Abstract
Under different circumstances, natural killer T (NKT) cells can cause a T helper (Th) 1 or a Th2 polarization of immune responses. We show here, however, that mouse NKT cells with an invariant Vα14 rearrangement (Vα14i NKT cells) rapidly produce both IL-4 and IFN-γ, and this pattern could not be altered by methods that polarize naive CD4+ T cells. Surprisingly, although cytokine protein was detected only after activation, resting Vα14i NKT cells contained IL-4 and IFN-γ mRNAs. Despite this finding, in vivo priming of mice with the glycolipid antigen recognized by Vα14i NKT cells resulted in a more Th2-oriented response upon antigen re-exposure. The Vα14i NKT cells from primed mice retain the ability to produce IL-4 and IFN-γ, but they are less effective at activating NK cells to produce IFN-γ. Our data therefore indicate that Vα14i NKT cells have a relatively inflexible immediate cytokine response, but that changes in their ability to induce IFN-γ secretion by NK cells may determine the extent to which they promote Th1 responses.
CD1d-reactive natural killer T (NKT) cells are capable of the rapid production of both IL-4 and IFN-γ. These cells express an invariant T cell antigen receptor (TCR) α chain comprised of a Vα14–Jα18 rearrangement and they recognize a nonpolymorphic class I antigen-presenting molecule, CD1d. Their response to CD1d is enhanced by the glycosphingolipid α-galactosylceramide (αGalCer) (1, 2). Because cells with this invariant Vα14 rearrangement and CD1d specificity do not always express NK1.1, we refer to these cells as Vα14 invariant (Vα14i) T cells (3).
The results from studies of animal models of autoimmune diseases (4–7), as well as in human autoimmune disease patients (8–11), support the hypothesis that Vα14i T cells and their human homologs may regulate the immune response. They are also involved in the prevention of tumor metastases (12, 13) and in augmenting the responses to several infectious agents (14–16). Consistent with a role in immune regulation, there is a selective decrease of IFN-γ compared with IL-4 production after repeated doses of αGalCer (17). This could be responsible for the beneficial effects of αGalCer treatment of diabetes-prone NOD mice and in experimental allergic encephalomyelitis models (18–22). Curiously, several investigators have observed an opposite effect of αGalCer priming, with IFN-γ levels increased (23), and in a number of cases the beneficial effect of αGalCer depended on increased IFN-γ rather than IL-4 (24, 25). The reason for the apparently diverse mechanisms by which activated Vα14i T cells exert these effects is unknown; cytokine polarization of the Vα14i T cells themselves may be responsible.
To examine the potential to polarize Vα14i T cells in vivo, we analyzed both the ex vivo cytokine production by Vα14i T cells from the liver and the spleen of mice given αGalCer and the systemic responses of these mice by measuring the cytokines released immediately in the blood. The data reveal that Vα14i T lymphocytes are resistant to cytokine polarization in vivo, which may be related to the presence of IL-4 and IFN-γ cytokine transcripts in these cells in the resting state.
Material and Methods
Mice and Treatments. BALB/c, IL-12Rβ2-/-, and IL-4Rα-/- mice on the BALB/c background were purchased from The Jackson Laboratory. C57BL/6 mice were offspring of stock originally obtained from The Jackson Laboratory. CD40-/- mice on the C57BL/6 background (26) were provided by S. Schoenberger (La Jolla Institute for Allergy and Immunology). CD28-/- mice on the C57BL/6 background have been described (27). All mice were maintained under specific pathogen-free conditions. Unless otherwise indicated, mice were injected i.v. with 2 μg of αGalCer, provided by the Pharmaceutical Research Laboratory of Kirin Brewery (Gunma, Japan). C57BL/6 and BALB/c mice demonstrated comparable Vα14i T cell responses to αGalCer, both by intracellular cytokine staining and serum cytokine levels (see Figs. 2 and 4). Miniosmotic pumps (Alzet 2001, Alza) were filled with 100 pg, 10 ng, 200 ng, or 4 μg of α-GalCer in 0.5% polysorbate-20 and 0.9% NaCl and implanted s.c. following the manufacturer's instructions. Mice were analyzed 7 days after implantation.
Cytokine Reporter Mice. The 4get (IL-4 GFP enhanced transcript) mice have been described (28). To generate IFN-γ reporter mice, a fragment containing exons 2–4 and 2.5 kb of 3′ untranslated sequence of the Ifng gene was inserted into the pgkTK vector containing the herpes simplex virus thymidine kinase gene for negative selection (29). A bicistronic reporter cassette, containing an encephalomyocarditis virus internal ribosome entry site element, modified as described (30), was cloned 5′ of enhanced yellow fluorescent protein (eYFP) followed by a bovine growth hormone polyadenylation signal (CLONTECH). A loxP-flanked neomycin resistance cassette, derived from pL2neo2 (31), was placed at the 3′ end to generate the final cassette, which was cloned into the BamHI and SalI sites in the mutated Ifng gene. This targeting construct was electroporated into PrmCre embryonic stem cells, which express Cre recombinase under control of the protamine promoter (32), and selection was achieved by using 400 μg/ml G418 and 2 μM gancyclovir. The neomycin resistance cassette is deleted in the male germ line by Cremediated recombination. Chimeric males were bred to WT C57BL/6 mice, and offspring were selected by using Southern blotting for the mutated knock-in Ifng allele and for deletion of the neomycin cassette. Mice were backcrossed four generations onto C57BL/6 and used as heterozygotes. Targeted mice were designated Yeti, for yellow-enhanced transcript for IFN-γ and will be described in more detail elsewhere (M.M., K. Mohrs, D.B.S., R. L. Reinhardt, and R.M.L., unpublished work).
Flow Cytometry and Cytokine Assays. Cells were stained with αGalCer/CD1d tetramers as described (33). mAbs used in this study for flow cytometry include FITC-, CyChrome-, or allophycocyanin (APC)-labeled anti-TCRβ clone H57-597, phycoerythrin (PE)- or APC-labeled anti-NK1.1 clone PK136, and PE-labeled pan-NK clone DX5 (PharMingen). For intracellular staining, cells were incubated with blocking 2.4G2 anti-FcγR mAb and neutravidin (Molecular Probes) and then surface stained. Cells were permeabilized by using Cytofix/Cytoperm (PharMingen) and stained by using FITC-labeled anti-IL-4 clone BVD4–1D11 (Caltag Laboratories, Burlingame, CA, and PharMingen) PE-labeled anti-IL-4 clone BVD4–1D11 and FITC-labeled anti-IFN-γ clone XMG1.2 (PharMingen) according to the manufacturer's protocol. Intracellular cytokine staining of NK cells was done by gating on DX5+ TCRβ- cells. Cytokines in the serum were evaluated in a sandwich ELISA using anti-IL-4 and IFN-γ mAbs (PharMingen).
