Abstract
Exposure to short-wavelength UV light (UVC) strongly induces p53 expression. In human RKO colorectal carcinoma cells, this increase was not due to elevated p53 mRNA abundance, cytoplasmic export of p53 mRNA, or UVC-triggered stabilization of the p53 protein. Instead, p53 translation was potently enhanced after UVC irradiation. The 3′ UTR of p53 was found to be a target of the RNA-binding protein HuR in a UVC-dependent manner in vitro and in vivo. HuR-overexpressing RKO cells displayed elevated p53 levels, whereas cells expressing reduced HuR showed markedly diminished p53 abundance and p53 translation. Our results demonstrate a role for HuR in binding to the p53 mRNA and enhancing its translation.
Keywords: UV light, embryonic lethal abnormal vision
In mammalian cells, the expression and activity of the tumor suppressor gene product p53 is induced in response to a variety of mitogenic and stressful stimuli such as DNA damage, hypoxia, and nutrient deprivation. In turn, p53 induces the transcription of many genes, including several that govern cell cycle arrest, apoptosis, and DNA repair such as Gadd45 (growth arrest and DNA damage-inducible 45), Bcl-2, PUMA (p53-up-regulated mediator of apoptosis), p21WAF1, and mdm2 (1, 2). In human cancer, p53 function has been found to be impaired either through mutation (described in more than half of all malignancies) or through aberrant regulatory events, underscoring its critical function as tumor suppressor (3). Given the central role that p53 plays in both normal cell division and tumor suppression, understanding the molecular pathways through which p53 expression and function are regulated has been the subject of intense investigation over the past decade (3).
After exposure to mitogenic and stressful agents, adaptive responses occur in the cell that culminate in the implementation of altered gene expression patterns. Such gene expression changes can be regulated at many levels, including transcription, splicing, mRNA transport, and stability, as well as protein translation and stability. p53 expression and function are extensively regulated through mechanisms that are stress-, species-, and cell type-specific (4). Although instances of transcriptional regulation of p53 expression have been described (5), it is widely accepted that p53 expression is primarily regulated through modulation of the steady-state levels of p53 in the cell, its subcellular localization, and its activity (1). In unstimulated cells, p53 expression is typically maintained at very low levels, mainly through continuous ubiquitin-mediated proteolysis, and can be rapidly induced by blocking this degradation process (3, 6). In addition, genotoxic agents have been shown to increase the stability and activity of p53 protein through posttranslational modification such as phosphorylation, acetylation, and altered intracellular localization (4). In recent years, however, evidence has emerged revealing that p53 mRNA stability and translation (4, 7–9) are also strong regulators of p53 expression, although the underlying regulatory events remain largely unknown.
Here, we set out to examine the mechanisms governing the elevation in p53 expression triggered by irradiation with short-wavelength UV light (UVC) in human RKO colorectal carcinoma cells. Our findings reveal that UVC irradiation potently enhanced p53 translation. We further demonstrate that the 3′ UTR of p53 is a target of the RNA-binding protein HuR in a UVC-dependent manner in vitro and in vivo. RKO cells displaying either elevated or reduced HuR levels showed either enhanced or diminished p53 expression, respectively, linked to altered p53 translation rates. These results demonstrate a role for HuR in binding to the p53 mRNA and enhancing its translation.
Materials and Methods
Cell Culture, Treatment, and Plasmids. Human RKO colorectal carcinoma cells (10) were maintained in minimum essential medium (GIBCO/BRL) supplemented with 10% FBS and antibiotics. For UVC treatment, medium was removed and saved, cells were rinsed with PBS and irradiated, and medium was restored. Unless otherwise specified, cells were irradiated with 15 J/m2 UVC and collected at the times indicated thereafter. S11 and zeo cells were generated after stable transfection with pZeoSV2 (Invitrogen) and pZeoSV2-HuR, respectively. Lactacystin was from Calbiochem. Plasmid pGL3-Luc-p53 3′ UTR was constructed by inserting the p53 3′ UTR (positions 1421–2629), into the XbaI site of pGL3-promoter (Promega).
Northern and Western Blot Analyses. Total RNA was isolated from intact cells. Nuclear and cytoplasmic RNA were prepared from the pellet and supernatant, respectively, obtained after lysis of cells in 10 mM Tris (pH 7.4), 1 mM KCl, 1 mM MgCl2, and 10% Triton X-100 and brief centrifugation (420 × g, 6 min, 4°C). All RNA extractions were carried out by using STAT-60 (Tel-Test, Friendswood, TX), and Northern blot analysis was performed as described (11). For detection of p53 and 18S transcripts, oligonucleotides GTGAACCATTGTTCAATATCGTCCGGGGACAGCATCAAATCATCCATTGCTTGGG and ACGGTATCTGATCGTCTTCGAACC, respectively, were end-labeled by using [α-32P]dATP and terminal transferase. Signals were quantified with a PhosphorImager (Molecular Dynamics). Calculation of p53 mRNA half-life was carried out by using actinomycin D-based assays, as described (11).
For Western blot analysis, 20-μg aliquots of either total, cytoplasmic, cytosolic, or polysome-bound protein were resolved by electrophoresis in SDS-containing polyacrylamide gels, transferred, and hybridized by using monoclonal antibodies that recognize p53 (DO-1, Santa Cruz Biotechnology), HuR (3A2, Santa Cruz Biotechnology), or β-actin (Abcam, Cambridge, U.K.). After incubation with appropriate secondary antibodies, signals were detected by enhanced chemiluminescence.
