Abstract
The action of cyclin-dependent kinases (CDKs) is regulated by phosphorylation, cyclin levels, the abundance of CDK inhibitors, and, as recently has been shown for cyclin B/cdc2, their localization. It is unclear how localization regulates the action of cyclin E/Cdk2 and its inhibitors. Here, we show that the closest known Xenopus laevis homolog of mammalian Cdk2 inhibitors p27Kip1 and p21CIP1, Xic1, is concentrated, ubiquitinated, and destroyed in the nucleus. Furthermore, Xic1 destruction requires nuclear import, but not nuclear export, and requires the formation of a transport-competent nuclear envelope, but not interactions between the lamina and chromatin. We show that (i) cyclin E/Cdk2 and Xic1 are transported into the nucleus as a complex and that Xic1 destruction requires the activity of cyclin E, (ii) that phosphorylation of Xic1 by cyclin E/Cdk2 bypasses the requirement for nuclear formation, and (iii) that the phosphorylation of Xic1 by cyclin E/Cdk2 is concentration dependent and likely realized through second-order interactions between stable cyclin E/Cdk2/Xic1 ternary complexes. Based on these results we propose a model wherein nuclear accumulation of the cyclin E/Cdk2/Xic1 complex triggers a concentration-dependent switch that promotes the phosphorylation of Xic1 and, consequently, its ubiquitination and destruction, thus allowing subsequent activation of cyclin E/Cdk2.
Keywords: cyclin-dependent kinase inhibitor, ubiquitin, proteolysis
In vertebrates, the G1/S transition requires the activity of cyclin E/Cdk2 (1–3). In turn, the abundance of cyclin E, the phosphorylation state of Cdk2, and the abundance of cyclin-dependent kinase (CDK) inhibitors such as p21Cip1 and p27Kip1 combine to regulate cyclin E/Cdk2 activity. p27Kip1 abundance is thought to be largely controlled by its stability (4), suggesting that the destruction of p27Kip1 is critical for the G1/S transition. p27Kip1 is a highly conserved protein. The closest known Xenopus homolog, p27Xic1, is thought to play a similar role in restraining the G1/S transition and p28Kix1 (a p27Xic1 isoform) is up-regulated during gastrulation at the time when the G1 phase first appears in development (5). The ability of these inhibitors to restrain activation of cyclin E/Cdk2-dependent activation of DNA replication is thus critical for determining the length of G1.
In Saccharomyces cerevisiae, passage through the G1/S transition is gated by the Cdc28 inhibitor p40Sic1. p40Sic1 is destroyed by ubiquitin-dependent proteolysis and targeted for ubiquitination by the Skp1–Cullin–F-box (SCF) ubiquitin ligase complex Cdc34/Cdc53/Skp1/Cdc4 (6–10). The SCF binds to p40Sic1 through the F-box protein Cdc4 only after p40Sic1 is phosphorylated by a G1-specific cyclin/Cdc28. Thus, the regulated phosphorylation of the inhibitor controls its stability.
The basic elements of this mechanism in yeast appear to be conserved for regulating p27 stability in vertebrates. In mammalian cells (11, 12), p27Kip1 is destroyed by a similar phosphorylation- and ubiquitin-dependent pathway and has been suggested to require Cdc34 (11). However, there may be important differences. In yeast, p40Sic1 inhibits an S phase-specific cyclin/CDK complex (Clb5/Cdc28), but is phosphorylated by a distinct G1-specific cyclin/CDK complex (Cln2/Cdc28). But in mammalian cells, p27Kip1 apparently inhibits and is phosphorylated by the same S phase-promoting complex, cyclin E/Cdk2 (14). Thus, p27Kip1 functions as both an inhibitor and a substrate of cyclin E/Cdk2. This dual function creates a conundrum: how can cyclin E/Cdk2, while bound to its inhibitor, phosphorylate the very same molecule, promote its destruction, and thereby be freed to phosphorylate other targets? In other words, is there a molecular switch that changes p27Kip1 from an inhibitor to a substrate?
Biochemical studies have begun to address how p27 may act as both inhibitor and substrate. In mammalian cell extracts, p27 is destroyed only when bound to cyclin E/Cdk2 (13). Therefore, the pool of p27 bound to cyclin E/Cdk2 is most critically regulated. At physiological ATP concentrations (≈2 mM), p27's substrate activity is favored, whereas lower ATP concentrations favor its inhibitory activity (14). Further, a p27 mutant that binds the cyclin subunit but not the CDK subunit is more readily phosphorylated (15). Thus, cyclin E/Cdk2 may phosphorylate p27 via an intermediate in which p27 is bound to the cyclin but is not yet inhibiting the kinase. However, changes in ATP levels are unlikely to explain the inhibitor-substrate transition inside the cell. Further, the transition rate to the tightly bound state is fast (1 min−1) and the off rate is slow (1/120 min−1) (14), such that the vast majority of cyclin E/Cdk2 and p27 likely exists as an inhibited trimeric complex throughout G1. Another mechanism must account for the inhibitor-substrate transition.