Quantitative RT-PCR. Total RNA was extracted from sorted cells with the TRIzol solution. Reverse transcription was carried out by using the First Strand cDNA Synthesis Kit (Novagen) and oligo(dT) priming. The amount of amplicon generated during PCR was monitored by using an Applied Biosystems PRISM 5700 apparatus. A specific probe labeled with both a reporter and a quencher dye was added into the TaqMan PCR mix (Perkin–Elmer) at the beginning of the reaction. The sequences of the primers and TaqMan probes used in this study were intron-spanning and have been published (28).
Results
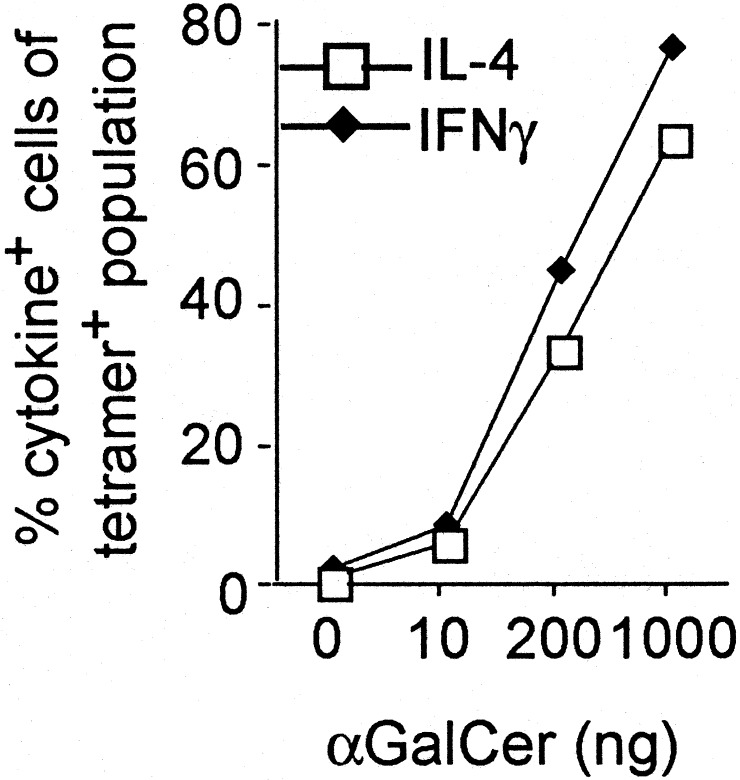
Antigen Dose Does Not Influence the Pattern of Cytokine Production. Because altering the dose of antigen can polarize naïve, conventional CD4+ T cells (34, 35), we analyzed the ability of αGalCer to elicit IL-4 and IFN-γ production at different doses. The CD1d tetramer allowed us to focus on Vα14i T cells exclusively and carry out the analysis ex vivo. Consistent stimulation of a significant fraction of the Vα14i T cells required 50–200 ng of αGalCer. Even at the lowest stimulating dose, however, both IL-4 and IFN-γ were detected by intracellular cytokine staining (Fig. 1) or analysis of the sera by ELISA (data not shown). At all doses that stimulated, responding lymphocytes produced both IL-4 and IFN-γ by intracellular cytokine staining (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Another method to polarize conventional CD4+ T cells is by the slow delivery of antigen by an osmotic pump placed under the skin (34, 36). When done with αGalCer, tetramer+ cells disappeared when the highest 4-μg dose was placed in the pump. Tetramer+ cells from mice that received lower doses, from 100 pg to 200 ng in the pumps, all produced both IL-4 and IFN-γ after restimulation (data not shown).
Fig. 1.
The effect of antigen dose on the cytokine response of Vα14i T cells. Percentage of CD1d tetramer+ cells in the liver that stained for intracellular IL-4 (□) or IFN-γ (⋄) as a function of the dose of αGalCer given to C57BL/6 mice 2 h previously. n = 2–3 for each condition, with at least two independent experiments performed for each case.
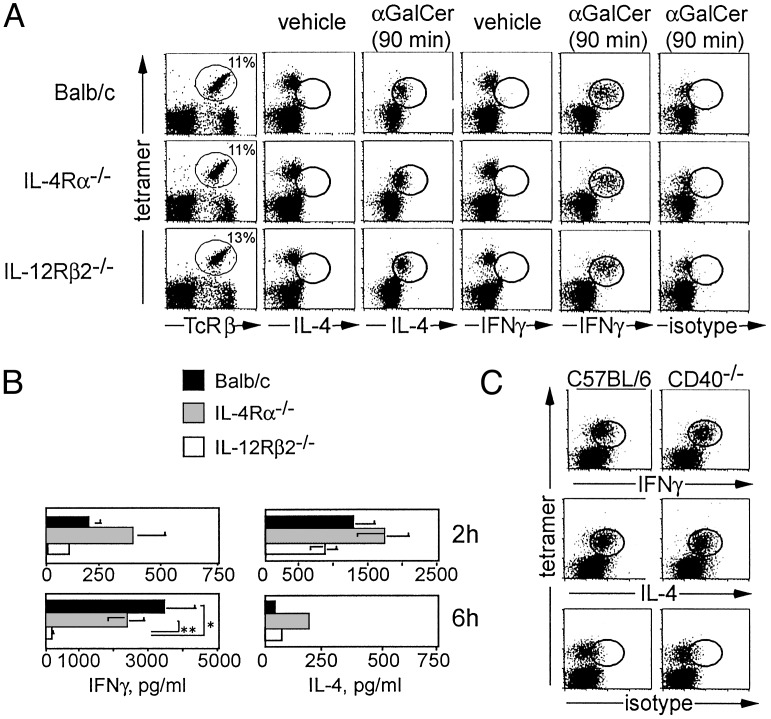
Vα14i T Cells Do Not Depend on Cytokine Receptor Signaling. To investigate the dependence of Vα14i T cells on IL-4 and IL-12 for their ability to produce cytokines, we analyzed mice with mutant cytokine receptor genes. Two hours after αGalCer administration, intracellular cytokine staining of CD1d tetramer+ T cells in the liver revealed that intracellular IL-4 was not reduced in cells from the IL-4Rα-/- mice (Fig. 2A). Likewise, IFN-γ was not reduced in Vα14i T cells from IL-12Rβ2-/- mice. Similar Vα14i T cell responses were observed in the spleen (data not shown). To measure the systemic cytokine response of Vα14i T cells after αGalCer stimulation, we analyzed sera 2 and 6 h after injection (Fig. 2B). Two hours after αGalCer injection, both the IL-4Rα-/- and IL-12Rβ2-/- mice had serum IL-4 and IFN-γ levels similar to that of the WT controls. By 6 h, the IL-4 levels found in all three strains were decreased. IFN-γ levels increased markedly in the serum of WT and IL-4Rα-/- mice by 6 h. In IL-12Rβ2-/- mice, however, serum IFN-γ was decreased >10-fold compared with IL-4Rα-/- or WT mice.