Preparation of Protein Fractions. Cytoplasmic, nuclear, and whole-cell fractions were prepared as described (11). For preparation of cytosolic and polysomal fractions, cytoplasmic lysates were layered onto a cushion of 30% sucrose in ice-cold buffer containing 20 mM Hepes (pH 7.4), 50 mM KOAc, 5 mM MgOAc, 1 mM DTT, 1 unit of RNasin per μl, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml, and 0.5 mM phenylmethylsulfonyl fluoride. After centrifugation (100,000 × g for 2 h,4°C), the supernatant (cytosolic fraction) was stored at -80°C; the pellet was resuspended in ice-cold buffer A containing 0.3 M NaCl, incubated on ice for 1 h, and centrifuged at 10,000 × g for 15 min at 4°C; and the resulting supernatant (polysomal extract) was stored at -80°C.
Preparation of Synthetic RNA Transcripts. Total RNA from RKO cells was reverse transcribed, and the cDNAs generated were used as templates for PCR amplification of the coding region and 3′ UTR of p53. The 5′ primers contained the T7 RNA polymerase promoter sequence (T7): CCAAGCTTCTAATACGACTCACTATAGGGAGA. To prepare the coding region (encompassing positions 252 to 1439), oligonucleotides (T7)ATGGAGGAGCCGCAGTCAGATCCTAGC and AGAATGTCAGTCTGAGTCAGGC were used. To prepare the 3′ UTR template (encompassing positions 1421 to 2629), oligonucleotides (T7)TGACTCAGACTGACATTCTCC and TGGCAGCAAAGTTTTATTGTAAAATAAGAGATCG were used. PCR-amplified products were resolved on agarose gels, purified, and used as templates for the synthesis of corresponding RNAs. For RNA electrophoretic mobility-shift assay (REMSA), PCR-amplified products were use for the synthesis of radiolabeled transcripts, as described (11) and used at a specific activity of 100,000 cpm/μl. For pull-down assays, PCR-amplified DNA was used as template to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-cytidine 5′-triphosphate (CTP), and purified as described (12).
RNA Protein-Binding Assays: REMSA, REMSA Supershift, and Pull-Down. REMSA analysis was carried out as described (11), by using 10 μg of cytoplasmic fractions. For REMSA supershift analysis, cytoplasmic protein aliquots were incubated with antibodies recognizing HuR, tristetraprolin (TTP), TIAR, TIA-1 (Santa Cruz Biotechnology), or AUF1 (a gift of G. Brewer, University of Medicine and Dentistry of New Jersey, Piscataway), or with IgG1 (BD Life Sciences), for 30 min on ice before addition to the binding reaction mixture; all subsequent steps were as described above for REMSA. For biotin pull-down assays, 6 μg of biotinylated transcripts were incubated with 120 μg of cytoplasmic lysate for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo), and pull-down material was analyzed by Western blotting.
Immunoprecipitation (IP) of endogenous HuR-mRNA complexes, used to assess the association of endogenous HuR with endogenous p53 mRNA, was performed as described (13). Twenty million RKO cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody [either mouse monoclonal anti-HuR antibody 3A2 (Santa Cruz Biotechnology) or IgG1]. RNA in IP material was used in RT-PCR reactions to detect the presence of p53 mRNA; the p53-coding region was amplified by using the primer pair described above, and the following amplification conditions: 1 min at 94°C, 1 min at 55°C, and 1 min at 68°C, for 35 cycles. PCR products were visualized by ethidium bromide staining of 1.5% agarose gels.
Analysis of Nascent p53. Newly translated p53 protein was measured by incubating 106 cells with 1 mCi (1 Ci = 37 GBq) l-[35S]methionine and l-[35S]cysteine (Easy Tag EXPRESS, NEN/Perkin–Elmer) per 60-mm plate for 20 min, whereupon cells were lysed by using TSD lysis buffer (50 mM Tris, pH 7.5/1% SDS/5 mM DTT), and lysates were immunoprecipitated by using either monoclonal anti-p53 antibody DO-1 (Santa Cruz Biotechnology) or IgG1 for 1 h at 4°C. After extensive washes in TNN buffer (50 mM Tris, pH 7.5/250 mM NaCl/5 mM EDTA/0.5% Nonidet P-40), immunoprecipitated material was resolved by 12% SDS/PAGE, transferred onto poly(vinylidene difluoride) filters, and visualized by using a PhosphorImager (Molecular Dynamics).
Small Interfering RNA (siRNA) Transient Transfection. The siRNA sequence targeting HuR (HuR4, AACACGCTGAACGGCTTGAGG) was derived from nucleotides 377–397 (GenBank accession no. BC003376). The sequence of the control (Ctrl) siRNA, AAGTGTAGTAGATCACCAGGC, does not match any known human gene. siRNA molecules were synthesized by using a kit from Ambion (Austin, TX) and were transfected at a concentration of 20 nM by using Olifectamine (Invitrogen). Cells were treated with UVC 48 h after transfection and collected for analysis 4 h later.