One such mechanism is suggested by the observation that triggering of DNA replication is tightly coupled to nuclear formation. Moreover, destruction of Xic1 in Xenopus egg extract requires addition of sperm chromatin (16). We find that Xic1 also can be both inhibitor and substrate of cyclin E/Cdk2 and that degradation requires association with cyclin E/Cdk2. Is the inhibitor-substrate transition of p27Xic1 coupled to nuclear transport and what nuclear-dependent or -independent mechanisms facilitate the transition?
To answer this question, we investigated the effect of nuclear function on Xic1 destruction. We find that Xic1 destruction requires nuclear formation and nuclear transport, that Xic1 and cyclin E accumulate in the nucleus after nuclear formation, and that Xic1 subsequently is ubiquitinated and destroyed in the nucleus, independent of nuclear export. We find that lamina-chromatin interactions required for DNA replication are not required for Xic1 destruction, confirming that Xic1 destruction principally requires nuclear import.
To explain the cyclin E/Cdk2 requirement for Xic1 destruction, we show that cyclin E/Cdk2 phosphorylation of Xic1 bypasses the nuclear requirement for Xic1 destruction, suggesting that the nuclear accumulation stimulates the phosphorylation of Xic1, and that ubiquitination and proteolysis can occur independent of nuclear formation. Finally, because cyclin E/Cdk2 is concentrated in the nucleus before DNA replication (17) we tested and confirmed the model that the effective activity of cyclin E/Cdk2 toward Xic1 depends on the second-order concentration of cyclin E/Cdk2 and Xic1 and likely mediated through interactions between ternary complexes. Based on these results we propose that the facilitated concentration of the cyclin E/Cdk2/Xic1 complex in the nucleus overcomes the inhibitory action of Xic1. This concentration-dependent switch then triggers the phosphorylation and consequent ubiquitination and destruction of Xic1, thereby fully activating cyclin E/Cdk2.
Materials and Methods
Preparation of Interphase Extracts.
Interphase extracts were prepared essentially as described (1) but the second spin was performed at 24,000 rpm in a TLS 55 rotor for 15 min at 4°. The golden middle fraction was used. In our hands, these extracts are more reproducibly competent for DNA replication than lower speed extracts.
Destruction and Transport Assays.
Destruction assays were conducted as described (16). 35S-labeled Xic1 (0.5 μl/10 μl extract), sperm (3,000/μl), and an energy regenerating system were mixed with extract. Reactions were incubated at room temperature for 2 h and stopped with sample buffer. Samples were resolved by SDS/PAGE, and proteins were transferred to immobilon-P transfer membrane and analyzed by using a Molecular Dynamics PhosphorImaging system.
In transport and destruction assays, reactions were initiated at room temperature and stopped with elution buffer (ELB) (50 mM KCL/10 mM Hepes, pH 7.7/2.5 mM MgCl2/250 mM sucrose) at indicated times. The diluted extract was immediately overlaid onto 0.5 M sucrose in ELB and spun 20 sec in a horizontal rotor (Beckman 152 centrifuge). The cytoplasmic fraction was removed from above the sucrose cushion and added to sample buffer. The cushion was carefully aspirated. The pellet fraction then was washed once with ELB, spun again, and resuspended in sample buffer. One-fifth of the cytoplasmic and all nuclear samples were resolved by SDS/PAGE, and the proteins were transferred to immobilon-P transfer membrane and analyzed by using a Molecular Dynamics PhosphorImaging system and immunoblotting.
In Vitro Phosphorylation Experiments.
Cyclin E/Cdk2 was purified from baculovirus and incubated with Xic1 for 30 min in kinase reaction buffer (100 mM NaCl/20 mM Hepes, pH 7.5/1 mM EDTA/5 mM MgCl2). Reactions were initiated by the addition of ATP (100 μM) and γ32P-ATP (1 μM). Reactions were stopped after 3 min with sample buffer. Equivalent volumes were resolved by SDS/PAGE and analyzed by PhosphorImaging.
DNA Replication Assays.
Reactions were conducted essentially as described (3) by using trichloroacetic acid precipitation of DNA onto glass fiber filters. Replication efficiency was typically greater than 70%.
Preparation of Recombinant Proteins.
Different types of Xic1 proteins [35S-labeled in vitro-translated (IVT) Xic1, glutathione S-transferase (GST)-Xic1, and myelin basic protein (MBP)-Xic1] behaved similarly in the assays described. 35S-labeled IVT Xic1 was prepared by using coupled in vitro transcription/translation from plasmid pCS2-Xic1. GST-Xic1 and MBP-Xic1 were purified from bacterial strain BL21 pLysS according to standard protocols. Xenopus cyclin E/Cdk2 complex was purified from SF9 cells coinfected with Xenopus cyclin E and Xenopus His-Cdk2 expressing viruses (multiplicities of infection of 15 and 10). Cells were harvested in buffer (50 mM Tris⋅HCl/100 mM KCl/20% glycerol/5 nM MgCl2/50 mM sodium phosphate/10 mM immidazole, pH 7.7), and the complex was purified on Ni2+-nitrilotriacetic acid resin. Peak fractions were pooled and dialyzed into XB (100 mM KCl/10 mM Hepes, pH 7.7) and 20% glycerol. LAP2 fragments were generously provided by Kathy Wilson, Johns Hopkins University School of Medicine, Baltimore (20).
