Abstract
Inactivation of the tumor suppressor p53 by missense mutations is the most frequent genetic alteration in human cancers. The common missense mutations in the TP53 gene disrupt the ability of p53 to bind to DNA and consequently to transactivate downstream genes. However, it is still not fully understood how a large number of the remaining mutations affect p53 structure and function. Here, we used a comprehensive site-directed mutagenesis technique and a yeast-based functional assay to construct, express, and evaluate 2,314 p53 mutants representing all possible amino acid substitutions caused by a point mutation throughout the protein (5.9 substitutions per residue), and correlated p53 function with structure- and tumor-derived mutations. This high-resolution mutation analysis allows evaluation of previous predictions and hypotheses through interrelation of function, structure and mutation.
The p53 tumor suppressor is a 393-aa transcription factor. In response to various types of genotoxic stresses, p53 transactivates a number of genes by binding to specific DNA sequences (1), thereby arresting cell cycle, repairing damaged DNA, or inducing apoptosis as the cell fates (2). The structure of the p53 core DNA-binding domain (residues 94–312) that binds directly to the DNA sequence has been resolved by x-ray crystallography, and both x-ray crystallography and NMR analysis have been used to deduce the structure of the tetramerization domain (residues 323–356), which is needed for optimum function (3–5). The p53 transactivity is regulated by posttranslational mechanisms such as phosphorylation, acetylation, and prolyl isomeration (6–10), or by protein–protein interaction (11). Through these mechanisms, p53 may select a subset of target promoters by changing its structure and affinity to bind to the DNA sequences with variations among the downstream genes. However, little is known about the underlying mechanism that regulates selectivity of the downstream genes and resulting cell fate.
Somatic TP53 mutations are the most common (≈50%) genetic alteration in human cancer (12), and a large number of TP53 mutations have been assembled in two major TP53 mutation databases (13, 14). The latest International Agency for Research on Cancer (IARC) TP53 mutation database contains 17,689 somatic mutations and 225 germ-line mutations. Among these, 97% of p53 mutations are clustered in the core DNA-binding domain and >75% of the mutations are missense mutations. So far, 1,135 distinct amino acid substitutions caused by missense mutations have been documented. There is little increase in the number of amino acid substitutions since 1999, suggesting that almost all types of naturally occurring pathogenic missense mutations have been already reported (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Therefore, the tumor-derived missense mutations may inactivate p53 by affecting its ability to bind to DNA, and unreported missense mutations might be nonpathogenic for tumorigenesis. Accordingly, a limited number of tumor-derived missense mutations have been shown to be functionally defective, but the majority of the >1,000 distinct types of amino acid substitutions are still unexamined.
Analysis of tumor-derived missense mutations and previously unreported missense mutations should advance our understanding of function–structure and function–mutation relationships, and thereby pathogenesis of tumor. In this study, we constructed 2,314 distinct p53 mutants that represented all possible amino acid substitutions caused by a point mutation throughout the protein (5.9 substitutions per residue) and were evaluated for their transactivities for many reporters. We describe here the correlation of p53 function with structure- and tumor-derived mutations.
Materials and Methods
Site-Directed Mutagenesis. To design oligonucleotide sequences for site-directed mutagenesis, we first substituted every first, second, and third nucleotide of all but the first ATG codon of p53 (codons 2–393) by another nucleotide, and then excluded nonsense mutations and silent mutations. When two substitutions encoded the same amino acid, we chose one of them according to whether the substitution is more frequently reported than the other(s) or more frequently used in natural human cDNA (15). According to the rule, we synthesized 2,314 distinct mutant oligonucleotides with one base mismatch (see Table 1, which is published as supporting information on the PNAS web site). The strategy to generate a mutant p53 cDNA are schematically shown in Fig. 6 and Tables 2 and 3, which are published as supporting information on the PNAS web site, and explained in the Fig. 6 legend.
Gap Repair Vectors and Yeast Transformation. The basic method of yeast transformation and gap repair assay was reported (16). Three different plasmids were used as gap vectors for the construction of the mutant p53 expression vectors in yeast (see Fig. 7, which is published as supporting information on the PNAS web site). The resulting 2,314 yeast transformants (YPH499-derived haploid cells) were stored at -80°C in 25 96-well plates.