Results and Discussion
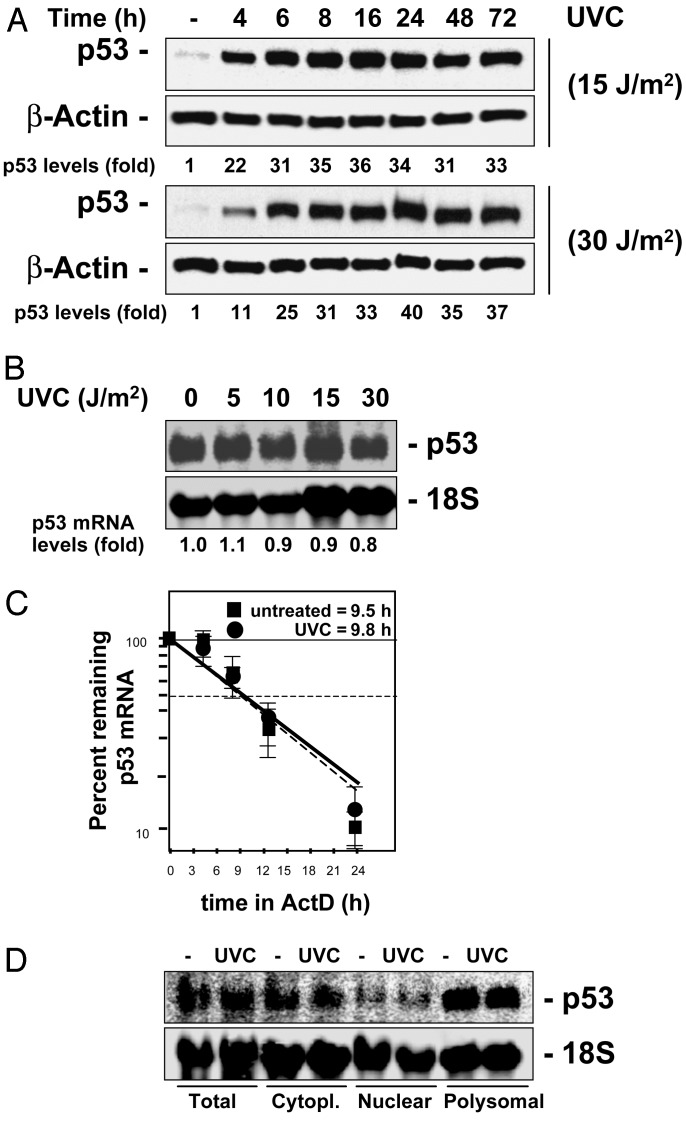
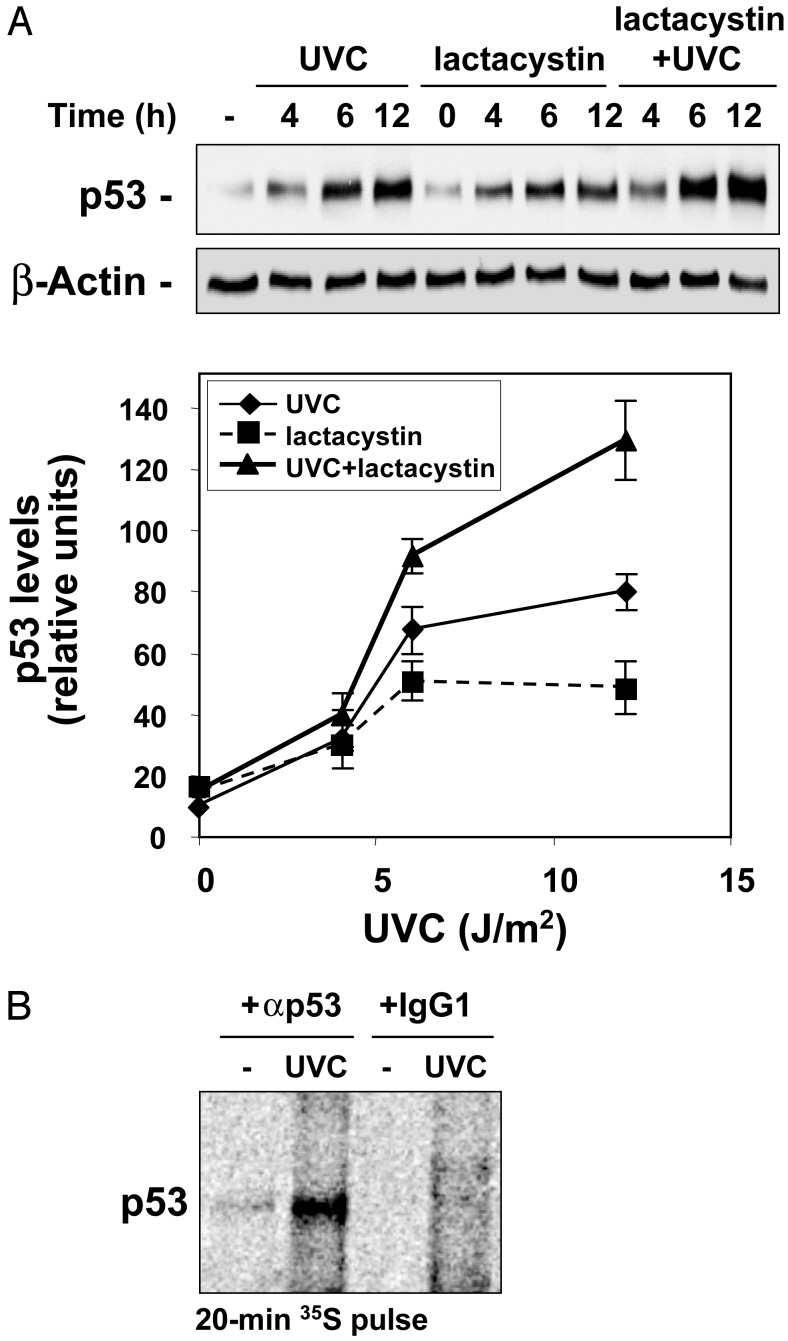
Here, we set out to examine the mechanisms underlying the elevation in p53 expression triggered by UVC. Treatment of human colorectal carcinoma RKO cells, which exhibit wild-type p53 function (10), with the relatively low UVC doses of 15 or 30 J/m2, increased p53 expression, peaking by 6 h and remaining elevated for up to 72 h (Fig. 1A). The rapid and robust increase in p53 expression was not accompanied by a concomitant elevation in p53 mRNA abundance (Fig. 1B), p53 mRNA stability (Fig. 1C), or relative levels of cytoplasmic p53 mRNA (Fig. 1D), indicating that the UVC-triggered induction of p53 was regulated by either translational or posttranslational events. UVC was previously shown to increase p53 protein stability by blocking its ubiquitin-mediated degradation through the proteasome (6). To assess the extent to which p53 protein increase was due to UVC-triggered protein stabilization in this system, we examined whether inhibition of the proteasome alone could recapitulate the effects of UVC on p53 induction in RKO cells. Cells were incubated with lactacystin, a potent and highly selective inhibitor of the proteasome, using doses that blocked proteasome activity completely and had been found to elicit maximal p53 induction (not shown). As shown, lactacystin treatment alone did cause an increase in p53 levels, but combined lactacystin and UVC treatments further induced p53 expression (Fig. 2A), indicating that inhibition of the proteasome was not the sole mechanism leading to p53 induction by UVC. We therefore investigated whether increased p53 translation might also contribute to elevating p53 expression after UVC irradiation by comparing the rate of new p53 synthesis between untreated and UVC-treated cells. RKO cells were incubated in the presence of L-[35S]methionine and L-[35S]cysteine for 20 min, whereupon newly translated p53 was visualized by IP. The brief incubation period was chosen to minimize the contribution of p53 degradation in our analysis. As shown in Fig. 2B, newly synthesized p53 was remarkably more abundant in UVC-irradiated cells, demonstrating that enhanced p53 protein translation is an essential mechanism leading to p53 induction in RKO cells after UVC exposure.
Fig. 1.
Expression of p53 in RKO cells irradiated with UVC. (A) Cells were exposed to either 15 or 30 J/m2 or left unirradiated (-), and whole-cell lysates were prepared for Western blotting at the times indicated. (B) Northern blot analysis of p53 expression 4 h after exposure to the indicated doses of UVC. Relative p53 abundance was calculated by densitometry and is expressed as fold increase relative to p53 levels in unirradiated cells. (C) Half-life of the p53 mRNA was calculated by addition of actinomycin D (ActD, 2 μg/ml) to RKO cells 4 h after either UVC irradiation (15 J/m2) or no treatment (untreated), as described (11). Relative p53 mRNA abundance is expressed as fold increase relative to p53 mRNA levels in unirradiated cells. (D) Total, cytoplasmic (Cytopl.), nuclear, and polysomal fractions were prepared for Northern blot analysis 4 h after exposure to UVC (15 J/m2); 18S signals reveal even loading of samples.
