Abstract
Langerhans cells (LCs) are suspected to be initial targets for HIV after sexual exposure (by becoming infected or by capturing virus). Here, productive R5 HIV infection of LC ex vivo and LC-mediated transmission of virus to CD4+ T cells were both found to depend on CCR5. By contrast, infection of monocyte-derived dendritic cells and transfer of infection from monocyte-derived dendritic cells to CD4+ T cells were mediated by CCR5-dependent as well as DC-specific ICAM-3-grabbing nonintegrin-dependent pathways. Furthermore, in 62 healthy individuals, R5 HIV infection levels in LCs ex vivo were associated with CCR5 genotype. Specifically, genotyping for ORFΔ32 revealed that LCs isolated from ORFΔ32/wt individuals were significantly less susceptible to HIV when compared with LCs isolated from ORFwt/wt individuals (P = 0.016). Strikingly, further genetic analyses of the A-2459G CCR5 promoter polymorphism in ORFΔ32/wt heterozygous individuals revealed that LCs isolated from -2459A/G + ORFΔ32/wt individuals were markedly less susceptible to HIV than were LCs from -2459A/A + ORFΔ32/wt individuals (P = 0.012). Interestingly, these genetic susceptibility data in LCs parallel those of genetic susceptibility studies performed in cohorts of HIV-infected individuals. Thus, we suggest that CCR5-mediated infection of LCs, and not capture of virus by LCs, provides a biologic basis for understanding certain aspects of host genetic susceptibility to initial HIV infection.
The most common mode of acquiring HIV infection is by sexual transmission across genital epithelial tissue (1). Studies in rhesus macaques exposed intravaginally to simian immunodeficiency virus (SIV) suggest that Langerhans cells (LCs), resident dendritic cells (DCs) of stratified squamous epithelia, are the first cells to encounter virus (2, 3). However, the mechanisms whereby these cells acquire virus remain controversial. Recently, DC-specific ICAM-3-grabbing nonintegrin (DC-SIGN) (CD209), a C-type lectin expressed on subepithelial DCs and monocyte-derived DCs (mDCs), has been shown to bind HIV gp120, and to mediate capture of HIV virions and protection from degradation (4–6). Viruses captured by this mechanism may not replicate within DCs, but may be transferred to CD4+ T cells where viral replication occurs (5). Because of this finding, a primary role for DC-SIGN in sexual transmission of HIV has been proposed. Several critical observations, however, do not support this model. First, DCs within epithelia do not express DC-SIGN (4–6). Second, DC-SIGN binds both CCR5-using viruses (i.e., R5 HIV) and CXCR4-using viruses (i.e., X4 HIV) (5), a feature that does not adequately explain preferential R5 HIV selection that occurs during sexual transmission of HIV (7–9).
An alternate model proposes that infection of DCs within epithelia, as opposed to capture of virus through DC-SIGN, is the major pathway that HIV utilizes to gain entry into the body (10). Although they do not express DC-SIGN, LCs express CD4 and HIV chemokine coreceptors, and these molecules have been shown to mediate infection of LCs (11–14). Immature LCs express surface CCR5, but not surface CXCR4, immediately after isolation from skin; CCR5 on LCs also mediates fusion with cells expressing the HIV envelope protein gp120 (11). Likewise, Patterson et al. (15) examined human cervix and found that CCR5 mRNA expression was at least 10-fold higher than CXCR4 mRNA expression in this tissue. These reports suggest that the HIV coreceptor expression pattern on LCs, or perhaps on other resident cells in genital tissue, confers a “gatekeeper” status on these cells at mucosal surfaces, allowing preferential entry of R5 HIV, but not X4 HIV.
Individuals who are homozygous for a 32-nt deletion in the CCR5 ORF (ORFΔ32/Δ32) comprise ≈1% of all whites, whereas individuals heterozygous for this deletion (ORFΔ32/wt) represent ≈20% of whites (16). Genetic studies in HIV+ cohorts have revealed that Δ32 homozygotes are highly resistant to HIV infection (16–21). However, there has been controversy about the risk of acquiring HIV infection in individuals who are ORFΔ32/wt. Some studies have found that ORFΔ32/wt individuals are less susceptible to HIV infection (18, 22, 23), whereas others failed to show protection (17, 19–21). This discrepancy may be related to the considerable variation in CCR5 surface expression among persons with ORFΔ32/wt, as is seen in ORFwt/wt individuals (24). This finding led to suggestions that additional polymorphisms in the CCR5 ORF and/or regulatory region might influence surface CCR5 expression. If true, then these polymorphisms, in addition to ORFΔ32, might be predicted to influence the acquisition of HIV infection (25–28).
In vitro studies originally identifying single-nucleotide polymorphisms (SNPs) in the CCR5 promoter have shown that alleles containing an adenine (A) nucleotide at the -2459 position [note: nucleotide numbering is relative to the initiating methionine (29), nucleotide 59029 of GenBank accession no. U95626] display higher CCR5 promoter activity than alleles with a guanine (G) nucleotide at this position (25, 30). The in vivo-relevance of these observations is supported by findings that HIV-infected individuals homozygous for the A allele (-2459A/A) progressed more rapidly to AIDS than those who were homozygous for the G allele (-2459G/G) (25, 26, 31, 32). Interestingly, ORFΔ32, which is known to confer protection against HIV infection, is tightly linked to the promoter -2459A allele, known to be associated with HIV disease progression (25). Although there is a growing awareness that complex polymorphic CCR5 haplotypes are associated with slower or rapid progression to AIDS (33–36), less is known about how CCR5 haplotype/haplotype (diplotype) combinations influence initial infection.
Here, LCs were isolated from healthy volunteers and infected with R5 HIV ex vivo. Concomitantly, CCR5 diplotypes were determined for all volunteers. Both LC infection and LC-mediated transfer of virus to CD4+ T cells were dependent on CCR5. LC infection levels also reflected CCR5 diplotype patterns. Interestingly, these data suggest a biologically plausible model for the observed restricted sexual transmission of R5 HIV, and for host genetic susceptibility to initial HIV infection.