Plasmids. For reporter plasmids in yeast system, we selected eight p53-binding sequences as transcriptional enhancer elements: two from human DNA fragments (the p21WAF1 and MDM2 promoters) and six from synthetic oligonucleotides (p53-binding elements of BAX, GADD45, 14-3-3σ, p53AIP1, Noxa, and p53R2) and inserted each of them into the enhancerless GFP or Ds-Red reporter plasmid. pAS03G, which contains two p53-responsive elements (p53RE, 5′-GAACATGTCCCAACATGTTG-3′ and 5′-AGACTGGGCATGTCTGGGCA-3′) and the flanking sequences of human WAF1 promoter upstream of the GAL1 promoter ΔUAS-EGFP expression cassette (17), was introduced into YPH499 (Stratagene). pKS01Rnull plasmid was constructed by inserting the GAL1 promoter ΔUAS and a fusion sequence of the 5′ coding region of HIS3 (51 bp) and full-length Ds-Red cDNA into pRS323 (18) at the EagI and BamHI sites. pKS05R contains two p53RE (5′-GGTCAAGTTCAGACACGTT-3′ and 5′-AGTTAAGTCCTGACTTGTCT-3′) and the flanking sequences of the MDM2 promoter sequence (GenBank accession no. U28935, nucleotides 686–791) at the EagI site of pKS01Rnull. The other pKS plasmids contain three tandem copies of the p53RE derived from the BAX, 14-3-3σ, p53AIP1, GADD45, Noxa, and p53R2 genes at the EagIsite of pKS01Rnull, and are called pKS07R, pKS09R, pKS11R, pKS13R, pKS15R, and pKS17R, respectively. The p53RE sequences in the pKS plasmids are as follows: pKS07R, 5′-AGACAAGCCTGGGCGTGGGC-3′;pKS09R,5′-AGGCATGTGCCACCATGCCC-3′; pKS11R, 5′-TCTCTTGCCCGGGCTTGTCG-3′; pKS13R, 5′-CAGCATGCTTAGACATGTTC-3′; pKS15R, 5′-AGGCTTGCCCCGGCAAGTTG-3′; and pKS17R, 5′-TGACATGCCCAGGCATGTCT-3′. These pKS plasmids were introduced into YPH500 (Stratagene). For p53 expression experiment in human cells, the wild-type p53 expression vector, pCR259-p53WT, was constructed by inserting a PCR-amplified wild-type TP53 cDNA into the EcoRI/EagI sites of pCR259 (Qbiogene, Carlsbad, CA). The mutant p53 cDNAs derived from yeast p53 expression vector were extracted by Zymoprep (Zymo Research, Orange, CA) yeast plasmid mini preparation kit and were amplified by PCR. The cDNAs were then introduced into EcoRI/EagI sites of the pCR259 plasmid. The mutant p53 expression vectors, pCR259-p53MT, were identical except for specific single-nucleotide substitutions in the TP53 cDNA. Luciferase reporter plasmids p21Ps-luc, pMDMPs-luc, pBAXPs-luc, and pSIGMAPs-luc for luciferase assay were described (17). Luciferase reporter plasmids p53R2Ps-luc and p53GADD45Ps-luc were constructed by inserting a 630-bp PCR fragment derived from p53R2 intron 1 (GenBank accession no. AP002907, nucleotides 58131–58761) and a 333-bp PCR fragment derived from GADD45 intron 3 (GenBank accession no. L24498, nucleotides 3645–3978), respectively, into an EcoRI site of pGL3E (17).
DNA Sequencing. The mutant p53 cDNA were amplified from yeast cells or p53 expression plasmids by PCR and were sequenced by using a DTCS DNA sequencing kit (Beckman Coulter) and an automated CEQ2000EX DNA sequencer (Beckman Coulter). The primers for DNA sequencing were shown in Table 4, which is published as supporting information on the PNAS web site.
Mating Assay. The 2,314 p53-expressing YPH499 (MATa) strains were inoculated from the frozen stock onto the surface of synthetic complete (SC) solid medium lacking leucine and tryptophan with autoclaved 96-pin replicators (Incyte Genomics) and were grown at 30°C for 2 days. YPH500 (MATα) strain, harboring each of the Ds-Red reporter plasmids was spread on the surface of yeast extract/peptone/dextrose (YPD) plates and grown at 30°C for 12 h. The p53-expressing YPH499 strains were then mated on the lawn of the YPH500-spread YPD plates with the autoclaved 96-pin replicators and further incubated at 30°C for 24 h. The mating cells were selected by touching the surface of the YPD plates with the tip of the replicators and inoculated on SC solid medium lacking leucine, tryptophan, and histidine. After incubation at 30°Cfor1or 2 days, the selected diploid (mating type a/α) cells were inoculated on the fresh medium.