Fig. 2.
The effect of cytokine receptor deficiency and CD40 on cytokine synthesis by Vα14i T cells. (A) Intracellular cytokine detection. The left column shows staining for CD1d tetramer+ TCRβ+ liver mononuclear cells from uninjected BALB/c, IL-4Rα-/-, and IL-12Rβ2-/- mice. Also shown is intracellular cytokine staining of liver mononuclear cells 90 min after i.v. administration of vehicle (columns 2 and 4) or 2 μgof αGalCer (columns 3, 5, and 6). Between 89% and 95% of CD1d tetramer+ TCRβ+ cells stained for intracellular IL-4 and IFN-γ. For vehicle-injected control mice and isotype control staining of αGalCer-injected mice, the percentage of CD1d tetramer+ TCRβ+ cells that were positive for either IL-4 and IFN-γ or for the isotype mAb was <3.5%. Three mice from each strain were injected. Two independent experiments were performed with similar results. (B) Serum cytokine levels. BALB/c controls, IL-4Rα-/-, and IL-12Rβ2-/- mice were analyzed 2 or 6 h after i.v. injection of 2 μg of αGalCer. At 2 h, BALB/c mice (n = 6), IL-4Rα-/- mice (n = 4), and IL-12Rβ2-/- mice (n = 6) were analyzed. At 6 h, BALB/c mice (n = 4), IL-4Rα-/- mice (n = 2), and IL-12Rβ2-/- mice (n = 4) were analyzed. Statistical analysis was performed by using t tests to compare cytokine levels for all strains at each time point, and significant differences are indicated. *, P = 0.02; **, P = 0.01. (C) Intracellular cytokine staining of liver lymphocytes 2 h after i.v. injection of 2 μgof αGalCer in C57BL/6 WT controls (n = 2) or CD40-/- mice (n = 2). Percentage of CD1d tetramer+ TCRβ+ cells that were positive for IL-4 and IFN-γ ranged from 87% to 94%, whereas staining with the isotype control mAb was <5%.
CD40/CD40L interactions have been reported to be required for IFN-γ production by cultures containing Vα14i NKT cells, whereas in the same study, CD28-B7 interactions were required for the production of both IL-4 and IFN-γ (37). Using intracellular cytokine staining, however, we determined that virtually all CD1d/αGalCer tetramer+ cells in the liver produced both IFN-γ and IL-4 in CD40-/- mice 2 h after αGalCer administration (Fig. 2C). Likewise, the production of IFN-γ (≈50 pg/ml) and IL-4 in the serum at 2 h after antigen in CD40-/- mice is similar to the levels produced by activated Vα14i T cells from WT mice. However, at 6 h, the level of IFN-γ is much reduced (Fig. 7, which is published as supporting information on the PNAS web site), possibly because of an inability of activated Vα14i T cells in the CD40-/- mice to induce IL-12 production by dendritic cells or other cell types (38). We conclude that CD40–CD40L interactions are not required for the immediate IFN-γ production by Vα14i T cells, although, as in the IL-12Rβ2-/- mice (Fig. 2B), they are required for the production of systemic IFN-γ in the serum. Similarly, Vα14i T cells from CD28-/- mice retained the ability to produce cytokines after αGalCer stimulation (data not shown). Therefore, activation of Vα14i T cells and their secretion of a mixed cytokine pattern can occur in the absence of the costimulatory molecules that are important for naive T cells.
Reporter Mice Reveal Cytokine mRNAs in Resting Vα14i T Cells. To confirm these findings, we used cytokine gene reporter mice that enable fluorescent detection of cytokine gene expression. The IL-4 reporter or 4get mice have a construct that enables IL-4 and enhanced GFP (eGFP) expression from a single bicistronic transcript. IFN-γ reporter mice, designated Yeti, have an internal ribosome entry site-eYFP cassette immediately downstream of the IFN-γ translation stop and upstream of the endogenous polyadenylation signal. Because the fluorescent marker proteins are trapped intracellularly, the reporter mice enable direct single-cell cytokine detection, while leaving the endogenous cytokine ORFs unmodified.