Fig. 2.
(A) Western blot analysis of p53 expression in RKO whole-cell lysates prepared at the times indicated after treatment with lactacystin (5 μM), UVC (15 J/m2), or a combination of both. Signals were quantitated by densitometry (graph). (B) Newly translated p53 was measured by incubating cells with l-[35S]methionine and l-[35S]cysteine for 20 min, followed by immunoprecipitation by using either anti-p53 antibody or IgG1, resolving immunoprecipitated samples by SDS/PAGE, and transferring for visualization of signals by using a PhosphorImager.
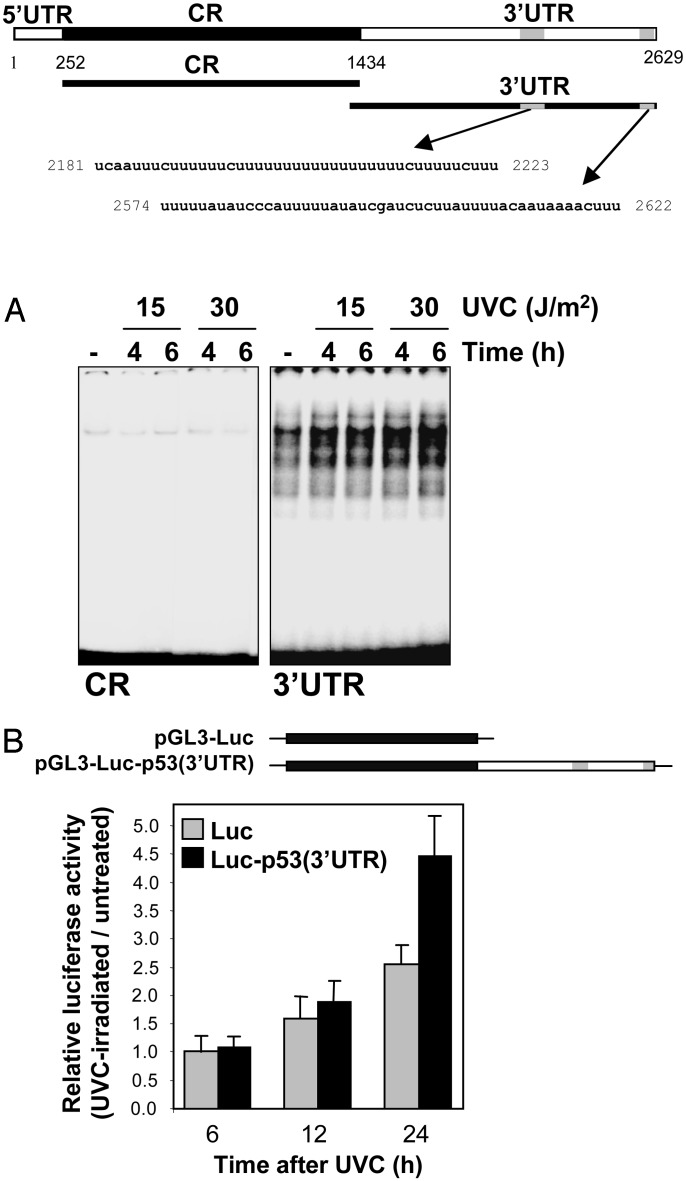
Earlier studies examining p53 translational control had implicated sequences in the 5′ and 3′ UTRs of the p53 transcript (14–16). We thus examined whether the p53 mRNA was a target of RNA-binding proteins in RKO cells, particularly if such association(s) occurred in a UVC-dependent manner. Depicted in Fig. 3 (schematic) are transcripts spanning different sections of the p53 mRNA that were synthesized in the presence of radiolabeled UTP for use in REMSA or biotinylated CTP for use in pull-down assays. REMSA analysis was carried out by using lysates that were prepared from either untreated or UVC-irradiated RKO cells. Transcripts corresponding to the 5′ UTR and coding region (CR) revealed almost undetectable binding to proteins present in cytoplasmic lysates, and complex abundance did not change in a UVC-dependent manner (Fig. 3A, and data not shown). By contrast, transcripts encompassing the p53 3′ UTR were capable of extensive association with cytoplasmic proteins, and the abundance of the complexes increased when using lysates prepared from UVC-treated cells (Fig. 3A). In control REMSA analyses, nuclear lysates revealed abundant binding to all transcripts tested and exhibited no UVC-dependent inducibility, similar to what was previously demonstrated with other transcripts (refs. 11 and 12, and data not shown). Evidence that the p53 3′ UTR indeed possessed translational enhancing properties came through the analysis of chimeric transcripts. Plasmid vectors expressing either luciferase mRNA (pGL3-Luc) or a chimeric mRNA comprising the luciferase coding region linked to the p53 3′ UTR [pGL3-Luc-p53(3′UTR)] were transiently transfected into RKO cells. Expression levels of chimeric transcripts were unaffected by UVC irradiation (not shown), and therefore assessment of luciferase activity served as a measure of mRNA translation, as described (17). The presence of the p53 3′ UTR was found to significantly enhance the translation of chimeric mRNAs after UVC irradiation (Fig. 3B).