Fluorescence Intensity. The yeast clones were replicated on the selective SC solid media and were grown at 37°C for 2 days. The plates were then directly processed to a 96-well formatted Fluoroskan Ascent FL fluorometer (Labsystems, Helsinki, Finland) to measure their fluorescence intensity (excitation: 485 nm, emission: 538 nm) for p53-dependent enhanced green fluorescent protein (EGFP) expression through the human p21WAF1-derived p53-binding sequence, and the fluorescence intensity (excitation: 544 nm, emission: 590 nm) for Ds-Red through other p53-binding sequences. At least two independent experiments were performed for each reporter and the fluorescence intensities were averaged. The averaged values were standardized for each p53-binding sequence, clustered, and visualized with the cluster and treeview programs.
Luciferase Assay. Saos-2 cells were cultured in 96-well assay plates (Corning) in RPMI medium 1640 with 10% FCS at 37°C. When the cells reached at ≈60-90% confluence, pCR259-p53WT or pCR259-p53MT (12.5–50 ng), and one of the six luciferase reporter plasmids (50–87.5 ng), were cotransfected into the cells by using an Effectene transfection reagent (Qiagen, Valencia, CA). After 24 h, luciferase activity was assayed by using a Steady-Glo luciferase assay system (Promega) and was measured by a Fluoroskan Ascent FL fluorometer. Six experimental values of the luciferase activities for the p53 mutants were compared with that of wild-type p53 and the statistical significance (P > 0.001) was examined by paired t test.
Susceptibility Scores. p53 mutants (2,314) were fractionated into p53 structural categories including domains, secondary structures, evolutionary amino acid conservation, DNA contact, and Zn binding, yielding the following three sets of susceptibility scores (i) for function. The ratio of the number of functionally inactive p53 mutants to the number of constructed mutants in each structural category was calculated for each promoter. If all p53 mutants in a structural category have no transactivity, the category is scored as 1. If all p53 mutants in a structural category have transactivity, the category is scored as 0; (ii) for somatic mutation. The ratio of the number of reported somatic mutants in the IARC database to the number of constructed p53 mutants in each structural category was calculated for each promoter. If none of the p53 mutants in a structural category have been listed in the database, the category is scored as 0. If all of the p53 mutants in a structural category have been listed, the category is scored as 1; and (iii) for mutation frequency. The ratio of the frequency of the amino acid substitution in the IARC somatic mutation database to the number of constructed p53 mutants in each structural category was calculated for each promoter. If the total frequency of the amino acid substitution in a structural category is 100, and the number of constructed mutants in the category is 20, the category is scored as 5. The data were summarized in Table 5, which is published as supporting information on the PNAS web site, and the standardized values were visualized with the cluster and treeview programs.
Drawing of p53 Peptide Structures. To draw the p53 core domain and oligomerization domain, we downloaded the two National Center for Biotechnology Information structure files 1TUP and 3SAK. These files were customized for our purpose and visualized with cn3d software (19) or the swiss-pdb viewer (20).
Results and Discussion
Construction of 2,314 Distinct p53 Missense Mutations. Based on a combination of a PCR-mediated megaprimer method and a yeast-based gap repair assay (see Materials and Methods and Figs. 6 and 7), we constructed 2,314 p53 mutants. This result was achieved by synthesizing 2,314 distinct oligonucleotides, each of which consisted of 26 nucleotides with one base mismatch at the 14th nucleotide from the 5′ end, which were distributed into 25 96-well microtiter plates (Fig. 1a). Full-length TP53 cDNAs with each missense mutation were then generated by two-step PCRs in the 96-well format with wild-type TP53 cDNA templates. Each of the mutations was constructed into a gap p53-expressing vector by homologous recombination in vivo in a yeast strain harboring a p53-responsive p21WAF1 reporter plasmid. We confirmed appropriate incorporation of missense mutations by direct DNA sequencing of all TP53 cDNAs and expression of the p53 protein. The resulting mutation library consists of 2,314 yeast clones containing distinct TP53 cDNAs with point mutations covering all possible amino acid substitutions caused by single-nucleotide substitutions throughout the full length of the TP53 ORF, except for the translation initiation ATG codon. The yeast clones expresses a corresponding mutant p53 protein under the control of the constitutive yeast ADH1 promoter. The library provides an average of 5.9 substitutions in each residue, and covers >95% of the tumor-derived p53 amino acid substitutions listed in the IARC database, as well as a large number of previously unreported substitutions.