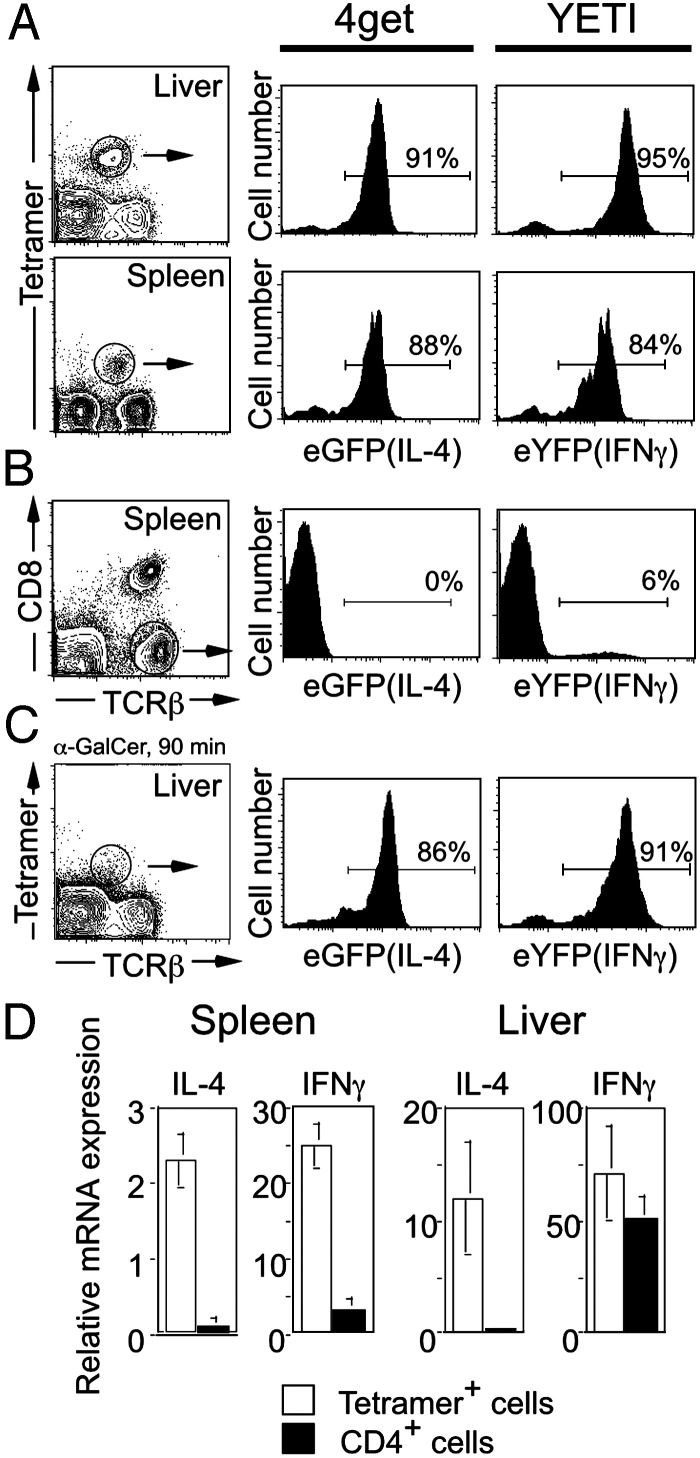
Vα14i T cells were studied in the liver and spleen of the two reporter mice before and after activation by αGalCer (Fig. 3). Unexpectedly, the vast majority of Vα14i T cells in the liver and spleen of mice not exposed to αGalCer were eGFP+ in 4get mice and eYFP+ in Yeti mice (Fig. 3A). Despite this finding, the IL-4 and IFN-γ proteins were not detected by intracellular cytokine staining in unstimulated Vα14i T cells from these gene-targeted mice (data not shown) or WT mice (Fig. 2 A and ref. 33). In the spleen, few conventional TCRβ+ cells were eGFP+ (<1%) or eYFP+ (6%) (Fig. 3B and ref. 30), demonstrating that the expression of the reporter genes is most prevalent in Vα14i T cells. After activation of Vα14i T cells in vivo with αGalCer, tetramer staining revealed evidence for TCR down-regulation, but little difference in the mean fluorescence intensities for eYFP or eGFP (Fig. 3C). Taken together, analysis using cytokine reporter mice suggests that Vα14i T cells, in contrast to most conventional T cells, express cytokine transcripts in the resting state, but express protein only after stimulation.
Fig. 3.
Resting Vα14i T cells contain cytokine mRNA. (A) Vα14i T cells in the liver and spleen of 4get (IL-4 locus) and Yeti (IFN-γ locus) mice were analyzed for their expression of eGFP (IL-4 mRNA) and eYFP (IFN-γ). Histogram panels are data from gated αGalCer/CD1d tetramer+ TCRβ+ cells. (B) Conventional T cells in the spleen do not express eGFP or eYFP. Histogram panels are gated on T cells (CD1d tetramer- CD8- TCRβ+). (C) Vα14i T cells in the liver of 4get and Yeti mice were analyzed for their expression of eGFP and eYFP after in vivo stimulation with αGalCer. Similar results were obtained from the spleen (data not shown). Histogram panels are gated on αGalCer/CD1d tetramer+ TCRβ+ cells. Each of the plots represents pooled cells from three mice. Two individual experiments were performed with similar results. (D) IL-4 and IFN-γ mRNA are present in Vα14i T cells from WT mice. Real-time PCR was performed with primers specific for IL-4 and IFN-γ by using cDNA prepared from Vα14i T cells and CD4+ T cells (CD1d tetramer-) sorted from the spleen and liver of C57BL/6 mice. Cytokine amplicons were normalized against the levels of hypoxanthine phosphoribosyltransferase amplified in each sorted population. Statistical analysis was performed by using t tests to compare the levels of cytokine messenger detected in tetramer+ cells versus CD4+ tetramer- cells. *, P < 0.05.
Cytokine mRNA in Vα14i T Cells from WT Mice. The results from the reporter mice raised the possibility that cytokine mRNAs might be present in Vα14i T cells and revealed by the unconstrained translation of the fluorescent proteins, mediated by translation initiated from the internal ribosome entry site element. To determine whether cytokine transcripts also were present in Vα14i T cells from WT mice, we sorted Vα14i T cells from the liver and spleen of C57BL/6 mice and performed real-time fluorogenic PCR to detect IL-4 and IFN-γ. Hypoxanthine phosphoribosyltransferase was used to normalize for differences in the amounts of starting cDNA for each sample. Vα14i T cells from the liver and spleen contained both IL-4 and IFN-γ mRNA (Fig. 3D). By contrast, CD4+ T cells from the spleen contained ≈10-fold less mRNA for both IL-4 and IFN-γ. CD4+ T cells from the liver did not contain IL-4 mRNA, but they did contain some IFN-γ mRNA, although less than Vα14i T cells. This finding is perhaps consistent with the presence of activated, T helper 1 effector cells at this site.