Materials and Methods
Study Participants. The Institutional Review Board of the National Cancer Institute approved all aspects of this study, and informed consent was obtained from all healthy individuals volunteering for the suction blister procedure. The 62 volunteers who were genotyped for this study included 41 males (66%) and 21 females (34%). Fifty-two individuals were self-described as whites, 7 as African Americans, and 3 as Asians (note, individuals in the latter two groups are discouraged from undergoing the blistering procedure because of increased risks of keloidal scarring and postinflammatory hyperpigmentation). Volunteers reported no histories of chronic disease (including no histories of HIV risk factors) and no use of medications on a regular basis.
Reagents. All of the murine mAbs were purchased from PharMingen, except for FITC-conjugated rat anti-p24 mAb (Beckman Coulter), mouse anti-CCR5 mAb (clone 45531, R & D Systems), mouse anti-Langerin mAb (clone DCGM4, Beckman Coulter), mouse anti-DC-SIGN mAb (clone 120507, R & D Systems), and 3′-azido-3′-deoxythymidine (AZT), mannan, and EGTA were purchased from Sigma. Robin Offord (University of Geneva, Geneva) kindly provided Na-nonanoyl[thioproline2, cyclohexylglycine3]RANTES (2–68) [PSC-RANTES, a newer more potent analogue of RANTES (regulated on activation normal T cell expressed and secreted)]. Nobutaka Fujii (Kyoto University, Kyoto) kindly provided AMD3100. Purified, pelleted, and titered HIV-1Ba-L (R5) (stock at 1.8 × 1010 virus particles per ml) was purchased from Advanced Biotechnologies (Columbia, MD).
HIV Infection of LCs ex Vivo. Epithelial tissue explants were prepared from suction blister roofs of healthy volunteers and LCs within these explants were infected as described (13). Briefly, 100-μl droplets containing HIV-1Ba-L at 1:100 dilution were placed on the inside surface of sterile plastic culture dish covers and explants were draped over droplets and incubated together at 37°C for 2 h. For some experiments, 50-μl droplets containing antibodies including mouse control IgG (40 μg/ml) or mAbs (40 μg/ml) directed against CD4, CCR5, CXCR4, DC-SIGN, or Langerin, or reagents including AZT (0.1–100 μM), PSC-RANTES (200 nM), AMD3100 (200 nM), mannan (200 μg/ml), or EGTA (10 mM) were placed on the inside surface of sterile plastic culture dish covers. Explants were draped over droplets for 20 min at 37°C; 50 μl of HIV-1Ba-L at 1:100 final dilution was then added to antibodies or reagents under the explants and incubated for an additional 2 h at 37°C. Explants were washed three times with sterile PBS, and then floated in six-well plates containing 4 ml of complete medium. In some experiments, culture media contained AZT (0.1–100 μM).
HIV Infection of mDCs in Vitro. mDCs were prepared and infected with HIV as described (37). One million mDCs suspended in 0.5 ml of complete medium with GM-CSF/IL-4 were preincubated with antibodies or reagents described above for 20 min at 37°C, and then HIV-1Ba-L (at 1:100 or 1:500 final dilution) was added to cultures. After a 2-h incubation at 37°C, DC were washed three times, resuspended in complete medium supplemented with GM-CSF/IL-4, and cultured for 3 additional days.
Assessment of HIV Infection in LCs or mDCs. LCs that had emigrated from explants and mDCs were collected 3 days after HIV exposure, preincubated in staining buffer for 10 min at room temperature to block nonspecific staining, and then incubated with 10 μg/ml phycoerythrin-conjugated mouse anti-human HLA-DR mAb for 30 min at 4°C. Cells were then incubated with Dead red (Molecular Probes) for 20 min at room temperature to label dead cells and fixed and permeabilized with Cytofix/ Cytoperm reagents (PharMingen) for 20 min at 4°C. Cells were then incubated with 10 μg/ml FITC-conjugated rat anti-p24 mAb diluted in Perm/Wash for 30 min at 4°C and examined by flow cytometry using a FACScan (Becton Dickinson) equipped with LYSIS II software (Becton Dickinson). Dead cells, i.e., Dead-red positive cells, were excluded from all analyses. “Crawlout” LCs were harvested on day 3 or day 4 and were processed for electron microscopy as described (13).
Assessment of HIV Transmission to CD4+ T Cells. LCs that had emigrated from explants 3 days after HIV exposure or HIV-exposed mDCs were washed and then 1 × 104 LCs or mDCs were cocultured with 1 × 106 autologous CD4+ T cells for 6 days. For detection of secreted p24 protein, supernatants were harvested at day 3 and day 5 from cocultures of HIV-exposed mDCs or emigrated LCs and CD4+ T cells, inactivated with 2% (vol/vol) Triton X-100 (Sigma) in PBS, and frozen at -20°C. Samples were thawed and examined for p24 protein content by ELISA.
PCR and Genotyping. Genomic DNA was isolated from PBMC (200 μl) of suction blister volunteers by using the QIAamp blood kit (Qiagen, Valencia, CA). Genotyping by analysis of fragment length polymorphisms (CCR5 ORFΔ32 and CCR2 64V/I) was performed as described (38, 39). CCR5 promoter polymorphism genotyping was performed by ligase detection reaction analyses and is outlined in detail in Table 1 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site.
Statistical Analysis. Statistical analyses performed to test associations between CCR5 and CCR2 genotype data and LC infection by HIV were performed by using Mann–Whitney U or Kruskal–Wallis tests. All statistical tests were performed by using STAT-VIEW 5.0.1 software (SAS Institute, Cary, NC).