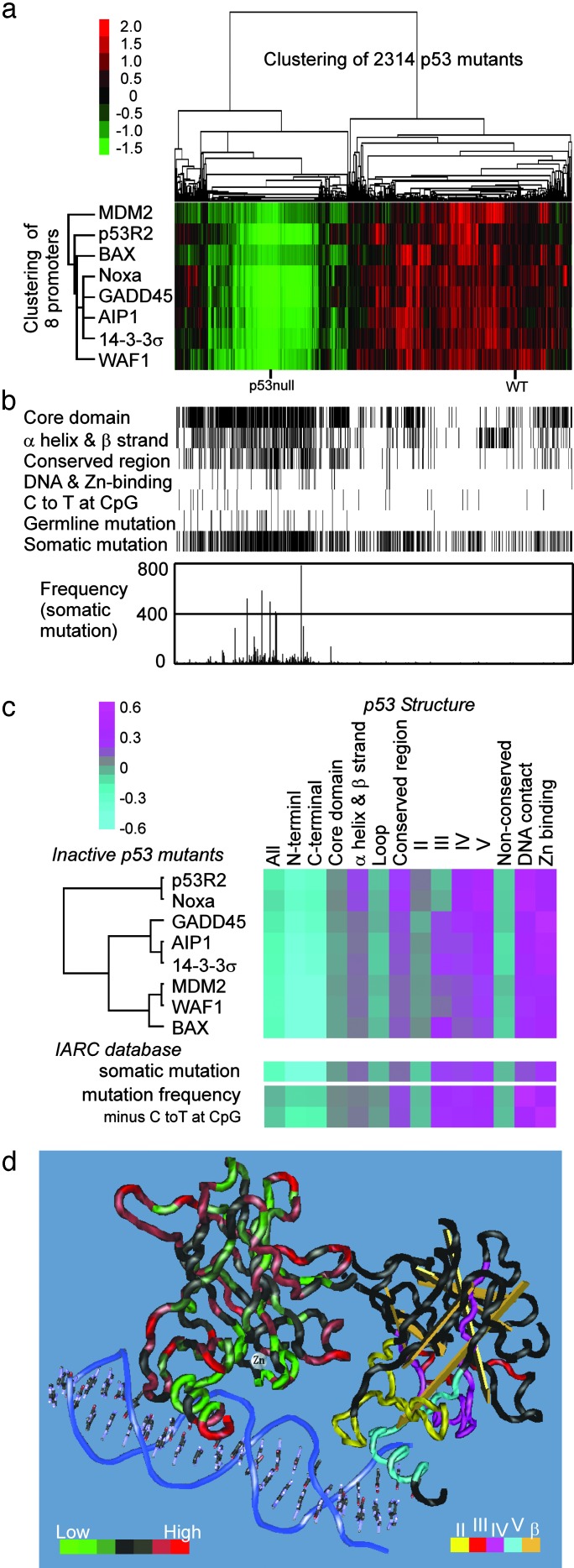
Fig. 1.
Construction of a p53 missense mutation library and a map of sequence-specific transactivities. (a) Schematic flow of construction of the mutant library. An example (P250A) of the 2,314 mutant p53 clones is shown. An oligonucleotide with a point mutation was used for PCR-based site-directed mutagenesis. The second PCR product was cotransformed with a linearized gap vector into a haploid yeast strain. The resulting yeast haploid cells (MATa) expressed a distinct p53 protein with an amino acid substitution corresponding to the original point mutation and also expressed EGFP, depending on the ability to transactivate through the p53BS of the human p21WAF1 promoter. The p53-expressing haploid cells were mated with other haploid cells (MATα) harboring a Ds-Red reporter plasmid with a p53BS other than the p21WAF1 promoter. (b) Schematic map of the p53 mutants showing transactivities for p21WAF1 promoter and mutation frequencies. (Bottom) The fluorescence intensities of the expressed EGFP (CLONTECH) for 2,314 clones were measured by a 96-well formatted fluorometer. Mean values derived from three independent experiments on the 2,314 clones were mapped from the NH2 terminus to the COOH terminus of the p53 protein. (Top) The frequency of each p53 missense mutation extracted from the IARC database (14) is indicated by the bar graph and 5 of the 10 most frequently reported mutations are labeled. (Middle) Primary structure of p53 showing the NH2-terminal portion (residues 1–95, purple) with the transactivation domain, the functionally defined DNA-binding domain (residues 96–286, yellow), and the COOH-terminal portion (residues 287–393, light blue) with the tetramerization domain. Residue numbers are italicized. Horizontal lines represent evolutionarily conserved regions I-V among vertebrates (29). The gray zone across the top and bottom indicates the functionally defined DNA-binding domain. (c) Fluorescence intensities of EGFP (p21WAF1) or Ds-Red (MDM2, BAX, 14–3-3σ, p53AIP1, GADD45, Noxa, and p53R2) were standardized and mapped from the NH2 terminus to the COOH terminus of the p53 protein. Relative fluorescence intensities are displayed from red (highest) to green (lowest). (d) The fluorescence intensities EGFP (p21WAF1) of the 2,314 clones were divided into 40 fractions and the frequency in each fraction is shown graphically. The NH2-terminal portion (purple), the sequence-specific DNA-binding domain (yellow), and the COOH-terminal portion (light blue) were also fractionated separately. The mean values of fluorescence intensities from wild-type p53 and p53 null clones are indicated as arrows.