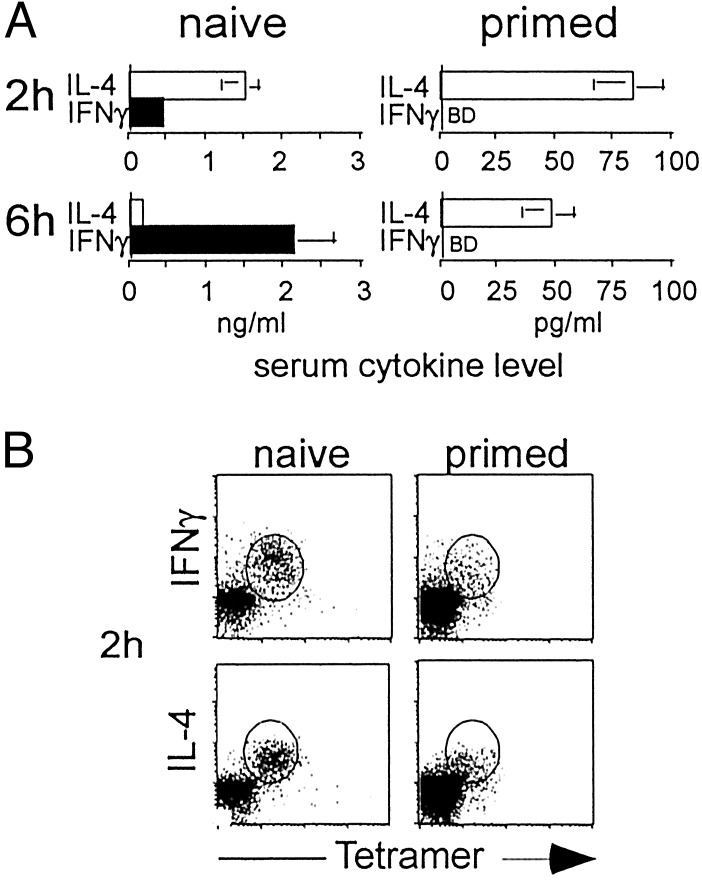
Absence of Cytokine Polarization in Primed Vα14i T Cells. The inability to polarize cytokine production by Vα14i T cells would appear to contradict our previous reports, in which the IL-4 to IFN-γ ratio was increased when spleen cell suspensions from mice previously primed with αGalCer were restimulated in vitro (17, 39). To analyze the effect of αGalCer priming in vivo on subsequent Vα14i T cell responses, cytokine levels in the blood of mice reimmunized with αGalCer were determined by ELISA (Fig. 4A). At 2 h, the production of IL-4 in the primed mice was reduced in proportion to the reduction in the percentage of tetramer+ cells found in these mice given αGalCer 1 week earlier (≈10-fold). IFN-γ was not detectable at 2 h after restimulation, but this finding is consistent with the low level of IFN-γ detected in the sera of naïve mice and a proportional decrease in the tetramer+ cells. At 6 h after injection, however, IFN-γ still could not be detected in the sera from the primed mice. This decrease was greater than the proportional decrease in Vα14i T cells, as a proportional 10- to 15-fold decrease below the level in the control mice still would have been nearly 10-fold above the detection limit. Therefore, primed mice re-exposed to αGalCer retained the ability to produce some systemic IL-4 rapidly, whereas IFN-γ could not be detected. The T helper 2 (Th2) trend in these data are similar to the in vitro results, although in the in vitro cultures IL-4 was not decreased (17). In these in vitro cultures, however, the dynamics of Vα14i T cell cytokine production could be altered, and furthermore, cells activated by the Vα14i T cells could have contributed to the IL-4 synthesis.
Fig. 4.
Cytokine production by Vα14i T cells in αGalCer-primed mice. (A) IL-4 and IFN-γ in the sera were measured by ELISA 2 or 6 h after injection of αGalCer into naïve mice or C57BL/6 mice primed 1 week earlier. BD, below detection. At 2 h, five naïve mice and five primed mice were analyzed. At 6 h, nine naïve mice and seven primed mice were analyzed. Statistical analysis was performed by using t tests to compare the levels of each cytokine at each time point for naïve versus primed mice. For all comparisons, P < 0.05. IFN-γ levels at 6 h in naïve versus primed mice, P = 0.01. (B) Intracellular cytokine staining of liver lymphocytes 2 h after i.v. injection of 2 μgof αGalCer in naïve or primed mice. Percentage of CD1d tetramer+ TCRβ+ cells that were positive for IL-4 and IFN-γ ranged from 83% to 94%. The percentage of CD1d tetramer+ TCRβ+ cells isolated from naïve mice ranged from 16% to 24% whereas the percentage of CD1d tetramer+ TCRβ+ cells in primed mice ranged from 2% to 5%. Data are representative of eight naïve mice and seven primed mice analyzed with similar results.
We also analyzed antigen-induced cytokine production in primed Vα14i T cells by intracellular cytokine staining. In agreement with previous results, the number of Vα14i T cells declined rapidly and became nearly undetectable within hours after αGalCer administration (33, 40). This population did not recover completely within the first week after antigen priming, although more rapid recoveries have been observed in other experiments (18). Slowed recovery of Vα14i T cells may relate to persistent antigen. Unexpectedly, however, intracellular cytokine staining of Vα14i T cells taken from primed mice reinjected with αGalCer showed both IFN-γ and IL-4 (Fig. 4B). Therefore, as in the IL-12Rβ2-/- and CD40-/- mice, the immediate response of Vα14i T cells in the primed mice did not reflect the relatively Th2 polarized serum response to αGalCer.
Regulated NK Cell Activation by Vα14i T Cells. Although IL-4 levels in the sera were maximal at 2 h, IFN-γ levels peaked later when the Vα14i T cells were no longer detected by flow cytometry. This lag suggested that most of the systemic IFN-γ in the blood might be produced by a different cell type. Previous reports have demonstrated that activated Vα14i NKT cells stimulate NK cells in an IFN-γ-dependent fashion to produce IFN-γ (41, 42). We therefore sought to determine whether the diminished levels of IFN-γ in the sera of αGalCer-primed mice and IL-12Rβ2-/- mice were caused by a failure of NK cells to produce IFN-γ after Vα14i T cell activation. Consistent with an earlier report (41), we found that ≈20–30% of the liver NK cells were positive for IFN-γ 6 h after injection of αGalCer in naïve mice (Fig. 5A). By contrast, NK cells of both αGalCer-primed mice and IL-12Rβ2-/- mice failed to produce detectable levels of IFN-γ. In fact, the IFN-γ detected in the sera at this time point was similar when IL-12Rβ2-/- mice were compared with mice treated with anti-asialo GM1 before αGalCer injection (Fig. 5B). This anti-asialo GM1 treatment specifically depleted NK cells, but did not affect the percentage of CD1d tetramer+ cells (data not shown). Despite this, sorted liver NK cells from the αGalCer-primed mice retain the ability to make IFN-γ in response to culture with IL-12 (Fig. 8, which is published as supporting information on the PNAS web site). Hence, although Vα14i T cells do not appear to be polarized by repeated exposures to αGalCer, or by the absence of IL-12 signaling, the systemic immune response in these mice becomes relatively Th2 polarized, which may be caused by a disruption in the Vα14i T cell-mediated activation of IFN-γ release by NK cells.
Fig. 5.
NK cells in αGalCer-primed mice and IL-12Rβ2-/- mice produce less IFN-γ.(A) Intracellular IFN-γ content of DX5+TCRβ- cells from the liver 6 h after αGalCer injection. Percentages of IFN-γ+ (boldface line) or isotype+ cells are indicated. One representative mouse is shown for each strain. For WT, n = 5; primed mice, n = 5; and IL-12Rβ2-/- mice, n = 4. (B) Comparison of IFN-γ levels in sera 6 h after αGalCer injection in WT, asialo-GM1 treated, and IL-12Rβ2-/- mice. One representative experiment of two is shown. Statistical analysis was performed by using t tests to compare IFN-γ levels in WT versus GM1-treated and IL-12Rβ2-/- mice. *, P = 0.003; **, P = 0.005.