Results
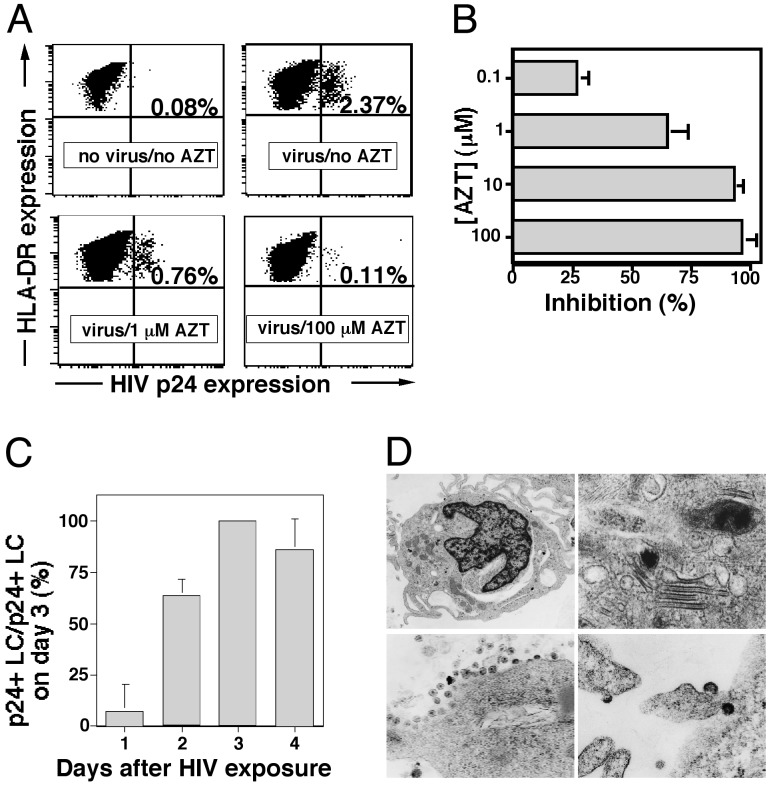
R5 HIV Replicates Within LCs. Recently, we established an ex vivo model whereby resident LCs within epithelial tissue explants are exposed to HIV and then allowed to emigrate from tissue, thus mimicking conditions that occur after mucosal exposure to HIV (13). Here, we found that AZT blocked HIV replication within LCs in a dose-dependent manner (Fig. 1 A and B). AZT would be predicted to have no effect on HIV virions captured by or bound to LC surfaces. AZT at the doses used had no effect on viability of LCs (data not shown). When examined on days 1, 2, 3, and 4 after HIV exposure, the number of HIV p24+ LCs progressively increased over the first several days, peaking on day 3 (Fig. 1C). These data also indicate ongoing active HIV replication within LCs. Furthermore, cells that emigrated from HIV-exposed explant tissue demonstrated classic ultrastructural features of LCs (e.g., Birbeck granules) and showed clear evidence of virions budding from cell surfaces (Fig. 1D). In addition to the HIV p24 staining and transmission electron microscopy (TEM), we have recently reported that HIV transcripts can be detected and quantified within crawl-out LCs, further confirming productive infection of these cells (40).
Fig. 1.
R5 HIV replicates within LCs. (A) LCs within skin explants were exposed to HIV in the presence of AZT, virus was washed from explants, and tissue was then cultured for 3 days in the presence of AZT. Representative FACS analyses of MHC class II and p24 mAb double-stained cells that have emigrated from HIVBa-L-exposed or uninfected epithelial tissue explants demonstrating varying numbers of productively infected LCs are shown. (B) Summary of experiments showing that AZT blocks HIV replication within LCs in a dose-dependent manner. Means ± SD derived from three separate experiments are shown. (C) Summary of experiments showing that the number of p24+ LC peaks 3 days after HIV exposure. Data from three separate experiments were normalized (number of p24+ LCs on day 3 = 100%) and expressed as means ± SD. (D) Representative transmission electron microscopy micrographs showing the typical dendritic morphology of LCs (Upper Left), the characteristic Birbeck granules of LC (Upper Right), immature HIV virions budding from the membrane of an LC (Lower Left), as well as mature HIV virions near the cell membrane of an LC (Lower Right).
Uninfected LCs stained with a FITC-conjugated anti-p24 mAb (Fig. 1 A) and HIV-infected LCs stained with a FITC-conjugated isotype control antibody (data not shown) always showed <0.10% positivity, confirming the specificity of the p24 direct staining. Keratinocytes, which are occasionally present in “crawl-out” cell preparations, never showed p24 positivity (data not shown). Last, CD3+ T cells or CD14+ monocytes were never detected in crawl-out cell populations (data not shown), consistent with what is known about the cell constituents of normal human epidermis.
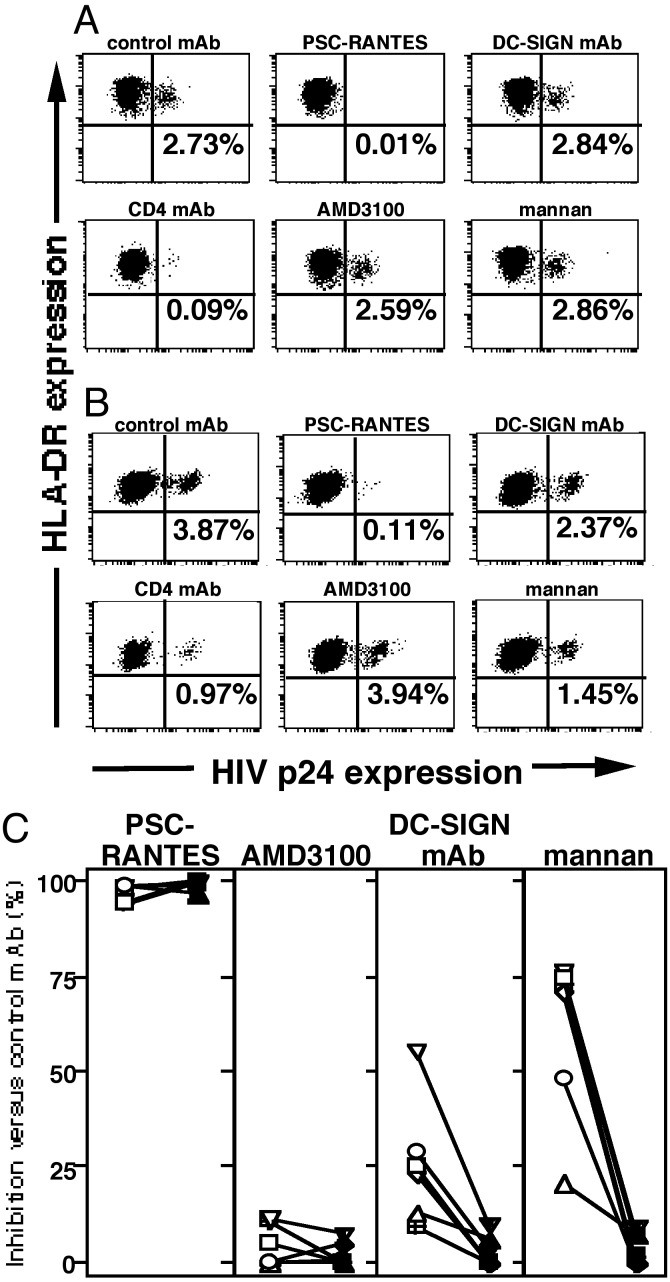
Productive LC Infection Is Mediated by CCR5 and CD4, but Not by C-Type Lectin–HIV Interactions. Although DC-SIGN predominantly mediates capture and endocytosis of HIV virions (5), this molecule is also coexpressed with surface CD4 and CCR5 and promotes CCR5-mediated infection of cells (41). Therefore, we tested whether C-type lectins were involved in HIV infection of LCs by using our tissue explant model system. Preincubation of epithelial sheets with neutralizing anti-DC-SIGN mAbs or anti-Langerin mAbs did not affect HIV infection levels in LCs (Fig. 2 A and C and data not shown). In addition, preincubation of LCs with mannan or EGTA, known inhibitors of C-type lectin binding, did not affect HIV infection levels in LC (Fig. 2 A and C and data not shown). Consistent with previously published results using AOP-RANTES (the original RANTES analogue; ref. 13), preincubation of epithelial sheets with PSC-RANTES, a RANTES analogue, completely blocked R5 HIV infection of LC (Fig. 2 A and C). As controls, epithelial sheets preincubated with AMD3100, which binds CXCR4 and inhibits X4 HIV infection (42), failed to block R5 HIV infection of LCs. Last, preincubation of explants with anti-CD4 mAb completely blocked HIV infection of LCs (Fig. 2 A and C).