Results
Xic1 Destruction Requires Transport-Competent Nuclei.
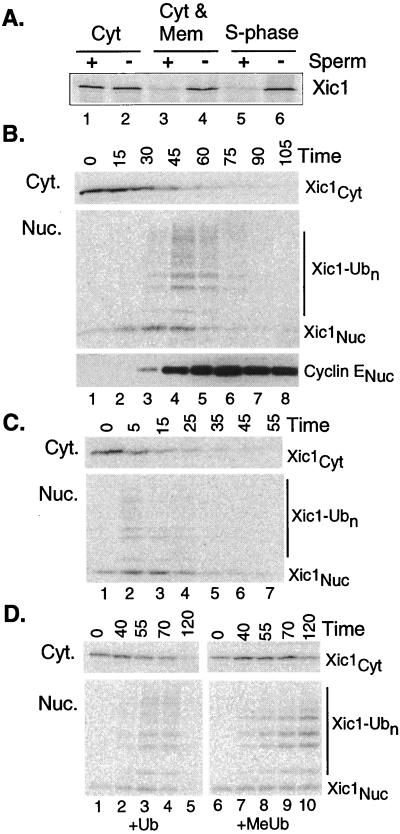
Sperm chromatin is required for Xic1 destruction (16). To study this requirement in more detail, we tested whether nuclear formation and nuclear transport are required for Xic1 destruction. In these assays, sperm chromatin templates are added to crude egg cytoplasm that includes the vesicular components required for nuclear assembly. The sperm rapidly decondenses (<5 min) and binds vesicles, which then fuse to form the nuclear membrane. Nuclear transport is established by ≈20 min, and DNA replication initiates after ≈30 min. Fig. 1A shows that Xic1 is destroyed in S-phase extracts (lanes 5 and 6) or a reconstituted mix of cytosolic and membrane fractions (lanes 3 and 4), but not in the cytosolic fraction alone (lanes 1 and 2). Thus, Xic1 is destroyed only in extracts with membranes in which the nuclear envelope may form. To test whether Xic1 must enter the nuclear compartment to be destroyed, we added the nuclear transport-blocker wheat germ agglutinin, which blocked Xic1 destruction (not shown), thus confirming our hypothesis. The ability of transport blockers to inhibit Xic1 degradation also supports the idea that the in vitro-assembled nuclei specifically import factors for Xic1 destruction and that they are not simply enclosed within assembling nuclei.
Figure 1.
Xic1 is rapidly transported into and ubiquitinated in the nucleus. (A) Destruction of Xic1 requires formation of nuclei. 35S-labeled IVT Xic1 was added to the indicated extract fraction(s) plus or minus sperm DNA. Reactions were processed and analyzed as described in Materials and Methods. Light microscopy confirmed that nuclei formed only in S-phase extract and in the reconstituted cytosolic and membrane fractions (Cyt and Mem). (B) Xic1 is ubiquitinated in the nucleus. Reactions were prepared as in A with sperm DNA, separated into the cytoplasmic and nuclear fractions at the indicated times, and processed as described in Materials and Methods. Samples were analyzed by PhosphorImaging or Western blotting with anti-cyclin E antibody. Subtypes of Xic1 are indicated: Xic1Cyt is the cytoplasmic fraction, Xic1-Ub0 is the unubiquitinated nuclear fraction, and Xic1-Ubn is the ubiquitinated nuclear fraction. The amount of added IVT Xic1 did not measurably affect the normal time course of DNA replication. (C) Xic1 destruction begins rapidly in preformed nuclei. Sperm and energy were mixed with interphase extract and incubated for 50 min to allow nuclei to form. After nuclear formation was confirmed by microscopy, IVT Xic1 and additional extract were added to the reaction (t = 0 min). Samples were removed at indicated times and processed as in B. (D) The modified forms of Xic1 are ubiquitinated. Reactions were prepared as in A and ubiquitin (Ub) or methylated ubiquitin (MeUb) were added and processed at indicated times as in B. To assess the overall effect of MeUb on destruction, comparison of the summed amount of cytoplasmic (Cyt) and nuclear (Nuc) Xic1 remaining was quantitated to be more than 7-fold greater in the sample with added MeUb.
Ubiquitination of Xic1 Occurs Within the Nucleus.
Although these results suggest that nuclear formation and transport are required for Xic1 destruction they do not show where Xic1 ubiquitination and destruction occur. It is possible, for example, that Xic1 enters the nuclear environment, perhaps to be phosphorylated, but is exported before ubiquitination and destruction. Recent work suggests that overexpressed p27Kip1 is destroyed after nuclear export (18). To determine where Xic1 is ubiquitinated and destroyed, we developed a nuclear transport and ubiquitination assay using egg extracts to analyze kinetically how Xic1 and cyclin E/Cdk2 are partitioned between the nuclear and cytoplasmic fractions.