Evaluation of 2,314 Missense Mutations on Sequence-Specific Transactivation. To evaluate the effect of each mutant on p53 function, the ability of the expressed mutant p53 to transactivate the EGFP reporter plasmid through the p53 binding sequence (p53BS) derived from the p21WAF1 promoter was quantitatively measured by the fluorescence intensity of EGFP, and mapped onto the p53 primary structure (Fig. 1b). Overall, one-third (36.0%) of the p53 mutants were functionally inactive. Approximately two-thirds (64.0%) of the mutants in the core domain of p53 were inactive, whereas activity was mostly unaffected by both NH2- and COOH-terminal mutations, except for a significant number of inactivating mutations within the tetramerization domain. The data clearly show marked differences in sensitivity to amino acid substitutions among the three p53 domains (see also Table 5) and support the previous speculation that both the NH2 and COOH termini are insensitive to amino acid substitutions. Combined the data with our single-nucleotide substitution model (see Fig. 8, which is published as supporting information on the PNAS web site), we conclude that this is the major reason why tumor-derived TP53 missense mutations are clustered within the DNA-binding domain and rarely observed in the NH2 and COOH termini. There were clear limits between residues 95 and 96 and between residues 286 and 287 (Fig. 1 b and c). This functionally defined DNA-binding domain (residues 96–286) corresponds well to the proteolysis-resistant and structurally compact core domain (subtilisin fragment, residues 102–292; thermolysin fragment, residues 92–30; refs 21 and 22).
When the transcriptional transactivities of the p53 mutants were fractionated (Fig. 1d), there were at least three subtypes: mutants with no activity (type N: 9.6%), mutants with significantly reduced but some residual activity (type R: 26.5%), and mutants with activity comparable with that of wild-type p53 (type W: 63.9%). The transactivities of the mutants for other p53BSs derived from MDM2, BAX, 14-3-3σ, p53AIP1, GADD45, Noxa, and p53R2 promoters were also determined quantitatively from the Ds-Red fluorescence intensity of diploid yeast clones obtained by mating the p53-expressing haploid cells with another haploid cell line harboring one of the Ds-Red reporter plasmids (Fig. 1a). The values for each p53BS were standardized and mapped onto the p53 primary structure (Fig. 1c). The results were similar to those with the p21WAF1 promoter. Overall, about one-third (19–41%, depending on the p53BS) of the p53 mutants were functionally inactive.
The Function–Structure and Function–Mutation Relationships. To gain an insight into the relationships among p53 function, protein structure, and mutation, the 2,314 missense mutants plus p53 null and wild-type p53 controls were clustered by unsupervised two-dimensional analysis (Fig. 2a). The transactivities of the p53 mutants were divided into two large clusters: the first cluster contained wild-type p53 and the mutants with functional activity for all or most of the promoters (56.3%), and the second cluster contained p53-null clones and the mutants lacking functional activity for all or most of the promoters (43.7%). Background information related to p53 structural features (residues in core domain, secondary structure, highly conserved regions, DNA-contact, and Zn-binding) and the IARC database (tumor-related germ-line mutations, somatic mutations, and their frequencies) for each amino acid substitution was then correlated to the clusters (Fig. 2b). Clearly, these structural features and the IARC information tend to concentrate in the second cluster, whereas C-to-G transition mutations at CpG sites, unrelated to p53 structure and function, show no such tendency. These results indicate a strong correlation of the p53 transactivation function with p53 structure- and tumor-related mutations.
Fig. 2.