Fig. 2.
Productive LC infection is mediated by CCR5 and CD4, but not by C-type lectin–HIV interactions. LCs (A) or mDCs (B) isolated from the same donor were preincubated with the indicated mAbs or reagents for 20 min at 37°C, exposed to HIVBa-L for 2 h, and cultured for 3 days. Cells were then stained with anti-p24 and anti-MHC class II mAbs. (C) Summary of the percent inhibition due to the indicated mAbs or reagents obtained in six separate experiments. Anti-CXCR4 mAb was used as negative reagent control as well (data not shown). mDCs (open symbols) and LCs (filled symbols) obtained from the same donors are joined by a line.
As further controls, mDCs were prepared from peripheral blood of a subset of volunteers who donated skin for the LC-infection studies. By contrast to the findings with LCs, there was partial inhibition of R5 HIV infection in mDCs when these cells were preincubated with neutralizing anti-DC-SIGN mAbs or with mannan (Fig. 2 B and C). As seen with LCs, preincubation of mDCs with PSC-RANTES or anti-CD4 mAbs inhibited subsequent R5 HIV infection of these cells, whereas preincubation with AMD3100 failed to block R5 HIV infection of mDCs (Fig. 2 B and C). These data clearly demonstrate differences in how HIV infects different types of DCs (in this case, resident epidermal LCs versus mDCs derived from peripheral blood).
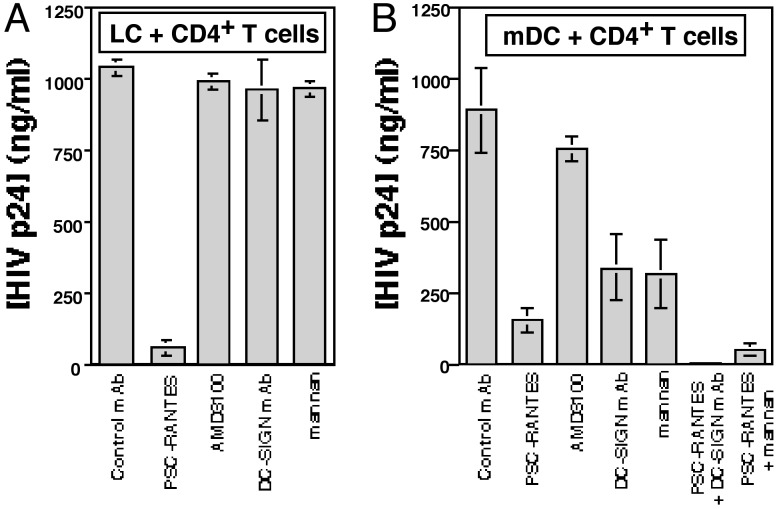
Blocking CCR5 on LCs, but Not C-Type Lectin–HIV Interactions, Blocks Transmission of Virus from LCs to CD4+ T Cells. DC-SIGN has been shown to mediate infection of CD4+ T cells that were cocultured with HIV-exposed mDCs (5). Thus, we next tested whether C-type lectins were involved in HIV transmission from LCs to CD4+ T cells. Although only relatively few productively infected LCs could be identified in crawl-out populations (Figs. 1 and 2), these cells could induce high levels of HIV infection when cocultured with resting CD4+ T cells (Fig. 3A). Strikingly, preincubation of epithelial sheets with PSC-RANTES completely blocked subsequent HIV transmission from LCs to CD4+ T cells, whereas LC-mediated transmission was not affected by preincubation of epithelial sheets with anti-DC-SIGN mAbs or mannan (Fig. 3A). Importantly, p24 protein content in coculture supernatants detected by ELISA correlated well with the percentage of p24+CD3+ T cells in cocultures, suggesting that the main source of secreted p24 protein in LC-T cell cocultures was CD3+ T cells (Fig. 3A and data not shown).
Fig. 3.
Blocking CCR5 on LCs, but not C-type lectin–HIV interactions, blocks transmission of virus from LCs to CD4+ T cells. LCs (A) or mDCs (B) were preincubated with the indicated mAbs or reagents for 20 min at 37°C, exposed to HIVBa-L for 2 h, and cultured for 3 days. (A) Emigrated LCs were collected 3 days later and then cocultured with autologous resting CD4+ T cells. (B) mDCs were infected with HIV for 3 days and then cocultured with autologous CD4+ T cells. HIV infection levels were assessed by measuring p24 protein in coculture supernatants (collected 5 days after coculture) by ELISA. Data shown are means plus SD of three separate experiments.