Discussion
The role of Vα14i T cells in preventing autoimmune disease and in protective immunity has been studied extensively. The production of IFN-γ appears to be the major mechanism for the beneficial effect of αGalCer stimulation of Vα14i T cells in the clearance of microbial pathogens (14, 43) and the response to tumors (25). By contrast, the αGalCer-mediated stimulation of IL-4 and/or IL-10 production underlies its beneficial effects in the prevention of several autoimmune diseases (18–22) and the induction of anterior chamber-associated immune deviation (44). It is not known how a single glycolipid antigen can lead to modulation of the immune response by such divergent pathways.
Our principal approach was to analyze the response immediately after activation of Vα14i T cells in vivo. Efforts to polarize Vα14i T cells by altering the dose, the route, or the timing of antigen failed to result in an alteration in the immediate cytokine production by these cells. Furthermore, ex vivo studies with cells from IL-4Rα-/- and IL-12Rβ2-/- mice revealed that the cytokine profile of Vα14i T cells did not depend on IL-4- or IL-12. It therefore appears that Vα14i T cells have relatively little plasticity in modulating their mixed cytokine response. The relatively invariant cytokine profile elicited from Vα14i T cells is consistent with their having a role in the early or innate-type immune response. NK cells also have a fixed cytokine profile, dominated by IFN-γ, although NK cell synthesis of IL-4 during their development (45) and under Th2 polarizing conditions in vitro (46) has been reported.
While this manuscript was in preparation, ex vivo analyses were reported using human Vα24i NKT cells, which are homologous to Vα14i T cells. In humans, the CD4+ subset produced more IL-4 than the CD4- subset (47, 48), a difference that was not observed in the mouse. Results from in vitro culture of adult human Vα24i NKT cells, however, also suggested that it may be difficult to polarize cytokines in these cells (49).
Previous reports suggested that repeated exposure to αGalCer favors Th2 responses (17, 39). In our analysis, there were several instances where the intracellular cytokine staining of activated Vα14i T cells did not reflect the cytokine levels measured in the sera, including in IL-12Rβ2-/- and CD40-/- mice. In these cases, Vα14i T cells retained the ability to produce both IL-4 and IFN-γ, although the systemic IFN-γ decreased at 6 h after αGalCer stimulation. This finding correlated with decreased IFN-γ production by NK cells from the IL-12Rβ2-/- mice, which are responsible for the bulk of the systemic IFN-γ production after αGalCer activation of Vα14i NKT cells. Although it was well established that IL-12 contributes to the production of IFN-γ after αGalCer injection (38, 41, 50), here we show that IL-12-mediated signaling is required for IFN-γ production by NK cells after Vα14i T cell activation, but not by Vα14i T cells themselves. Our data do not formally prove, however, that NK must express IL-12Rβ for the production of IFN-γ. It remains possible that the absence of IL-12Rβ on Vα14i T cells affects IFN-γ production by NK cells in an indirect manner. Our results with the CD40-/- mice are consistent with studies demonstrating the importance of CD40/CD40L interactions between Vα14i T cells and dendritic cells for inducing IL-12 production by these antigen-presenting cells (51). We suggest that the observed decrease in IFN-γ production reported in CD40-/- mice after in vitro activation of Vα14i T cells (37) also may be caused by a failure of NK cells in the mixed-in cultures that contained many cell types, rather than to cytokine polarization of Vα14i NKT cells. Like CD40, the NK1.1 molecule has also been proposed to bias Vα14i NKT cells to produce IFN-γ (52). Further studies will be required to investigate the role of other costimulatory or accessory molecules has on cytokine production by Vα14i NKT cells and their downstream effectors.
The systemic response also was Th2 polarized in mice reimmunized with αGalCer. The decreased production of IFN-γ by NK cells after αGalCer restimulation might reflect the quantitative decrease in Vα14i T cells we observed. It remains possible, however, that qualitative changes in the Vα14i T cells, such as the predominance of relatively immature, recent thymic emigrants, is responsible. Additionally, αGalCer priming may cause changes in antigen-presenting cell function (53). Our study involved repeat exposure to αGalCer with 1 week between initial and final exposure. More than one restimulation with αGalCer, or different time periods between αGalCer administrations, might affect the cytokine production of Vα14i NKT cells. Regardless, in all of the cases we have studied, the ability of Vα14i T cells to induce systemic IFN-γ release correlated with their ability to activate NK cells.
We have made the intriguing observation that resting Vα14i T cells contained mRNA for both IL-4 and IFN-γ. The results from RT-PCR analysis of Vα14i T cells from WT mice corroborated the data from the cytokine gene reporter mice. Despite this, analysis of cells from WT and reporter mice for intracellular cytokines failed to detect any IL-4 or IFN-γ protein in Vα14i T cells until they were stimulated. The presence of cytokine transcripts in resting Vα14i T cells is correlated with their immediate effector capacity, and it is likely to contribute to their rapid cytokine secretion. In addition to Vα14i T cells, other effector T cells may contain some untranslated cytokine mRNAs. Consistent with this finding, it was recently reported that memory CD8+ T cells have untranslated RANTES mRNA (54).
Our results are in contrast to those from a number of studies suggesting that Vα14i T cells behave in a more conventional manner, requiring costimulation and cytokine signaling to regulate their cytokine production (37, 55–57). The inherent technical limitations of in vitro culture conditions may have been responsible, and in some cases some contaminating NK cells or other cell types could have secreted much of the cytokine analyzed. Additionally, the tracking and/or isolation of Vα14i NKT cells, using for example, anti-NK1.1 in conjunction with anti-TCRβ mAbs, does not precisely identify the αGalCer/ CD1d-reactive population. Finally, sorting on the basis of NK1.1 (52), TCR, or other markers could activate Vα14i T cells in an unphysiologic way.
Altered peptide ligands can alter the cytokine profile of conventional CD4+ T cells (35), and consistent with this, an analog of αGalCer with a shortened alkyl chain on the sphingosine has been reported to preferentially induce IL-4 secretion by NKT cells (58). Much of this analysis was carried out on unfractionated spleen cells stimulated in vitro, however, and further studies will be required to determine whether this analog directly alters the quality of the response by Vα14i T cells. Finally, although Vα14i T cells contain IL-4 and IFN-γ mRNAs, and the response of these cells in vivo could not be polarized in our studies, it remains possible that such polarized populations can be generated. If so, we speculate that this could be caused in part by posttranscriptional control of cytokine gene expression and control of transcription. Regardless, such polarized cells may have an important regulatory role that could be exploited for the generation of novel immune therapies.