Coupled nuclear assembly–Xic1 destruction reactions containing Xenopus egg extract and trace amounts of 35S-labeled Xic1 are initiated by addition of sperm. At various times, nuclear and cytoplasmic fractions are separated by rapid centrifugation, resolved by SDS/PAGE, and analyzed by autoradiography for Xic1 and Western blotting for cyclin E, Cdk2, and other proteins. The kinetics of Xic1 ubiquitination and destruction are best understood in the context of chromatin and nuclear formation in extracts. In the first 15–20 min after sperm addition, chromatin decondenses, and many chromatin-associated proteins including the origin recognition complex (ORC), Cdc6, and the minichromosome maintenance (MCM) proteins, assemble onto chromatin (data not shown); however, Xic1 and cyclin E remain exclusively in the cytoplasm (see Fig. 1B, Xic1Cyt). After ≈20 min nuclear vesicles bind to chromatin and fuse to form a double membrane containing nuclear pore complexes, and nuclear transport is established. By 30 min, Xic1 and cyclin E rapidly accumulate in the nuclear fraction (Fig. 1B, lane 3) and after Xic1Nuc and cyclin E) and DNA replication begins (not shown). Shortly after cyclin E and Xic1 begin to accumulate in the nuclear fraction, slower migrating forms of Xic1 appear (Xic1-Ubn). These higher forms are rapidly degraded until the overall level of Xic1 is reduced to a background level. The appearance of the slower migrating forms of Xic1 in the nuclear fraction suggests that Xic1 is ubiquitinated in the nucleus.
To confirm that the 30-min time lag before ubiquitinated forms appear (seen in Fig. 1C) is the result of the process of nuclear formation, we added Xic1 to extracts containing preformed S-phase nuclei (see Materials and Methods). Here, the bulk of Xic1 is transported, ubiquitinated, and destroyed in about 15 min (Fig. 1C), confirming that nuclear formation is the rate-limiting step and showing the rapid rate of ubiquitination. In fact, unless we were careful to avoid prematurely mixing radiolabeled Xic1 with the nuclei, ubiquitinated forms appeared almost immediately. Addition of methylated ubiquitin (MeUb), a ubiquitin chain terminator stabilized the upper forms in the nuclear fraction (Fig. 1D). The accumulation of ubiquitinated forms in the nuclear, but not the cytoplasmic fraction, indicates that Xic1 is ubiquitinated in the nucleus.
Association with Cyclin E/Cdk Is Required for Xic1 Destruction.
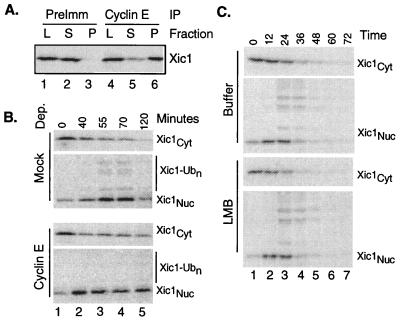
It previously was shown that p21 blocks the destruction of Xic1 (16), suggesting that CDK activity is required. Cyclin E/Cdk2 apparently is required for the destruction of p27Kip1 in human tissue culture cells (14). To test whether cyclin E/Cdk2 is required for Xic1 destruction in Xenopus extracts, we immunodepleted cyclin E from interphase extracts and performed the Xic1 transport and destruction assay. Fig. 2B shows that in the absence of cyclin E, Xic1 enters the nucleus, but is neither ubiquitinated nor destroyed. Presumably, in these conditions Xic1 is not bound to the cyclin/Cdk complex and, being only 27 kDa, freely diffuses in and out of the nucleus. Here, formation of transport-competent nuclei is the only limiting factor for Xic1 nuclear accumulation and accumulation is only partial. In the control reaction, there is a longer delay before Xic1 reaches its maximal concentration in the nucleus, suggesting that active transport of the cyclin/Cdk2/Xic1 complex after nuclear formation is also a limiting factor. Here, nuclear accumulation of Xic1 is more complete, consistent with active transport. In Cdc34-depleted extracts, Xic1 accumulates in the nucleus at a rate and extent similar to the control depletion, presumably in complex with cyclin E/Cdk2, even though it is not ubiquitinated (data not shown). Thus, it is not merely the nuclear destruction of Xic1 that depletes the cytoplasmic pool, it appears to be the active transport of the cyclin/Cdk/inhibitor complex.
Figure 2.
Xic1 is destroyed inside the nucleus in a cyclin E-dependent manner. (A) Cyclin E and Xic1 form a complex in extract. IVT Xic1 was added to extract. After 30 min the reactions were stopped by dilution into buffer with preimmune or anti-cyclin E sera. After 1 h protein A-Sepharose beads were added. After 20 min the beads were isolated and washed. Load (L), supernatant (S), and pellet (P) fractions were resolved by SDS/PAGE. (B) Imunodepletion of cyclin E blocks ubiquitination and destruction but not nuclear accumulation of Xic1. Mock or anti-cyclin E-depleted (3) extracts were assayed for Xic1 transport and destruction as in Fig. 1. (C) The addition of 1 μM LMB does not alter the kinetics of the nuclear accumulation, ubiquitination, and destruction of Xic1.