Similar types of transmission experiments were performed by using mDCs isolated from peripheral blood of LC donors. In contrast to findings with LCs, preincubation of mDCs with either PSC-RANTES, anti-DC-SIGN mAb, or mannan partially inhibited subsequent HIV transmission to CD4+ T cells (Fig. 3B), suggesting multiple methods of viral transmission were operative in mDCs. Additional experiments revealed that preincubation with combinations of PSC-RANTES plus anti-DC-SIGN mAb or PSC-RANTES plus mannan completely blocked HIV transmission from mDCs to CD4+ T cells (Fig. 3B). These data strongly suggest that R5 HIV interacts with LCs primarily through a CCR5-dependent and DC-SIGN-independent infection pathway, whereas mDCs interact with R5 HIV through both a CCR5-mediated infection pathway and an infection-independent DC-SIGN-mediated capture pathway.
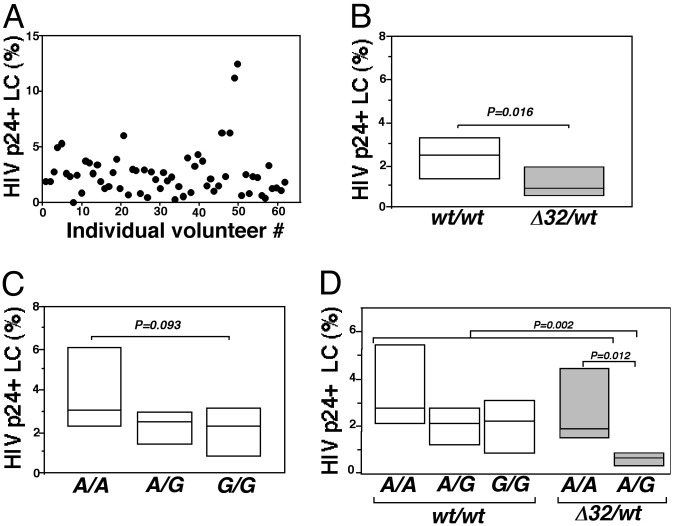
Compound CCR5 Polymorphisms Influence R5 HIV Infection Levels in LCs. Consistent with previous findings (13), R5 HIV infection levels in LCs isolated from 62 different individuals were highly variable (range 0.02–11.20%, mean 2.67%; Fig. 4A). Because ex vivo R5 HIV susceptibility of donor LCs was notably heterogeneous, and after determining that R5 HIV infection of LCs depended on CCR5, we then asked whether infection levels in LCs were influenced by CCR5 genotype. Two CCR5 loci were genotyped by using post-PCR fragment-length polymorphism and ligase chain reaction analyses in 62 volunteers and these genetic data were analyzed blinded to the LC-infection results. Specifically, the CCR5 promoter SNP -2459 (containing either A or G) and the CCR5 ORF for the 32-nt deletion (ORFΔ32) were examined. No individuals were homozygous for ORFΔ32 (see Table 2, which is published as supporting information on the PNAS web site). Consistent with previous linkage data (25), all individuals with a Δ32 allele also had an A nucleotide at the -2459 SNP. No evidence for linkage of -2459G and Δ32 was observed (see Table 2).
Fig. 4.
Compound CCR5 polymorphisms influence R5 HIV infection levels in LCs. (A) Plot of the percentage of p24+ LCs (with background staining subtracted) obtained from 62 individuals on separate days showing the extent of the individual variability in R5 HIV infection levels is shown. (B) LCs isolated from ORFΔ32 heterozygotes are less susceptible to R5 HIV infection than LCs isolated from ORFwt/wt individuals. (C) LCs isolated from individuals with one or two -2459G promoter allele(s) tend to be less susceptible to R5 HIV than individuals who are homozygous for -2459A. (D) Among individuals who were heterozygous for ORFΔ32, the additional presence of a single -2459G allele conferred strong protection against R5 HIV infection. The horizontal line represents the median percentage of p24+ LCs (background staining subtracted) in each genotypic group. The box indicates the 25%/75% quartiles for each group. For each individual, different researchers at different institutions performed HIV infection experiments and CCR5 genotyping independent of one another.
Interestingly, the percentage of p24+ LCs isolated from ORFΔ32/wt individuals was significantly lower than the percentage of p24+ LCs isolated from ORFwt/wt individuals (ORFΔ32/ wt, n = 12; ORFwt/wt, n = 50; Mann–Whitney U test, P = 0.016; Fig. 4B). As four different haplotypes have been identified that contain the -2459G allele (haplotypes A–D) and three different haplotypes have been identified that contain the -2459A allele [haplotypes E, F1, and G1; excludes haplotypes containing CCR264I (F2) and ORFΔ32 (G2) (33)], it is important to note that no single -2459G haplotype was consistently associated with a low percentage of p24+ LCs and no single -2459A haplotype was consistently associated with a high percentage of p24+ LCs. Although significant differences in HIV infection levels in LCs among individuals who were homozygous for the wild-type ORF with either -2459A/A,-2459A/G, or -2459G/G promoter genotypes were not detected, LCs isolated from -2459G/G individuals tended to be less susceptible to HI V than LCs isolated from -2459A/A individuals (-2459G/G, n = 13; -2459A/A, n = 15; Mann–Whitney U test, P = 0.093; Fig. 4C). The observed differences associated with genotype were likely to be specific, because other factors, such as race and sex, were not correlated with LC HIV infection levels (data not shown).
We next asked whether diplotypes (as described above) influenced susceptibility of LCs to HIV infection. Strikingly, LCs from -2459A/G + ORFΔ32/wt individuals (functionally hemizygous, expressing CCR5 only from the -2459G promoter allele) were very difficult to infect, showing nearly 4-fold less susceptibility to HIV than LCs isolated from -2459A/A + ORFΔ32/wt individuals (functionally hemizygous, expressing CCR5 from the -2459A promoter allele) (-2459A/G + ORFΔ32/wt, n = 7; -2459A/A + ORFΔ32/wt, n = 5; Mann–Whitney U test, P = 0.012; Fig. 4D). Further analysis revealed that LC infection levels in individuals with the diplotype of -2459A/G + ORFΔ32/wt were markedly lower than levels in the other four diplotypes combined (Kruskal–Wallis test, P = 0.002). These data are consistent with the finding that CCR5 promoter activity is greater when driven by a -2459A allele than when the promoter is driven by a -2459G allele (25).