Fig. 3.
Binding of cytoplasmic proteins to the p53 3′ UTR is linked to p53 translational up-regulation. (Upper) Schematic of p53 mRNA, depicting U-rich and AU-rich sequences (gray), as well as transcripts (CR and 3′ UTR) used for analysis. (A) REMSA (11) by using either CR or 3′ UTR p53 radiolabeled transcripts and cytoplasmic lysates prepared 4 and 6 h after treatment of RKO cells with either 15 or 30 J/m2 UVC. (B) Plasmids pGL3-Luc and pGL3-Luc-p53(3′UTR) were used in transient transfections; 24 h after transfection, cells were either irradiated (15 J/m2) or left untreated, and the relative luciferase activity was calculated 6, 12, and 24 h later.
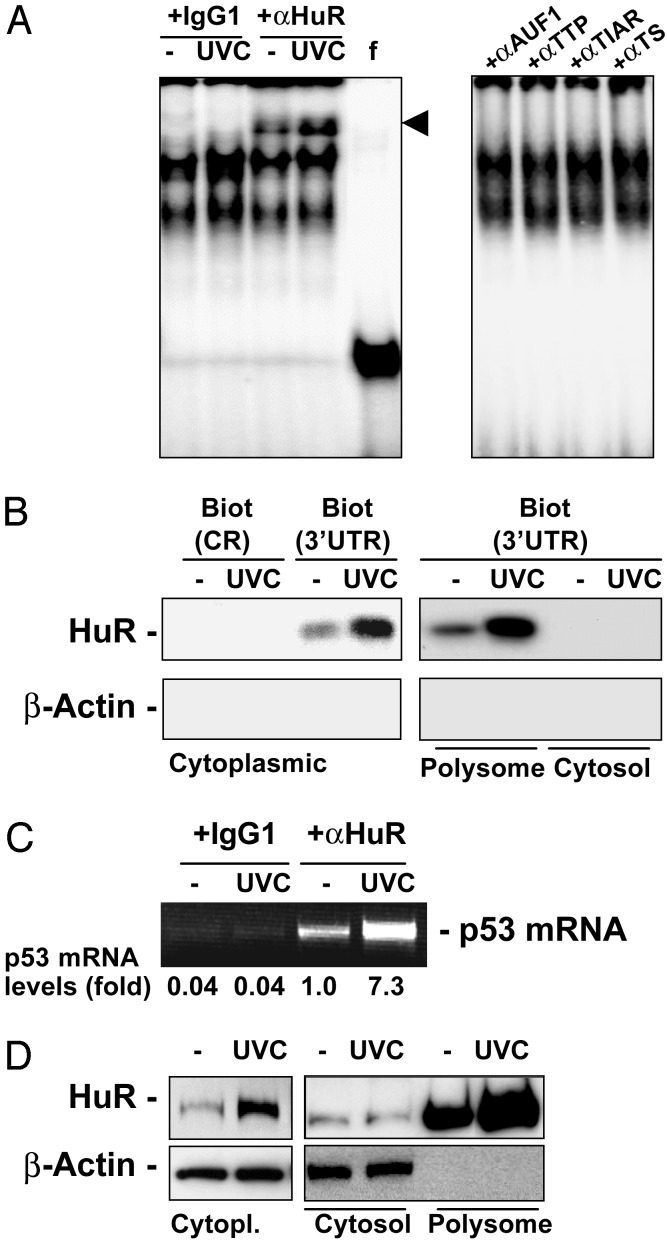
Given previous reports linking members of the ELAV RNA-binding protein family to translational regulation (18), our earlier reports of a UVC-dependent increase in HuR function in RKO cells (11), and the existence of U-rich and AU-rich elements, akin to other HuR-target sequences, in the p53 3′ UTR (Fig. 3 Upper, gray shading), we directly examined whether HuR, the only ELAV member shown to be expressed in RKO cells (11), might be involved in the translational regulation of p53. To directly investigate the potential association of HuR with the p53 3′ UTR in a UVC-dependent fashion, we carried out three types of assays. First, we used a p53 3′ UTR transcript to perform REMSA supershift analysis with anti-HuR antibodies. As shown in (Fig. 4A), HuR indeed forms a complex with the p53 3′ UTR, as revealed by the appearance of slower electrophoretic mobility bands (arrowhead) in anti-HuR antibody incubations, but not in control antibody (IgG1) incubations. Furthermore, the intensity of the supershift increased when using lysates from UVC-treated cells. No supershifts were observed when assessing the presence of additional AU-rich element RNA-binding proteins implicated in regulating either mRNA turnover or translation [AUF1, TTP, TIAR, and TIA-1 (19–21)]. Second, use of biotinylated p53 transcripts in pull-down assays using streptavidin-coated beads revealed an association of the p53 3′ UTR with HuR present in cytoplasmic and polysomal preparations, in greater abundance when lysates were prepared from UVC-treated cells. However, no such complexes were found when using biotinylated p53(CR) or cytosolic lysates (Fig. 4B). Finally, evidence for the in vivo association of endogenous p53 mRNA and HuR in the cell was obtained through immunoprecipitation of HuR under conditions that preserved its association with target mRNAs, by using a previously described method (13). RT-PCR analysis revealed the presence of endogenous p53 mRNA in the material