These conclusions require that the majority of Xic1 forms a complex with cyclin E/Cdk2 in extract. To verify this requirement, we immunoprecipitated cyclin E from extract to which exogenous IVT Xic1 was added and examined the soluble and precipitated fractions by SDS/PAGE and autoradiography. Fig. 2A shows that more than 75% of the added Xic1 coprecipitates with cyclin E (lanes 5 and 6), whereas none coprecipitates in a control reaction with preimmune sera (lanes 2 and 3).
Nuclear Export Is Not Required for Xic1 Destruction.
It recently was shown in mouse fibroblasts that ectopic expression of the Jab1 protein induces the transport of ectopically expressed p27 from the nucleus into the cytoplasm (18). After nuclear export, p27 is destroyed. In these overexpression experiments, the nuclear export inhibitor leptomycin B (LMB) blocked p27 destruction. The effect of LMB on endogenous p27 destruction with or without Jab1 overexpression was not determined. To test whether nuclear export is required for Xic1 destruction, we added LMB to the transport and destruction assay to a concentration that blocks nuclear export of cyclin B in oocytes (19) and the association of the cyclin B nuclear export sequence with Crm1 in oocyte (19) or egg extracts (data not shown, see Materials and Methods). Despite this addition, the kinetics of nuclear accumulation, ubiquitination, and destruction of Xic1 was not delayed (Fig. 2C). Indeed, in many experiments, Xic1 destruction was moderately accelerated in the presence of LMB, suggesting that nuclear export is a back reaction competing with Xic1 destruction, further indicating that Xic1 ubiquitination and destruction occur inside the nucleus.
Disruption of the Nuclear Lamina Blocks DNA Replication but Not Destruction.
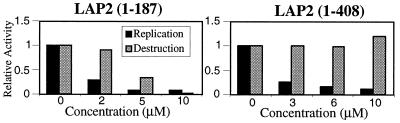
We considered whether the nuclear lamina also might promote Xic1 destruction. To test this possibility, we perturbed the nuclear assembly-ubiquitination reaction with fragments of human LAP2, an integral nuclear membrane and lamin- and chromatin-binding protein. The addition of 2.5–30 μM of LAP2 fragment 1–187, a region sufficient for chromatin binding, blocks lamin assembly, nuclear import, nuclear membrane fusion, and, thus DNA replication (20). In contrast, addition of 1–3 μM of fragment 1–408, the region sufficient for binding to both chromatin and the lamina, does not block lamin assembly or nuclear import, but inhibits nuclear expansion. Higher concentrations of fragment 1–408 (> 6 μM) blocked DNA replication.
We found that fragment 1–187 blocked DNA replication and Xic1 destruction (Fig. 3 Left) in a dose-dependent manner at concentrations similar to those reported. This result was expected because this fragment disrupts nuclear membrane formation. In our hands, fragment 1–408 blocks DNA replication at modest concentrations and, as expected, at high concentrations. However, even at high concentrations Xic1 destruction is unperturbed (Fig. 3 Right). This result indicates that whereas both nuclear formation and the function of the nuclear lamina are required for overall DNA replication, destruction of Xic1 requires only formation of the enclosed environment.
Figure 3.
Disruption of the nuclear lamina does not block Xic1 destruction. Human LAP2 fragments 1–408 and 1–187 (see Results) were added to Xic1 destruction and replication assays (see Materials and Methods). Destruction activity was defined as the fraction of Xic1 degraded as determined by quantification on the PhosphorImager. Values were normalized to the unperturbed samples.
Cyclin E/Cdk Phosphorylation of Xic1 on Threonine 205 Bypasses the Nuclear Requirement for Xic1 Destruction.
Degradation of Xic1 apparently requires basic steps of phosphorylation, ubiquitination, and proteolysis. We were interested in which of these basic steps depends on nuclear formation and first tested whether the concentrating effect of the nucleus on Xic1 destruction could be mimicked by Xic1 phosphorylation.
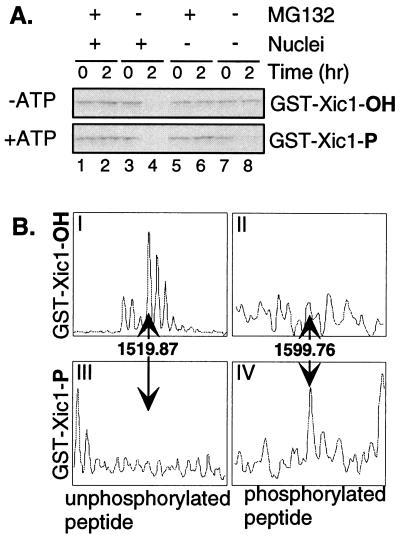
To examine this possibility, we prepared prephosphorylated Xic1 protein by incubating a GST-Xic1 fusion protein with cyclin E/Cdk2 in the presence or absence of ATP. After 30 min at room temperature these reactions were added to extract in the presence or absence of the proteasome inhibitor MG132 and the presence or absence of sperm DNA. Fig. 4A shows that if ATP is added and GST-Xic1 is phosphorylated before addition to extract (+ATP: bottom tier), the protein is reproducibly destroyed even in the absence of nuclei (compare lanes 7 and 8). In this experiment, more than 90% of Xic1 is phosphorylated (see below). However, if GST-Xic1 is not prephosphorylated (−ATP: top tier), nuclei are required for its destruction as shown earlier (Fig. 1A). In each case, Xic1 destruction is blocked by MG132, confirming the destruction is proteosome-mediated. Note that although Xic1 binds cyclin E/Cdk2 in the reaction without ATP, it is not destroyed. Therefore, binding is not adequate to bypass the nuclear requirement; phosphorylation is required.