In addition to CCR5 ORFΔ32 and CCR5 -2459 loci, SNP analyses at CCR2 64I/V and CCR5-2733, -2554, -2459, -2135,-2132, -2086, and -1835 were performed in all 62 individuals. No association was observed between the heterozygous CCR2 64I/V genotype and reduced R5 HIV infection in LCs (I/V, n = 17; V/V, n = 43; Mann–Whitney U test, P = 0.451; no individuals were CCR2 64I/I). Complete CCR2-CCR5 SNP analyses revealed 19 different diplotypes among the 62 volunteers. At this level of complexity, no statistically significant associations were observed between LC infection levels ex vivo and CCR2-CCR5 polymorphism (data not shown). Finally, because -2459G is in complete linkage disequilibrium with -2135T and -2459A is in complete linkage disequilibrium with -2135C (34), is it is important to acknowledge that it may not be sufficient to focus solely on the SNP at position -2459 at the exclusion of other CCR5 promoter polymorphisms to fully understand and explain in vitro susceptibility to R5 HIV infection as studied here.
Discussion
Recent data (2) generated from rhesus macaque experiments provide the strongest evidence to date that intraepithelial LCs are the first cells to become infected after intravaginal exposure to SIV. Although there is no direct evidence that LCs are the initial cells infected with HIV in humans, genetic data generated in large cohorts make it clear that CCR5 genotype is a major determinant for initial HIV susceptibility (16–19). Whether CCR5 influences initial infection at the level of the LCs, at the level of the epithelial cell (43, 44), at early replication steps in lymphoid tissue, or at multiple sites, however, is less clear. The former theories can be described as primary gatekeeper models, whereas the latter process represents a viral fitness model (40).
Our current results lend strong support for a model where CCR5+ LCs can function as primary gatekeepers. Using an ex vivo explant model for LC infection (13), R5 HIV infection of LCs and LC-mediated transmission of virus to cocultured CD4+ T cells were entirely dependent on CCR5. Specifically, both LC infection and LC-mediated transmission of virus were blocked by preincubating LCs with PSC-RANTES or anti-CD4 mAbs, but not by preincubation of LCs with anti-Langerin mAbs, anti-DC-SIGN mAbs, mannan, or EGTA (Figs. 2 and 3). By contrast, the latter three reagents partially inhibited R5 HIV infection of mDC and mDC-mediated transmission of virus to T cells (Figs. 2 and 3), consistent with functional DC-SIGN expressed on surface mDCs. Of note, these data do not lend support to the theory that DC-SIGN, Langerin, or some other unidentified C-type lectin act as HIV ligands that facilitate transmission of virus from LCs to T cells (5, 6, 45, 46). Our data also clearly show that HIV-C-type lectin interactions do not contribute to productive HIV infection of LCs. Thus, these results support the model that CCR5-mediated infection of LCs is the major pathway involved in sexual transmission of HIV.
Based on this model, microbicides that interfere with virus binding to CCR5 on LCs could lead to substantial protection from acquiring HIV infection (47). This protection may be analogous to natural protection conferred by the homozygous ORFΔ32 genotype. Accordingly, we are currently studying chemically modified RANTES analogues for their abilities to block R5 HIV infection of LCs. Previously, AOP-RANTES was shown to inhibit R5 HIV infection of LCs (13). Here, PSC-RANTES (T.K. and A.B., unpublished work) was also able to block infection in LCs, as well as LC-mediated transfer of infection to T cells (Figs. 2 and 3). PSC-RANTES is now undergoing further evaluation as a potential microbicide.
Although host genotype is known to influence susceptibility to HIV infection, the cellular and tissue mechanisms that control this process are unknown. Because LCs are potential initial targets for HIV, we asked whether CCR5 genotype influenced R5 HIV infection levels in these cells. LCs isolated from heterozygous ORFΔ32 individuals were less susceptible to HIV infection than LCs isolated from individuals without this mutant allele (Fig. 4B). This finding suggests that individuals with a single ORFΔ32 allele express reduced levels of CCR5 on the surface of their LCs, an assumption that would be consistent with data obtained from similar experiments performed with T cells (24).
Interestingly, on further analysis of the ORFΔ32 heterozygotes, LCs isolated from individuals with the -2459A/G + ORFΔ32/wt diplotype were found to be markedly less susceptible to R5 HIV infection when compared with LCs isolated from individuals with the -2459A/A + ORFΔ32/wt diplotype (Fig. 4D). The former diplotype occurs in ≈10% of whites, whereas the latter diplotype occurs in ≈6% of whites. Based on our LC data, -2459A/G + ORF/Δ32/wt individuals would be predicted to have considerable (but not complete) protection from acquiring HI V infection, whereas -2459A/A + ORFΔ32/wt individuals would not.
Indeed, genetic epidemiologic data in three cohorts of homo-sexual men have recently been reported by Tang et al. (36) that are consistent with our ex vivo findings. These researchers reported that the lowest risks both for acquiring HIV infection and for progression to AIDS were observed in individuals with a -2459A/A + ORFΔ32/wt diplotype, whereas the highest risks for both initial infection and progression to AIDS were observed in individuals with a -2459A/A + ORFΔ32/wt/wt diplotype. Of additional interest, HIV+ persons with a -2459A/A + ORFΔ32/wt diplotype demonstrated the lowest levels of plasma viremia. Furthermore, our data linking ex vivo R5 HIV susceptibility to CCR5 genotype are also consistent with additional cohort data correlating CCR5 genotype and HIV disease progression as reported by two other studies (33, 35). Thus, our current ex vivo infection findings with LCs provide a plausible biologic explanation for observed epidemiologic data obtained from large cohorts of individuals regarding host genetic susceptibility to HIV infection.
One strength of our study is that HIV infection levels in primary cells in vitro were correlated with host cell genotype in a relatively large number of individuals. R5 HIV was used for infection, and the ORFΔ32 locus, as well as all possible SNPs in the CCR5 promoter reported at frequencies ≈1% in human populations, were examined (34). Polymorphisms at the ORFΔ32 locus and the -2459 promoter locus were associated with the most significant differences in HIV infection levels. Other polymorphisms were examined, but were not found to be significantly associated with R5 HIV infection levels in LCs. CCR5 diplotypes reported here are predicted to influence expression of CCR5 protein on cell surfaces significantly, and likely through this mechanism regulate HIV infection levels within these cells. In particular, based on previous work (25), CCR5 protein expression is predicted to be highest when driven by a -2459A + ORFwt allele, intermediate when driven by a -2459G + ORFwt, and absent when driven by a -2459A + ORFΔ32 allele. Indeed, we have recently found (48) that CCR5 surface expression on monocytes correlates with these compound genotypes as predicted. Although CCR5 surface expression on LCs was not measured (due to limited numbers of cells), R5 HIV infection levels in LCs correlated well with predicted protein levels. Taken together, these data clearly indicate that compound CCR5 polymorphisms influence productive R5 HIV infection levels in LCs, and this in turn influences the ability of LCs to transmit virus to CD4+ T cells.