Relationships among p53 function, structure, and mutation. (a) Unsupervised two-dimensional analysis of 2,314 p53 missense mutants plus p53 null and wild-type p53 controls. Fluorescence intensities of EGFP (p21WAF1) or Ds-Red (MDM2, BAX, 14-3-3σ, p53AIP1, GADD45, Noxa, and p53R2) were standardized and visualized by the cluster and treeview programs. Each column represents a p53 mutant and each row a p53 promoter. Relative fluorescence intensities are displayed from red (highest) to green (lowest). (b) Structural and IARC mutation information for the 2,314 mutants in a: residues in p53 core domains, four evolutionarily conserved regions in the core domain, secondary structure (α-helix and β-strand), DNA-contact, Zn-binding, germ-line mutation, and somatic mutation. Mutants caused by C-to-T transition at CpG sites are also noted. The frequency of somatic mutation for each mutant is shown graphically. (c) Comparison of relationship between p53 function and structure with relationship between p53 mutation and structure is shown. Susceptibility scores for p53 function, somatic mutation, and mutation frequency were calculated (see Materials and Methods and Table 5) and were visualized with the cluster and treeview programs. Relative susceptibility scores are displayed from pink (highest) to light blue (lowest). (d) Three-dimensional structure of the p53 core DNA-binding domain (National Center for Biotechnology Information 1TUP file) visualized by cn3d version 4.0 (19). The transactivities of the p53 mutants for the p21WAF1 promoter in each residue were averaged and were shown on the left (1TUP molecule B). The transactivities relative to wild-type activity are divided into seven fractions (<0.1, 0.1–0.15, 0.15–0.2, 0.2–0.25, 0.25–0.35, 0.35–0.6, and >0.6) and displayed from green (lowest) to red (highest). Structural information (highly conserved regions II-V and β-strands) was shown on the right (1TUP molecule C) using the indicated colors.
To confirm the correlation of p53 structure with function and tumor-related mutations, susceptibility for the p53 transactivity, somatic mutation, or mutation frequency in each p53BS was calculated for the several structural categories (see Materials and Methods and Table 5) and the standardized values were visualized in Fig. 2c. The DNA-binding domain is clearly more susceptible to inactivation by amino acid substitution than are the NH2- and COOH-terminal domains. Within the DNA-binding domain, the secondary structures (the two α-helices or the 11 β-strands) were more susceptible to amino acid substitution than were the loops (L1–3 and other short loops) connecting α-helix and β-strand or two β-strands. The four evolutionarily conserved regions are more susceptible than the nonconserved regions. Among the regions, regions IV and V, which are spatially close to DNA (Fig. 2d) are more susceptible than regions II and III, which are supportive structures for p53 binding to DNA. Furthermore, approximately half of the mutants in regions IV and V were type N, whereas most of the mutants in regions II and III were type R or type W (see above). DNA-contact residues and Zn-binding residues showed the strongest susceptibility among the categories. Overall patterns of the susceptibility scores for p53 function were similar among the distinct p53BSs, as were the susceptibility scores for IARC somatic mutation. These results indicate a strong correlation of p53 structure with transactivation function and with tumor-related mutations, and also an interrelation between p53 function and mutation. The cluster analysis divided the eight p53BSs into three subgroups. The first group (MDM2, p21WAF1, and BAX) was most similar to the susceptibility for mutation frequency. The second group (GADD45, p53AIP1, and 14-3-3σ) was also similar to the first, but showed higher susceptibility in Zn-binding residues. The third group (p53R2 and Noxa) was less similar to the second and showed lower susceptibility in region III. The difference in the similarities among the subgroups might mean the different contributions of the downstream genes for tumor suppression, although there are no supportive evidences at present. The average transactivities of the p53 mutants for the p21WAF1 promoter were mapped in the 3D structure of the p53 DNA-binding domain (Fig. 2d).