immunoprecipitated by using anti-HuR antibodies, but not when nonspecific antibodies (IgG1) were used (Fig. 4C). As earlier reported, whole-cell HuR levels did not change with UVC irradiation, and HuR was primarily nuclear at all times examined (11, 12). By contrast, its cytoplasmic presence (Fig. 4D, Cytopl.) increased on UVC irradiation, and was almost exclusively found in association with the polysomal fraction (Fig. 4D, Polysome). These findings demonstrate that HuR binds to the p53 3′ UTR mRNA in a UVC-dependent manner, and that HuR-p53 mRNA complexes colocalize with the cell's polysomes.
Fig. 4.
HuR binds to the p53 3′ UTR in a UVC-dependent manner. (A) REMSA supershift using either IgG1 or an anti-HuR antibody (11) (Left), or antibodies recognizing the RNA-binding proteins shown (Right). f, free probe, digested with RNase T1 but left without incubating with cytoplasmic lysates; arrowhead, supershift of HuR-containing complexes. (B) Detection of HuR after pull-down by using biotinylated p53 transcripts CR and 3′ UTR (described in Fig. 3 Upper). After incubations with proteins present in cytoplasmic, cytosolic, or polysomal fractions of cells that were either left untreated or treated with 15 J/m2 UVC, the presence of HuR and β-actin was detected by Western blotting. (C) IP by using either anti-HuR antibody or IgG1 under conditions that preserve the association of RNA-binding proteins with target mRNAs was followed by RT-PCR analysis to detect endogenous p53 mRNA; PCR products were resolved by electrophoresis in 1.5% agarose gels stained with ethidium bromide. (D) Subcellular localization of HuR in cytoplasmic (Cytopl.), cytosolic (Cytosol), and polysomal (Polysome) fractions prepared from cells that were either left untreated or UVC-treated.
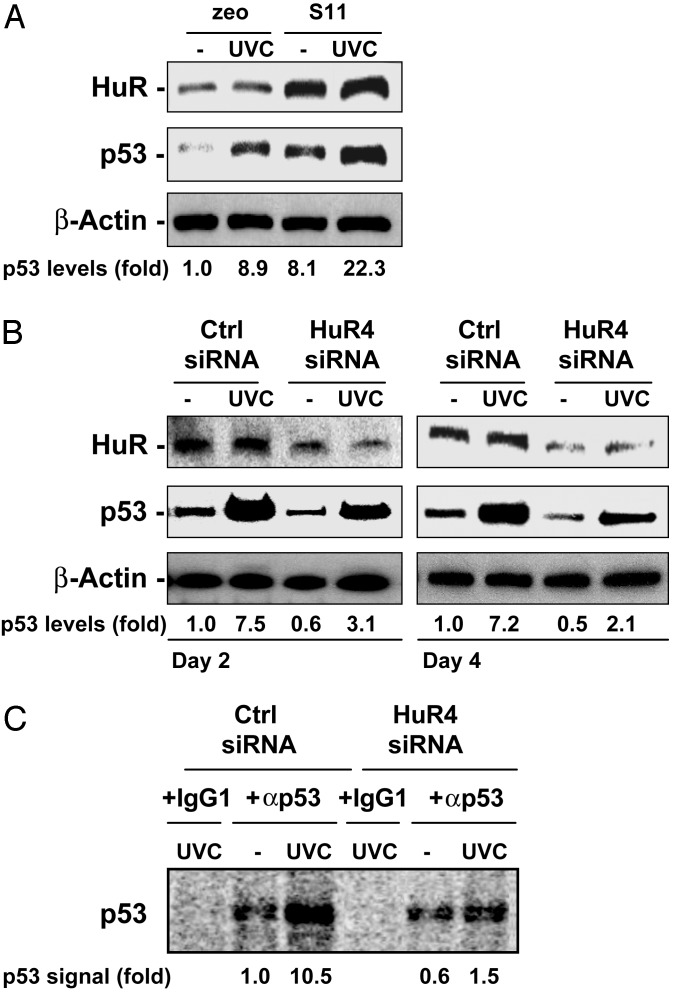
Direct analysis of HuR's role in p53 expression was carried out through experiments in which HuR expression levels in RKO cells were modified. RKO cells stably overexpressing HuR (S11) revealed heightened p53 levels, both basally and in response to UVC (Fig. 5A). Conversely, reduction of HuR levels through HuR-directed siRNA (HuR4; routinely reaching 20–30% of control “Ctrl” siRNA transfections) was found to markedly decrease p53 levels and to attenuate p53 induction after UVC treatment (Fig. 5B). To directly assess whether HuR influenced p53 translation, siRNA-transfected cells expressing lower HuR, as well as control populations, were pulse-labeled in the presence of L-[35S]methionine and L-[35S]cysteine for 20 min, and nascent p53 was immunoprecipitated. As shown, p53 translation was potently induced by UVC in control siRNA-transfected cells, whereas cells expressing reduced HuR exhibited strikingly lower p53 production (Fig. 5C). These data reveal that HuR enhances p53 translation in a UVC-dependent manner.