Supplementary Material
Acknowledgments
We thank Mark C. Udey, Philip M. Murphy, and Donald Mosier for helpful discussions, and Ana Hancox for technical assistance. This work was supported in part by National Institutes of Health Grants AI43645 and AI51649.
Abbreviations: LC, Langerhans cell; DC, dendritic cell; mDC, monocyte-derived DC; DC-SIGN, DC-specific ICAM-3-grabbing nonintegrin; SNP, single-nucleotide polymorphism; AZT, 3′-azido-3′-deoxythymidine; RANTES, regulated on activation normal T cell expressed and secreted.
References
- 1.Royce, R. A., Sena, A., Cates, W. & Cohen, M. S. (1997) N. Engl. J. Med. 336 1072-1078. [DOI] [PubMed] [Google Scholar]
- 2.Hu, J., Gardner, M. B. & Miller, C. J. (2000) J. Virol. 74 6087-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spira, A. I., Marx, P. A., Patterson, B. K., Mahoney, J., Koup, R. A., Wolinsky, S. M. & Ho, D. D. (1996) J. Exp. Med. 183 215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turville, S. G., Arthos, J., MacDonald, K., Lynch, G., Naif, H., Clark, G., Hart, D. & Cunningham, A. L. (2001) Blood 98 2482-2488. [DOI] [PubMed] [Google Scholar]
- 5.Geijtenbeek, T. B., Kwon, D. S., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Middel, J., Cornelissen, I. L., Nottet, H. S., KewalRamani, V. N., Littman, D. R., et al. (2000) Cell 100 587-597. [DOI] [PubMed] [Google Scholar]
- 6.Turville, S. G., Cameron, P. U., Handley, A., Lin, G., Pohlmann, S., Doms, R. W. & Cunningham, A. L. (2002) Nat. Immunol. 3 975-983. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, L. Q., MacKenzie, P., Cleland, A., Holmes, E. C., Brown, A. J. & Simmonds, P. (1993) J. Virol. 67 3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu, T., Mo, H., Wang, N., Nam, D. S., Cao, Y., Koup, R. A. & Ho, D. D. (1993) Science 261 1179-1181. [DOI] [PubMed] [Google Scholar]
- 9.van't Wout, A. B., Kootstra, N. A., Mulder-Kampinga, G. A., Albrecht-van Lent, N., Scherpbier, H. J., Veenstra, J., Boer, K., Coutinho, R. A., Miedema, F. & Schuitemaker, H. (1994) J. Clin. Invest. 94 2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piguet, V. & Blauvelt, A. (2002) J. Invest. Dermatol. 119 365-369. [DOI] [PubMed] [Google Scholar]
- 11.Zaitseva, M., Blauvelt, A., Lee, S., Lapham, C. K., Klaus-Kovtun, V., Mostowski, H., Manischewitz, J. & Golding, H. (1997) Nat. Med. 3 1369-1375. [DOI] [PubMed] [Google Scholar]
- 12.Reece, J. C., Handley, A. J., Anstee, E. J., Morrison, W. A., Crowe, S. M. & Cameron, P. U. (1998) J. Exp. Med. 187 1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamura, T., Cohen, S. S., Borris, D. L., Aquilino, E. A., Glushakova, S., Margolis, L. B., Orenstein, J. M., Offord, R. E., Neurath, A. R. & Blauvelt, A. (2000) J. Exp. Med. 192 1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoeteweij, J. P., Golding, H., Mostowski, H. & Blauvelt, A. (1998) J. Immunol. 161 3219-3223. [PubMed] [Google Scholar]
- 15.Patterson, B. K., Landay, A., Andersson, J., Brown, C., Behbahani, H., Jiyamapa, D., Burki, Z., Stanislawski, D., Czerniewski, M. A. & Garcia, P. (1998) Am. J. Pathol. 153 481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, R., Paxton, W. A., Choe, S., Ceradini, D., Martin, S. R., Horuk, R., MacDonald, M. E., Stuhlmann, H., Koup, R. A. & Landau, N. R. (1996) Cell 86 367-377. [DOI] [PubMed] [Google Scholar]
- 17.Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Smith, M. W., Allikmets, R., Goedert, J. J., Buchbinder, S. P., Vittinghoff, E., Gomperts, E., et al. (1996) Science 273 1856-1862. [DOI] [PubMed] [Google Scholar]
- 18.Samson, M., Libert, F., Doranz, B. J., Rucker, J., Liesnard, C., Farber, C. M., Saragosti, S., Lapoumeroulie, C., Cognaux, J., Forceille, C., et al. (1996) Nature 382 722-725. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y., Paxton, W. A., Wolinsky, S. M., Neumann, A. U., Zhang, L., He, T., Kang, S., Ceradini, D., Jin, Z., Yazdanbakhsh, K., et al. (1996) Nat. Med. 2 1240-1243. [DOI] [PubMed] [Google Scholar]
- 20.Michael, N. L., Chang, G., Louie, L. G., Mascola, J. R., Dondero, D., Birx, D. L. & Sheppard, H. W. (1997) Nat. Med. 3 338-340. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman, P. A., Buckler-White, A., Alkhatib, G., Spalding, T., Kubofcik, J., Combadiere, C., Weissman, D., Cohen, O., Rubbert, A., Lam, G., et al. (1997) Mol. Med. 3 23-36. [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman, T. L., MacGregor, R. R., Burger, H., Mick, R., Doms, R. W. & Collman, R. G. (1997) J. Infect. Dis. 176 1093-1096. [DOI] [PubMed] [Google Scholar]
- 23.Marmor, M., Sheppard, H. W., Donnell, D., Bozeman, S., Celum, C., Buchbinder, S., Koblin, B. & Seage, G. R., 3rd. (2001) J. Acquired Immune Defic. Syndr. 27 472-481. [DOI] [PubMed] [Google Scholar]