Function–Structure Relationship in the Tetramerization Domain of p53. Function–structure relationship was also investigated in the COOH-terminal portion of p53. Specific residues in the α-helix and β-strand of the tetramerization domain were sensitive to inactivation by amino acid substitutions (Fig. 3a). These 15 residues are Phe-328, Leu-330, Ile-332, and Arg-333 in the β-strand, Gly-334 in a turn, and Arg-337, Phe-338, Phe-341, Arg-342, Leu-344, Asn-345, Ala-347, Leu-348, Leu-350, and Lys-351 in the α-helix. Leu-330, Ile-332, Gly-334, Arg-337, Phe-341, Leu-344, and Ala-347, especially, were highly susceptible to functional inactivation because they were inactivated by at least three distinct substitutions. Substitution of the hydrophobic residues (Phe, Leu, Ile, or Ala) that form a hydrophobic core in the domain (5) by nonhydrophobic residues or by structure-destroying proline tends to inactivate p53 function (data not shown). Among the charged residues (Arg or Lys), Arg-337 was highly sensitive, and any of six substitutions were functionally defective. When these residues were mapped on the 3D structure of this domain (Fig. 3 b–d), it became clear that many of these residues locate spatially face-to-face and seem to be involved in monomer–monomer interaction to form a dimer through β-strand–β-strand and α-helix–α-helix interactions, and in dimer–dimer interactions to form a tetramer through an α-helix–α-helix interaction. Our data clearly underscore the structure-based studies predicting the functional importance of these residues (5, 23) and the significance of tetramer formation for p53 transactivation. Combined with the data for the core structure described above, our study supports a strong interrelation between p53 function and structure in the two structural domains.
Fig. 3.
Schematic representation of the transactivity of p53 mutants within the tetramerization domain. (a) The transactivities of 204 p53 mutants within residues 323–356 were mapped on the primary structure. (b–d) Residues affected by at least one type of amino acid substitution were mapped on the 3D structure (National Center for Biotechnology Information 3SAK file visualized by swiss-pdb viewer; ref. 20) of the tetramerization domain. The backbone structure (ribbon) is green (affected residues) or red (unaffected residues). (b) Right side view. The side chains of ten affected residues in theα-helix are shown. (c) Top view. The side chains of five affected residues in the β-strand are shown. (d) Front view. The side chains of three affected arginine residues are shown.
Classification and Interpretation of the Clustered Mutations. Among the 2,314 p53 mutants, 1,266 (54.7%) of them were easy explainable by the function–mutation hypothesis: 905 mutants (39.1%) showed intact transactivity on all eight promoters and have never been reported in tumors, whereas 361 mutants (15.6%) showed inactive transactivity on all of the eight promoters and have been reported at least once in tumors. There are three types of exceptions. First, 373 mutants (16.1%) have been reported in tumors at least once, even though the mutants retained wild-type activity on all promoters. There are several possible explanations. (i) Such mutants are functionally silent but were accidentally expanded during clonal selection of