Fig. 5.
Modulation of HuR levels affects p53 translation and steady-state levels. (A) Western blot analysis of HuR and p53 expression levels in RKO cells that had been stably transfected with either an insert less plasmid (zeo) or an HuR-expressing plasmid (S11). (B) RKO cells were transiently transfected with either a control (Ctrl) siRNA or siRNA directed to HuR (HuR4), and exposed to 15 J/m2 UVC 48 h later. HuR expression levels were analyzed by Western blotting using whole-cell lysates that were prepared 4 h after UVC. (C) Newly translated p53 was assessed as explained in the legend of Fig. 2B. Forty-eight hours after transfection with either control or HuR4 siRNAs, RKO cultures were irradiated or left untreated; 4 h later, they were incubated with l-[35S]methionine and l-[35S]cysteine for 20 min. Lysates were prepared and immunoprecipitated by using either anti-p53 antibody or IgG1, then processed as described in Fig. 2B.
Together, these findings demonstrate that HuR binds to the p53 mRNA in vitro and in vivo. Despite HuR's established role in the stabilization of target mRNAs, HuR was not found to influence p53 mRNA turnover in this stress paradigm (Fig. 1C) but instead enhanced p53 translation. A role for HuR in translational regulation has been previously proposed, based on the effects of other ELAV proteins on translational control, and the association of HuR with polysomes (18, 22, 23). Two recent works link HuR to the translational regulation of p27 expression through binding to the 5′ UTR of the p27 mRNA, one demonstrating an inhibitory role for HuR in p27 translation (24), another consistent with an HuR-mediated enhancement in p27 translation (25). Our results support a model whereby HuR, through its association with the p53 3′ UTR, positively influences p53 translation. These findings recapitulate the enhanced translation of neurofilament M previously reported for Hel-N1, which also occurs via a 3′ UTR-binding site, but does not involve mRNA stabilization (18). The precise mechanisms mediating HuR's enhanced translation of p53 are unclear, but may be linked to a mechanism of recruitment of mRNAs to active polysomes. For example, neurofilament M (NF-M) and Glut-1 mRNAs shift their positions on polysome gradients from the monosome peaks to the active polysome regions after transfection of HuB into hNT2 cells and 3T3L1 cells, respectively. Although the observed increase in half-life of target mRNAs by exogenously expressed HuR is in keeping with its major function, being mRNA stabilization (reviewed in refs. 22 and 28), those results are consistent also with a role of HuR in translational activation based on direct interactions with mRNA targets. Thus, the apparent stabilization of HuR's mRNA targets could be a function of their physical sequestration during recruitment to active polysomes via a mechanism intended to activate their translation (26).
The relationship between mRNA turnover and translation remains controversial. In some instances, translation of a given mRNA has been shown to render it unstable (27), whereas other studies support a link between mRNA stabilization and translation (23). In the present investigation, no differences in p53 mRNA abundance or stability were observed, and only an HuR-mediated enhancement in p53 translation was apparent (Fig. 5). To date, HuR has been shown to bind to many mRNAs and promote their stabilization (for review, see ref. 28), but no formal studies addressing potential links between HuR-mediated stabilization and translation of target transcripts have been reported. Should HuR prove to have a general function in enhancing the translation of target mRNAs, it will lend support to an emerging model whereby HuR binds to a given mRNA, likely assists in its nuclear export, protects it from degradation in the cytoplasm, and directs it to ribosomes, enhancing its translation in concert with that of other HuR-bound mRNAs (22, 28).
It is striking that several RNA-binding proteins, including HuR, respond to UVC, heat shock, and other perturbations by shifting their location to the cytoplasm where their target mRNAs can be stabilized and translated (11, 14, 29). In this study, we report that translation of the p53 mRNA, a target of HuR, was significantly increased after DNA damage by low levels of UVC irradiation. It is possible that posttranscriptional regulation of certain key proteins is specifically stimulated after DNA damage so that transcription can pause while DNA repair events proceed. This mechanism would prevent the production of aberrant transcripts or proteins with deleterious consequences for the cell, but also allow previously synthesized mRNAs that encode important growth regulatory proteins to be preserved, thereby permitting the protein products to maintain homeostasis during a period of DNA repair. HuR and other ELAV proteins would be ideal candidates for responding to genotoxins and other damaging agents because they interact with mRNAs encoding important growth regulatory factors and participate in their production at the posttranscriptional level (22, 28). In conclusion, such a broad posttranscriptional function for HuR and other ELAV proteins will have particular significance after exposure of mammalian cells to toxic agents: it will help ensure that key proteins such as p53 are in place through posttranscriptional means as the cell assesses the damage and prepares to undergo either growth arrest or apoptosis.