Figure 4.
Prephosphorylation of Xic1 on Thr-205 by cyclin E/Cdk2 bypasses the nuclear requirement for Xic1 destruction. (A) Prephosphorylated Xic1 is destroyed even in the absence of nuclei. GST-Xic1 was incubated with baculovirus-purified cyclin E/Cdk2 in the presence (lower tier) or absence of ATP (upper tier) for 30 min. Destruction assays were conducted plus or minus sperm and MG132 as indicated. Reactions were processed as in Fig. 1A except that membranes were immunoblotted with anti-Xic1 antibody. (B) Cyclin E/Cdk2 phosphorylates Xic1 on Thr-205. Phosphorylated (III and IV) and unphosphorylated (I and II) GST-Xic1 were prepared by kinase reactions with or without ATP (see Materials and Methods). Samples were digested with trypsin and analyzed by matrix-assisted laser desorption ionization–time of flight MS.
Human p27Kip1 requires phosphorylation of threonine 187 for its destruction (14). This phosphorylation site is located within the C-terminal QT domain. Xic1 also has a C-terminal QT domain highly homologous to that of p27Kip1, containing a homologous CDK phosphorylation site, T205 (Fig. 4B) (21), and five other potential CDK phosphorylation sites. To determine the in vitro cyclin E/Cdk2 phosphorylation sites, we analyzed tryptic digests of Xic1 phosphorylated in vitro by cyclin E/Cdk2 by MS. In the unphosphorylated sample, we observed a strong peak (≈1,519 Da) corresponding to an unmodified peptide containing threonine 205 (Fig. 4BI), but no peak corresponding to the phosphorylated peptide (Fig. 4BII). In the phosphorylated sample we observed a peak (≈1,599 Da) corresponding to the phosphorylated peptide (Fig. 4BIV), but not the unphosphorylated peptide (Fig. 4BIII). We did not observe peaks corresponding to phosphorylated forms of any of the other potential Cdk phosphorylated peptides, although we did detect the unphosphorylated peptides containing each of these sites (not shown). These data suggest that cyclin E/Cdk2 phosphorylation of Xic1 on T205 is sufficient to bypass the nuclear requirement for Xic1 destruction. Nonetheless, other mechanisms including phosphorylation-independent mechanisms may be capable of triggering Xic1 degradation.
The Rate of Phosphorylation of Xic1 by Cyclin E/Cdk2 Is Second-Order with Respect to Xic1.
We suspected that the active concentration of the cyclin E/Cdk2/Xic1 complex in the nucleus favors Xic1 phosphorylation. To examine this possibility, we investigated the mechanism by which cyclin E/Cdk2 phosphorylates Xic1 in vitro.
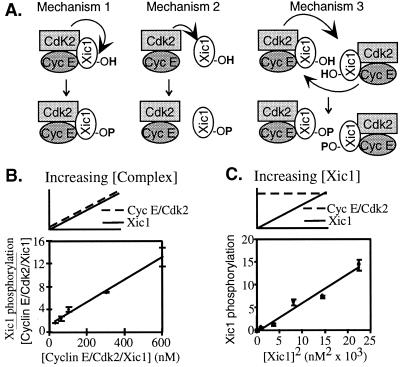
Cyclin E/Cdk2/Xic1 might phosphorylate Xic1 by at least three mechanisms. First, the cyclin E/Cdk2 complex might directly phosphorylate the inhibitor to which it is tightly bound (cis-phosphorylation) (Fig. 5A Left). Second, free cyclin E/Cdk2 might rapidly phosphorylate free Xic1 (trans-phosphorylation) (Fig. 5A Center). Third, a ternary cyclin E/Cdk2/Xic1 complex might phosphorylate the inhibitor associated with another similar ternary complex (also trans-phosphorylation) (Fig. 5A Right). A combination of mechanisms is also possible. The first mechanism is independent of cyclin E/Cdk2 and Xic1 concentration, whereas the second and third mechanisms depend on their concentration.
Figure 5.
Phosphorylation of Xic1 by cyclin E is second order with respect to concentration. (A) Schematic models describing mechanisms by which cyclin E/Cdk2 might phosphorylate Xic1. Mechanism 1 is an intracomplex interaction (first order), whereas mechanisms 2 and 3 are intercomplex interactions (second order). (B) Cyclin E/Cdk2 and MBP-Xic1 were mixed in equimolar amounts. After 30 min at room temperature, the reaction was diluted to the indicated concentrations, and kinase reactions were initiated, processed, and analyzed as described in Materials and Methods. Values for Xic1 phosphorylation were normalized by concentration and fit to a linear curve (R2 = 0.99). (C) Varying concentrations of MBP-Xic1 were incubated with a fixed concentration of cyclin E/Cdk2 (400 nM). After 30 min the Xic1 phosphorylation reactions were initiated, processed, and analyzed as in A except that the counts were plotted versus the concentration squared through and fit to a linear curve (R2 = 0.97).