Supplementary Material
Acknowledgments
We thank Alan Saluk and Cliff McArthur (Scripps Research Institute) for help with cell sorting, Ninetta Flores for expert animal care, Stephen Schoenberger for providing the CD40-/- mice, and Phillipa Marrack for critical review of this manuscript. This project was funded by National Institutes of Health Grants RO1 CA52511 (to M.K.) and AI30663 (to R.M.L.), a grant from the Human Frontiers of Science Program, and a fellowship from the Cancer Research Institute (to L.G.). R.M.L. is an Ellison Medical Foundation Senior Scholar in Global Infectious Diseases. This is manuscript no. 450 from the La Jolla Institute for Allergy and Immunology.
Abbreviations: αGalCer, α-galactosylceramide; NKT, natural killer T; TCR, T cell antigen receptor; eGFP, enhanced GFP; eYFP, enhanced yellow fluorescent protein; Th2, T helper 2.
References
- 1.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Motoki, K., Ueno, H., Nakagawa, R., Sato, H., Kondo, E., et al. (1997) Science 278 1626-1629. [DOI] [PubMed] [Google Scholar]
- 2.Burdin, N., Brossay, L., Koezuka, Y., Smiley, S. T., Grusby, M. J., Gui, M., Taniguchi, M., Hayakawa, K. & Kronenberg, M. (1998) J. Immunol. 161 3271-3281. [PubMed] [Google Scholar]
- 3.Kronenberg, M. & Gapin, L. (2002) Nat. Rev. Immunol. 2 557-568. [DOI] [PubMed] [Google Scholar]
- 4.Gombert, J. M., Herbelin, A., Tancrede-Bohin, E., Dy, M., Carnaud, C. & Bach, J.-F. (1996) Eur. J. Immunol. 26 2989-2998. [DOI] [PubMed] [Google Scholar]
- 5.Zeng, D., Lewis, D., Dejbakhsh-Jones, S., Lan, F., Garcia-Ojeda, M., Sibley, R. & Strober, S. (1999) J. Exp. Med. 189 1073-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond, K. J. L., Poulton, L. D., Palmisano, L. J., Silveira, P. A., Godfrey, D. I. & Baxter, A. G. (1998) J. Exp. Med. 187 1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falcone, M., Yeung, B., Tucker, L., Rodriguez, E. & Sarvetnick, N. (1999) J. Exp. Med. 190 963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson, S. B., Kent, S. C., Patton, K. T., Orban, T., Jackson, R. A., Exley, M., Porcelli, S., Schatz, D. A., Atkinson, M. A., Balk, S. P., et al. (1998) Nature 391 177-181. [DOI] [PubMed] [Google Scholar]
- 9.Sumida, T., Sakamoto, A., Murata, H., Makino, Y., Takahashi, H., Yoshida, S., Nishioka, K., Iwamoto, I. & Taniguchi, M. (1995) J. Exp. Med. 182 1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illes, Z., Kondo, T., Newcombe, J., Oka, N., Tabira, T. & Yamamura, T. (2000) J. Immunol. 164 4375-4381. [DOI] [PubMed] [Google Scholar]
- 11.van der Vliet, H. J., von Blomberg, B. M., Nishi, N., Reijm, M., Voskuyl, A. E., van Bodegraven, A. A., Polman, C. H., Rustemeyer, T., Lips, P., van den Eertwegh, A. J., et al. (2001) Clin. Immunol. 100 144-148. [DOI] [PubMed] [Google Scholar]
- 12.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Sato, H., Kondo, E., Harada, M., Koseki, H., Nakayama, T., et al. (1998) Proc. Natl. Acad. Sci. USA 95 5690-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth, M. J., Thia, K. Y., Street, S. E., Cretney, E., Trapani, J. A., Taniguchi, M., Kawano, T., Pelikan, S. B., Crowe, N. Y. & Godfrey, D. I. (2000) J. Exp. Med. 191 661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami, K., Kinjo, Y., Yara, S., Koguchi, Y., Uezu, K., Nakayama, T., Taniguchi, M. & Saito, A. (2001) Infect. Immun. 69 213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duthie, M. S., Wleklinski-Lee, M., Smith, S., Nakayama, T., Taniguchi, M. & Kahn, S. J. (2002) Infect. Immun. 70 36-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nieuwenhuis, E. E., Matsumoto, T., Exley, M., Schleipman, R. A., Glickman, J., Bailey, D. T., Corazza, N., Colgan, S. P., Onderdonk, A. B. & Blumberg, R. S. (2002) Nat. Med. 8 588-593. [DOI] [PubMed] [Google Scholar]
- 17.Burdin, N., Brossay, L. & Kronenberg, M. (1999) Eur. J. Immunol. 29 2014-2025. [DOI] [PubMed] [Google Scholar]
- 18.Hong, S., Wilson, M. T., Serizawa, I., Wu, L., Singh, N., Naidenko, O. V., Miura, T., Haba, T., Scherer, D. C., Wei, J., et al. (2001) Nat. Med. 7 1052-1056. [DOI] [PubMed] [Google Scholar]
- 19.Sharif, S., Arreaza, G. A., Zucker, P., Mi, Q. S., Sondhi, J., Naidenko, O. V., Kronenberg, M., Koezuka, Y., Delovitch, T. L., Gombert, J. M., et al. (2001) Nat. Med. 7 1057-1062. [DOI] [PubMed] [Google Scholar]
- 20.Jahng, A. W., Maricic, I., Pedersen, B., Burdin, N., Naidenko, O., Kronenberg, M., Koezuka, Y. & Kumar, V. (2001) J. Exp. Med. 194 1789-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh, A. K., Wilson, M. T., Hong, S., Olivares-Villagomez, D., Du, C., Stanic, A. K., Joyce, S., Sriram, S., Koezuka, Y. & Van Kaer, L. (2001) J. Exp. Med. 194 1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mars, L. T., Laloux, V., Goude, K., Desbois, S., Saoudi, A., Van Kaer, L., Lassmann, H., Herbelin, A., Lehuen, A. & Liblau, R. S. (2002) J. Immunol. 168 6007-6011. [DOI] [PubMed] [Google Scholar]
- 23.Cui, J., Shin, T., Kawano, T., Sato, H., Kondo, E., Toura, I., Kaneko, Y., Koseki, H., Kanno, M. & Taniguchi, M. (1997) Science 278 1623-1626. [DOI] [PubMed] [Google Scholar]
- 24.Kakimi, K., Lane, T. E., Chisari, F. V. & Guidotti, L. G. (2001) J. Immunol. 167 6701-6705. [DOI] [PubMed] [Google Scholar]