Acknowledgments
We thank X. Yang and W. Wang for helpful discussions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ELAV, embryonic lethal abnormal vision; UVC, short-wavelength UV light; REMSA, RNA electrophoretic mobility-shift assay; IP, immunoprecipitation; siRNA, small interfering RNA; CR, coding region.
References
- 1.Vousden, K. H. (2002) Biochim. Biophys. Acta 1602 47-59. [DOI] [PubMed] [Google Scholar]
- 2.Yu, J., Wang, Z., Kinzler, K. W., Vogelstein, B. & Zhang, L. (2003) Proc. Natl. Acad. Sci. USA 100 1931-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan, K. M., Phillips, A. C. & Vousden, K. H. (1997) Curr. Opin. Cell Biol. 13 332-337. [DOI] [PubMed] [Google Scholar]
- 4.Giaccia, A. J. & Kastan, M. B. (1998) Genes Dev. 12 2973-2983. [DOI] [PubMed] [Google Scholar]
- 5.Noda, A., Toma-Aiba, Y. & Fujiwara, Y. (2000) Oncogene 19 21-31. [DOI] [PubMed] [Google Scholar]
- 6.Maki, C. G. & Howley, P. M. (1997) Mol. Cell. Biol. 17 355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, H., You, S., Farris, J., Foster, L. K. & Foster, D. N. (2001) Oncogene 20 3306-3310. [DOI] [PubMed] [Google Scholar]
- 8.Kim, H., You, S., Foster, L. K., Farris, J. & Foster, D. N. (2001) Oncogene 20 5118-5123. [DOI] [PubMed] [Google Scholar]
- 9.Chu, E., Copur, S. M., Ju, J., Chen, T. M., Khleif, S., Voeller, D. M., Mizunuma, N., Patel, M., Maley, G. F., Maley, F. & Allegra, C. J. (1999) Mol. Cell. Biol. 19 1582-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kessis, T. D., Slebos, R. J., Nelson, W. G., Kastan, M. B., Plunkett, B. S., Han, S. M., Lorincz, A. T., Hedrick, L. & Cho, K. R. (1993) Proc. Natl. Acad. Sci. USA 90 3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, W., Furneaux, H., Cheng, H., Caldwell, M. C., Hutter, D., Liu, Y., Holbrook, N. J. & Gorospe, M. (2000) Mol. Cell. Biol. 20 760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, W., Lin, S., Caldwell, C. M., Furneaux, H. & Gorospe, M. (2000) EMBO J. 19 2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenenbaum, S. A., Lager, P. J., Carson, C. C. & Keene, J. D. (2002) Methods 26 191-198. [DOI] [PubMed] [Google Scholar]
- 14.Mosner, J., Mummenbrauer, T., Bauer, C., Sczakiel, G., Grosse, F. & Deppert, W. (1995) EMBO J. 14 4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, L. & Benchimol, S. (1997) EMBO J. 16 4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, L., Ma, W. & Benchimol, S. (1999) Oncogene 18 6419-6424. [DOI] [PubMed] [Google Scholar]
- 17.Galbán, S., Fan, J., Martindale, J. L., Cheadle, C., Hoffman, B., Woods, M. P., Temeles, G., Brieger, J., Decker, J. & Gorospe, M. (2003) Mol. Cell. Biol. 23 2306-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antic, D., Lu, N. & Keene, J. D. (1999) Genes Dev. 13 449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carballo, E., Lai, W. S. & Blackshear, P. J. (1998) Science 281 1001-1005. [DOI] [PubMed] [Google Scholar]
- 20.Gueydan, C., Droogmans, L., Chalon, P., Huez, G., Caput, D. & Kruys, V. (1999) J. Biol. Chem. 274 2322-2326. [DOI] [PubMed] [Google Scholar]
- 21.DeMaria, C. T., Sun, Y., Long, L., Wagner, B. J. & Brewer, G. (1997) J. Biol. Chem. 272 27635-27643. [DOI] [PubMed] [Google Scholar]
- 22.Keene, J. D. (1999) Proc. Natl. Acad. Sci. USA 96 5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain, R. G., Andrews, L. G., McGowan, K. M., Pekala, P. H. & Keene, J. D. (1997) Mol. Cell. Biol. 17 954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kullmann, M., Gopfert, U., Siewe, B. & Hengst, L. (2002) Genes Dev. 16 3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millard, S. S., Vidal, A., Markus, M. & Koff, A. (2000) Mol. Cell. Biol. 20 5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazumder, B., Seshadri, V. & Fox, P. L. (2003) Trends Biochem. Sci. 28 91-98. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay, D., Houchen, C. W., Kennedy, S., Dieckgraefe, B. K. & Anant, S. (2003) Mol. Cell 11 113-126. [DOI] [PubMed] [Google Scholar]
- 28.Brennan, C. M. & Steitz, J. A. (2001) Cell. Mol. Life Sci. 58 266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallouzi, I. E., Brennan, C. M. & Steitz, J. A. (2001) RNA 7 1348-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]