, Flanagan J. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 38.Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos G, Bertoli E, Tafani F, Zylicz M. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- 39.Lu Z, Cyr D M. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 40.Cupp-Vickery J R, Vickery L E. Protein Sci. 1997;6:2028–2030. doi: 10.1002/pro.5560060923. [DOI] [PMC free article] [PubMed] [Google Scholar]