- 25.Smyth, M. J., Crowe, N. Y., Hayakawa, Y., Takeda, K., Yagita, H. & Godfrey, D. I. (2002) Curr. Opin. Immunol. 14 165-171. [DOI] [PubMed] [Google Scholar]
- 26.Kawabe, T., Naka, T., Yoshida, K., Tanaka, T., Fujiwara, H., Suematsu, S., Yoshida, N., Kishimoto, T. & Kikutani, H. (1994) Immunity 1 167-178. [DOI] [PubMed] [Google Scholar]
- 27.Shahinian, A., Pfeffer, K., Lee, K. P., Kundig, T. M., Kishihara, K., Wakeham, A., Kawai, K., Ohashi, P. S., Thompson, C. B. & Mak, T. W. (1993) Science 261 609-612. [DOI] [PubMed] [Google Scholar]
- 28.Grogan, J. L., Mohrs, M., Harmon, B., Lacy, D. A., Sedat, J. W. & Locksley, R. M. (2001) Immunity 14 205-215. [DOI] [PubMed] [Google Scholar]
- 29.Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T. & Mulligan, R. C. (1991) Cell 65 1153-1163. [DOI] [PubMed] [Google Scholar]
- 30.Mohrs, M., Shinkai, K., Mohrs, K. & Locksley, R. M. (2001) Immunity 15 303-311. [DOI] [PubMed] [Google Scholar]
- 31.Gu, H., Zou, Y. R. & Rajewsky, K. (1993) Cell 73 1155-1164. [DOI] [PubMed] [Google Scholar]
- 32.O'Gorman, S., Dagenais, N. A., Qian, M. & Marchuk, Y. (1997) Proc. Natl. Acad. Sci. USA 94 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda, J. L., Naidenko, O. V., Gapin, L., Nakayama, T., Taniguchi, M., Wang, C. R., Koezuka, Y. & Kronenberg, M. (2000) J. Exp. Med. 192 741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guery, J. C., Galbiati, F., Smiroldo, S. & Adorini, L. (1996) J. Exp. Med. 183 485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Constant, S. L. & Bottomly, K. (1997) Annu. Rev. Immunol. 15 297-322. [DOI] [PubMed] [Google Scholar]
- 36.Foucras, G., Gapin, L., Coureau, C., Kanellopoulos, J. M. & Guery, J. C. (2000) J. Exp. Med. 191 683-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayakawa, Y., Takeda, K., Yagita, H., Van Kaer, L., Saiki, I. & Okumura, K. (2001) J. Immunol. 166 6012-6018. [DOI] [PubMed] [Google Scholar]
- 38.Kitamura, H., Iwakabe, K., Yahata, T., Nishimura, S., Ohta, A., Ohmi, Y., Sato, M., Takeda, K., Okumura, K., Van Kaer, L., et al. (1999) J. Exp. Med. 189 1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh, N., Hong, S., Scherer, D. C., Serizawa, I., Burdin, N., Kronenberg, M., Koezuka, Y. & Van Kaer, L. (1999) J. Immunol. 163 2373-2377. [PubMed] [Google Scholar]
- 40.Osman, Y., Kawamura, T., Naito, T., Takeda, K., Van Kaer, L., Okumura, K. & Abo, T. (2000) Eur. J. Immunol. 30 1919-1928. [DOI] [PubMed] [Google Scholar]
- 41.Carnaud, C., Lee, D., Donnars, O., Park, S. H., Beavis, A., Koezuka, Y. & Bendelac, A. (1999) J. Immunol. 163 4647-4650. [PubMed] [Google Scholar]
- 42.Eberl, G. & MacDonald, H. R. (2000) Eur. J. Immunol. 30 985-992. [DOI] [PubMed] [Google Scholar]
- 43.Kawakami, K., Kinjo, Y., Yara, S., Uezu, K., Koguchi, Y., Tohyama, M., Azuma, M., Takeda, K., Akira, S. & Saito, A. (2001) Infect. Immun. 69 6643-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonoda, K. H., Exley, M., Snapper, S., Balk, S. P. & Stein-Streilein, J. (1999) J. Exp. Med. 190 1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loza, M. J. & Perussia, B. (2001) Nat. Immunol. 2 917-924. [DOI] [PubMed] [Google Scholar]
- 46.Peritt, D., Robertson, S., Gri, G., Showe, L., Aste-Amezaga, M. & Trinchieri, G. (1998) J. Immunol. 161 5821-5824. [PubMed] [Google Scholar]
- 47.Gumperz, J. E., Miyake, S., Yamamura, T. & Brenner, M. B. (2002) J. Exp. Med. 195 625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, P. T., Benlagha, K., Teyton, L. & Bendelac, A. (2002) J. Exp. Med. 195 637-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadowaki, N., Antonenko, S., Ho, S., Rissoan, M. C., Soumelis, V., Porcelli, S. A., Lanier, L. L. & Liu, Y. J. (2001) J. Exp. Med. 193 1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Y. F., Tomura, M., Ono, S., Hamaoka, T. & Fujiwara, H. (2000) Int. Immunol. 12 1669-1675. [DOI] [PubMed] [Google Scholar]
- 51.Tomura, M., Yu, W. G., Ahn, H. J., Yamashita, M., Yang, Y. F., Ono, S., Hamaoka, T., Kawano, T., Taniguchi, M., Koezuka, Y. & Fujiwara, H. (1999) J. Immunol. 163 93-101. [PubMed] [Google Scholar]
- 52.Arase, H., Arase, N. & Saito, T. (1996) J. Exp. Med. 183 2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naumov, Y. N., Bahjat, K. S., Gausling, R., Abraham, R., Exley, M. A., Koezuka, Y., Balk, S. B., Strominger, J. L., Clare-Salzer, M. & Wilson, S. B. (2001) Proc. Natl. Acad. Sci. USA 98 13838-13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson, B. J., Murakami, M., Mitchell, T. C., Kappler, J. & Marrack, P. (2002) Immunity 17, 605-615. [DOI] [PubMed] [Google Scholar]
- 55.Leite-De-Moraes, M. C., Moreau, G., Arnould, A., Machavoine, F., Garcia, C., Papiernik, M. & Dy, M. (1998) Eur. J. Immunol. 28 1507-1515. [DOI] [PubMed] [Google Scholar]
- 56.Hameg, A., Gouarin, C., Gombert, J. M., Hong, S., Van Kaer, L., Bach, J. F. & Herbelin, A. (1999) J. Immunol. 162 7067-7074. [PubMed] [Google Scholar]
- 57.Leite-De-Moraes, M. C., Hameg, A., Pacilio, M., Koezuka, Y., Taniguchi, M., Van Kaer, L., Schneider, E., Dy, M. & Herbelin, A. (2001) J. Immunol. 166 945-951. [DOI] [PubMed] [Google Scholar]
- 58.Miyamoto, K., Miyake, S. & Yamamura, T. (2001) Nature 413 531-534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.