To distinguish the second and third mechanisms from the first, we incubated equimolar amounts of purified cyclin E/Cdk2 and purified MBP-Xic1 in the absence of ATP so that the components could reach their binding equilibrium without undergoing phosphorylation. Because the on-rate is much faster than the off-rate, the great majority of each species is within the ternary complex (data not shown). This reaction was diluted to a range of final concentrations from less than the concentration of endogenous cyclin E in the extract (≈60 nM) to many times above this concentration, thereby mimicking the active concentration of the complex in the nucleus. Addition of ATP initiated the phosphorylation of cyclin E and Xic1. Reactions were stopped after 3 min and resolved by SDS/PAGE, and the extent of phosphorylation was quantified. In Fig. 5B, the total phosphorylation of Xic1 is normalized by Xic1 concentration and plotted versus the concentration. This operation yields a linear plot (R2 = 0.99), as would be expected for a second-order reaction, and confirms that increasing concentration increases the intrinsic ability of cyclin E/Cdk2 to phosphorylate Xic1. Therefore, cyclin E/Cdk2 apparently phosphorylates Xic1 by a trans mechanism.
Although this result suggests that Xic1 phosphorylation occurs in trans, it does not determine whether the phosphorylation is mediated through free cyclin E/Cdk2 and Xic1 (mechanism two) or through the ternary complex (mechanism three). To distinguish between these two trans-acting mechanisms, we incubated varying concentrations of Xic1 with a fixed concentration of cyclin E/Cdk2 (400 nM) for 30 min to allow for binding. The phosphorylation reactions then were initiated with ATP. In these reaction conditions, if the phosphorylation occurred through the interaction of the individual components as more Xic1 is added, the phosphorylation of Xic1 would be first-order with respect to the concentration of Xic1. However, if the phosphorylation of Xic1 depends on the formation of the ternary complex cyclin E/Cdk2/Xic1, a second-order interaction is introduced. Consequently, as more Xic1 is added more of the trans-acting ternary complex would form and the phosphorylation of Xic1 would be second-order with respect to the concentration of Xic1. Fig. 5C shows that between 0 and 150 nM plotting the extent of Xic1 phosphorylation versus the square of the concentration of Xic1 yields a linear relationship with R2 equal to 0.97. If we plot Xic1 phosphorylation versus the concentration (a first-order interaction) R2 equals 0.87 (consistent with a linear fit to a quadratic). Thus, the data much better fits a second-order dependence on Xic1 concentration. This result suggests that ternary complexes of cyclin E/Cdk2/Xic1 phosphorylate members of other like complexes. Similar results were obtained with purified Xic1 generated by proteolytic cleavage from a GST fusion protein (not shown).
Discussion
We report several observations concerning how the nucleus facilitates the destruction of Xic1, the closest Xenopus homolog to p27Kip1. First, we show that Xic1 is ubiquitinated and destroyed inside transport-competent nuclei. In contrast to an earlier study of p27Kip1 (18), we show that nuclear export is not required for Xic1 ubiquitination or proteolysis. Second, we show that Xic1 destruction requires cyclin E/Cdk2 activity and that cyclin E/Cdk2 forms a complex with Xic1 in extract. Third, we find that disruption of lamina-chromatin interactions with a fragment of the lamina-associated protein LAP2 does not perturb Xic1 destruction, suggesting that destruction of Xic1 principally requires the formation of a transport-competent nuclear compartment. Fourth, we demonstrate that phosphorylation of Xic1 by cyclin E/Cdk2 bypasses the nuclear requirement for its destruction, suggesting that phosphorylation is the nuclear-dependent step for Xic1 destruction. Last, we find that the phosphorylation of Xic1 by cyclin E/Cdk2 is second-order with respect to the concentration of Xic1.
Based on these results we propose a model by which the active concentration of the cyclin E/Cdk2/Xic1 complex in the nucleus initiates the phosphorylation and destruction of Xic1. This model offers an answer for how nuclear formation contributes to the destruction of Xic1 and, ultimately, the regulation of DNA replication. The model also suggests a means by which CDK inhibitors may function as inhibitors or substrates in distinct cellular compartments.
Nuclear Formation, Xic1 Destruction, and DNA Replication.
The observations that nuclear assembly precedes the initiation of DNA replication and that disruption of the nuclear architecture blocks DNA replication as well as many biochemical reconstitution experiments have emphasized the role of the nuclear structure in DNA replication. Ongoing efforts are beginning to elucidate how nuclear formation confers competence for replication.
In this context it was surprising that a soluble extract prepared from crushed nuclei, which is incapable of forming nuclei, is competent to replicate chromosomal DNA (22). This result has been interpreted to indicate that there is no absolute structural requirement for DNA replication. However, it is vital to note that these extracts are prepared from aphidicolin-blocked nuclei that have already fired their origins. Consequently, some of the nuclear requirement for replication before initiation may be bypassed in these extracts.