- 24.Wu, L., Paxton, W. A., Kassam, N., Ruffing, N., Rottman, J. B., Sullivan, N., Choe, H., Sodroski, J., Newman, W., Koup, R. A. & Mackay, C. R. (1997) J. Exp. Med. 185 1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott, D. H., Zimmerman, P. A., Guignard, F., Kleeberger, C. A., Leitman, S. F. & Murphy, P. M. (1998) Lancet 352 866-870. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M. P., Dean, M., Smith, M. W., Winkler, C., Gerrard, B., Michael, N. L., Lee, B., Doms, R. W., Margolick, J., Buchbinder, S., et al. (1998) Science 282 1907-1911. [DOI] [PubMed] [Google Scholar]
- 27.Kostrikis, L. G., Neumann, A. U., Thomson, B., Korber, B. T., McHardy, P., Karanicolas, R., Deutsch, L., Huang, Y., Lew, J. F., McIntosh, K., et al. (1999) J. Virol. 73 10264-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mummidi, S., Ahuja, S. S., McDaniel, B. L. & Ahuja, S. K. (1997) J. Biol. Chem. 272 30662-30671. [DOI] [PubMed] [Google Scholar]
- 29.Carrington, M., Dean, M., Martin, M. P. & O'Brien, S. J. (1999) Hum. Mol. Genet. 8 1939-1945. [DOI] [PubMed] [Google Scholar]
- 30.Mummidi, S., Bamshad, M., Ahuja, S. S., Gonzalez, E., Feuillet, P. M., Begum, K., Galvis, M. C., Kostecki, V., Valente, A. J., Murthy, K. K., et al. (2000) J. Biol. Chem. 275 18946-18961. [DOI] [PubMed] [Google Scholar]
- 31.An, P., Martin, M. P., Nelson, G. W., Carrington, M., Smith, M. W., Gong, K., Vlahov, D., O'Brien, S. J. & Winkler, C. A. (2000) AIDS 14 2117-2122. [DOI] [PubMed] [Google Scholar]
- 32.Ometto, L., Bertorelle, R., Mainardi, M., Zanchetta, M., Tognazzo, S., Rampon, O., Ruga, E., Chieco-Bianchi, L. & De Rossi, A. (2001) J. Infect. Dis. 183 814-818. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez, E., Bamshad, M., Sato, N., Mummidi, S., Dhanda, R., Catano, G., Cabrera, S., McBride, M., Cao, X. H., Merrill, G., et al. (1999) Proc. Natl. Acad. Sci. USA 96 12004-12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez, E., Dhanda, R., Bamshad, M., Mummidi, S., Geevarghese, R., Catano, G., Anderson, S. A., Walter, E. A., Stephan, K. T., Hammer, M. F., et al. (2001) Proc. Natl. Acad. Sci. USA 98 5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangano, A., Gonzalez, E., Dhanda, R., Catano, G., Bamshad, M., Bock, A., Duggirala, R., Williams, K., Mummidi, S., Clark, R. A., et al. (2001) J. Infect. Dis. 183 1574-1585. [DOI] [PubMed] [Google Scholar]
- 36.Tang, J., Shelton, B., Makhatadze, N. J., Zhang, Y., Schaen, M., Louie, L. G., Goedert, J. J., Seaberg, E. C., Margolick, J. B., Mellors, J. & Kaslow, R. A. (2002) J. Virol. 76 662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawamura, T., Gatanaga, H., Borris, D. L., Connors, M., Mitsuya, H. & Blauvelt, A. (2003) J. Immunol. 170 4260-4266. [DOI] [PubMed] [Google Scholar]
- 38.Salkowitz, J. R., Purvis, S. F., Meyerson, H., Zimmerman, P., O'Brien, T. R., Aledort, L., Eyster, M. E., Hilgartner, M., Kessler, C., Konkle, B. A., et al. (2001) Clin. Immunol. 98 200-211. [DOI] [PubMed] [Google Scholar]
- 39.Smith, M. W., Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Lomb, D. A., Goedert, J. J., O'Brien, T. R., Jacobson, L. P., Kaslow, R., et al. (1997) Science 277 959-965. [DOI] [PubMed] [Google Scholar]
- 40.Ball, S. C., Abraha, A., Collins, K. R., Marozsan, A. J., Baird, H., QuiñonesMateu, M. E., Penn-Nicholson, A., Murray, M., Richard, N., Lobritz, M., et al. (2003) J. Virol. 77 1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, B., Leslie, G., Soilleux, E., O'Doherty, U., Baik, S., Levroney, E., Flummerfelt, K., Swiggard, W., Coleman, N., Malim, M. & Doms, R. W. (2001) J. Virol. 75 12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donzella, G. A., Schols, D., Lin, S. W., Este, J. A., Nagashima, K. A., Maddon, P. J., Allaway, G. P., Sakmar, T. P., Henson, G., De Clercq, E. & Moore, J. P. (1998) Nat. Med. 4 72-77. [DOI] [PubMed] [Google Scholar]
- 43.Meng, G., Wei, X., Wu, X., Sellers, M. T., Decker, J. M., Moldoveanu, Z., Orenstein, J. M., Graham, M. F., Kappes, J. C., Mestecky, J., et al. (2002) Nat. Med. 8 150-156. [DOI] [PubMed] [Google Scholar]
- 44.Fotopoulos, G., Harari, A., Michetti, P., Trono, D., Pantaleo, G. & Kraehenbuhl, J. P. (2002) Proc. Natl. Acad. Sci. USA 99 9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soilleux, E. J. & Coleman, N. (2001) Blood 98 1987-1988. [DOI] [PubMed] [Google Scholar]
- 46.Hartgers, F. C., Figdor, C. G. & Adema, G. J. (2000) Immunol. Today 21 542-545. [DOI] [PubMed] [Google Scholar]
- 47.Lehner, T. (2002) Trends Immunol. 23 347-351. [DOI] [PubMed] [Google Scholar]
- 48.Salkowitz, J. R., Meyerson, H., Valdez, H., Mosier, D. E., Harding, C. F., Zimmerman, P. A. & Lederman, M. M. (2003) Clin. Immunol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.