Our results suggest that in addition to actively concentrating the mechanistic factors responsible for DNA replication, the nucleus concentrates the cyclin E/Cdk2/Xic1 complex to promote the phosphorylation of Xic1. This process may be part of the mechanism by which the nucleus facilitates destruction of Xic1, the subsequent activation of cyclin E/Cdk2, and the initiation of DNA replication. This mechanism would be active before the initiation of DNA replication.
From Inhibitor to Substrate.
Our results also address the question of how a CDK inhibitor becomes a CDK substrate at the appropriate moment. The biochemical analysis of Roberts and coworkers (14) demonstrates that p27 can interact with cyclin E/Cdk2 transiently through the cyclin subunit before adopting a tightly bound inhibitory state in which it also binds the CDK subunit. Those authors propose that during this initial interaction, p27 can be phosphorylated. However, they also show that the transition to the inhibitory state is rapid (about 1 min), and the off-rate slow (about 2 h), suggesting that the cyclin E/Cdk2/inhibitor complex exists mostly in the tightly bound state. Therefore, the critical physiological question is how the population of the inhibitor that is tightly bound becomes phosphorylated.
As our work and previous work suggest (14), there is kinase activity associated with the inhibited complex even after equilibrium is reached. Two possibilities explain this observation: first, the more transient CDK interaction alternates between inhibitory and noninhibitory states, thereby allowing for rare phosphorylation of p27; second, a small subpopulation of free kinase may phosphorylate the inhibitor through the transitory interaction described above. Either case is a trans interaction and the rate of trans-phosphorylation will depend on the concentration of both enzyme and substrate. If the complex were only capable of phosphorylating the inhibitor to which it is bound there would be no concentration dependence. Our results confirm that the inhibitor complex is phosphorylated in a concentration-dependent manner. This concentration dependence likely enables cyclin E/Cdk2 to overcome the inhibitory effect of its inhibitor, thereby tipping the balance so that the inhibitor becomes a substrate.
Generating the Switch.
Once phosphorylation occurs and the proteolysis pathway is operational a positive feedback loop is established. As the proteolysis machinery destroys the inhibitor a subpopulation of the kinase is activated and able to rapidly phosphorylate more of the inhibitor, leading to more destruction and activation. Therefore, in this scenario, a concentration-dependent switch triggers cyclin E/Cdk2 activation. Advantages to this model are that it evokes only the physiological observation that the cyclin complex is concentrated in the nucleus and the enzymatic details of the cyclin E/Cdk2 p27 interaction.
Multiple Mechanisms for Xic1 Destruction?
One prediction of this model is that mutation of critical phosphorylation sites in Xic1 would block Xic1 destruction. In fact, we find that mutation of the six putative serine-proline (SP) or threonine-proline (TP) CDK phosphorylation sites to alanine-proline (AP) does not completely disrupt Xic1 destruction. Nevertheless, as indicated above, cyclin E/Cdk2 phosphorylation of Xic1 bypasses the nuclear requirement. Therefore, there appears to be multiple mechanisms by which Xic1 is destroyed. The mechanism we describe here is phosphorylation-dependent and normally is facilitated by nuclear concentration, but does not strictly depend on the nuclear compartment because this requirement can be bypassed by phosphorylation. The other mechanism is phosphorylation-independent, but apparently occurs normally within the nucleus because we don't observe any cytoplasmic degradation. We currently are working to reconcile these mechanisms, but an interesting possibility is that in the early embryo the phosphorylation dependence is reduced and that one reflection of the appearance of a G1 phase at the time of gastrulation is an increase in the phosphorylation-dependence for p27 destruction. Indeed, the mechanisms that regulate p27 destruction, including phosphorylation, may be among the most important determinants of the length of G1.
Abbreviations
- CDK
cyclin-dependent kinase
- GST
glutathione S-transferase
- IVT
in vitro-translated
- MBP
myelin basic protein
- MeUb
methylated ubiquitin
- LMB
leptomycin B
References
- 1.Fang F, Newport J W. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- 2.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson P K, Chevalier S, Philippe M, Kirschner M W. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 5.Shou W, Dunphy W G. Mol Biol Cell. 1996;7:457–469. doi: 10.1091/mbc.7.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 7.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 8.Verma R, Feldman R M, Deshaies R J. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 10.Mathias N, Johnson S L, Winey M, Adams A E, Goetsch L, Pringle J R, Byers B, Goebl M G. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 12.Alessandrini A, Chiaur D S, Pagano M. Leukemia. 1997;11:342–345. doi: 10.1038/sj.leu.2400581. [DOI] [PubMed] [Google Scholar]
- 13.Montagnoli A, Pagano M. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 15.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yew P R, Kirschner M W. Science. 1997;277:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]
- 17.Hua X H, Yan H, Newport J. J Cell Biol. 1997;137:183–192. doi: 10.1083/jcb.137.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomoda K, Kubota Y, Kato J. Nature (London) 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gant T M, Harris C A, Wilson K L. J Cell Biol. 1999;144:1083–1096. doi: 10.1083/jcb.144.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su J Y, Rempel R E, Erikson E, Maller J L. Proc Natl Acad Sci USA. 1995;92:10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter J, Sun L, Newport J. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]