tumor cells. In fact, there are many examples where tumors with such mutants also contain additional inactivating mutations (data not shown). (ii) There might be a minor tumor-suppressive p53 function unrelated directly to transactivity, and such mutants disrupt this function. (iii) There might be unexamined downstream genes unable to be transactivated by the mutants. Second, 39 mutants (1.7%) have never been reported in tumors although the mutants were inactive on all promoters. We speculate that the mutants are pathogenic but genetically infrequent, and that they were not detected because most of them are located in the NH2- or COOH-terminal portions, which have not been widely examined (13).
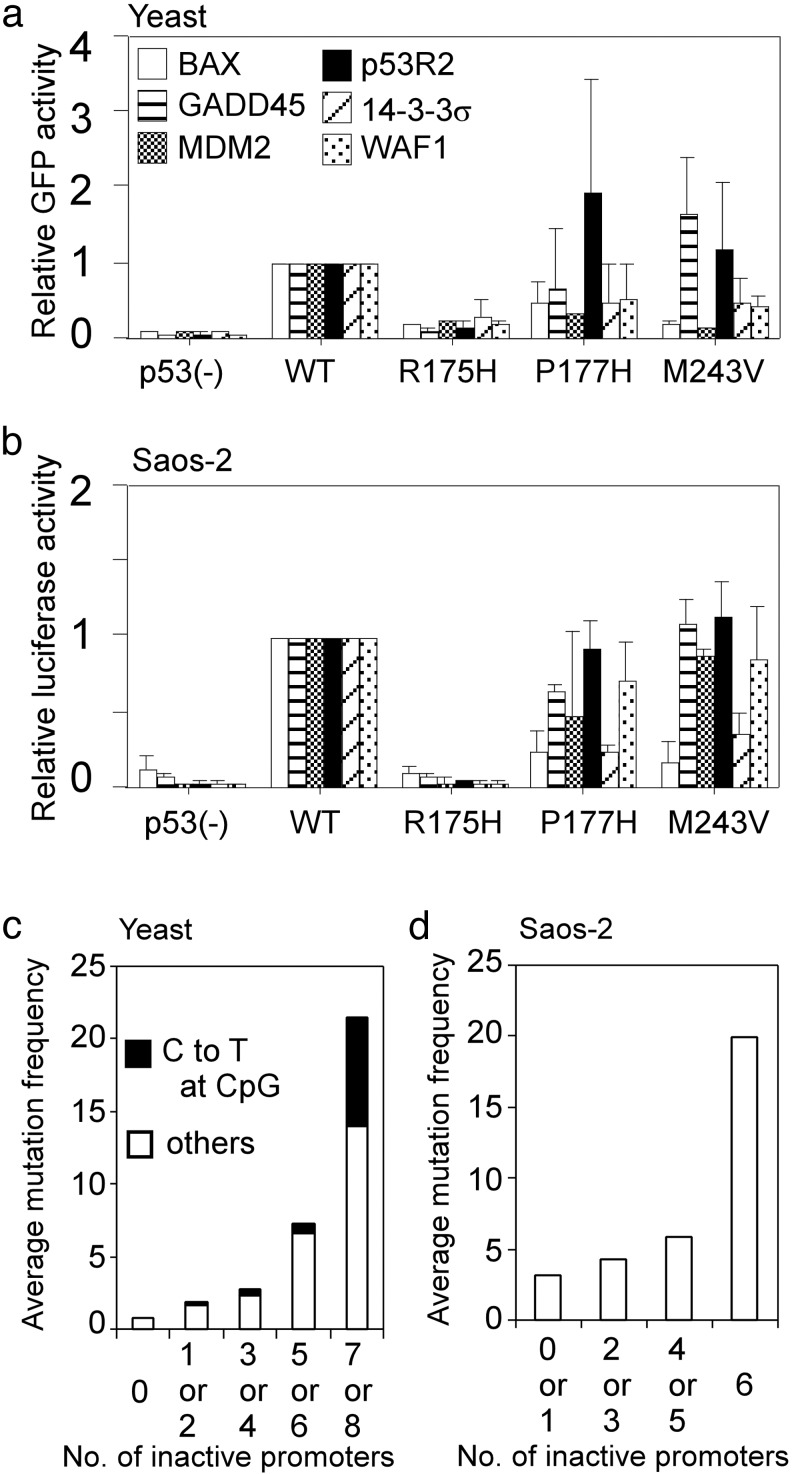
Finally, the remaining 635 mutants (27.5%) were inactive only for a limited number of promoter(s) (see Table 6, which is published as supporting information on the PNAS web site). These types of mutants are valuable for studying the possible relationship between promoter selectivity and tumorigenesis. To examine whether the promoter selectivity in yeast is analogous in human cells, we expressed some of the partially inactive mutants (P177H and M243V) as well as completely inactive R175H mutant in human osteosarcoma cell line, Saos-2 cells, and compared their ability to transactivate luciferase reporter gene through six distinct promoters with that in the yeast reporter system (Fig. 4 a and b). In both P177H and M243V mutants, spectra of transactivities were similar between the two cell systems although transactivities for the MDM2 and the WAF1 promoters were more sensitive in yeast than that in human tumor cells. In addition, S121F mutant, that fails to transactivate MDM2 and induce apoptosis better than wild-type p53 in human tumor cells (24), selectively lost transactivity for MDM2 in yeast (data not shown).
Fig. 4.
Comparison of p53 transactivation function in yeast with that in human cells. (a) Transactivities of three representative p53 mutants (R175H, P177H, and M243V) through the indicated six p53-binding sequences were shown as relative GFP (EGFP or Ds-Red) intensities against wild-type (WT) p53 activities. (b) The three mutants, which were the same as a were examined for their abilities to transactivate the luciferase reporter gene through the six corresponding promoters in Saos-2 cells. Bar, SD. (c) Average mutation frequency on number of inactivated promoters. Based on the yeast experiment, the 2,314 mutants were fractionated on the number of inactivated promoters for eight distinct p53-binding sequences. (d) Average mutation frequency on number of inactivated promoters based on the transactivities of 89 selected mutants in Saos-2 cells. Note that we did not choose mutants that theoretically occur by C-to-T transition at CpG sites as well as the top 10 hot spot mutants to avoid strong bias. In both c and d, mutation frequency for each mutant was derived from the IARC database and was averaged in each fraction.
Because the average mutation frequency depends on the number of inactive promoters, even if C-to-T transitions at CpG sites, which introduce a considerable bias unrelated to function, are excluded (Fig. 4c), these mutants are likely to have partial function in tumor suppression. Similar results were observed in the human cell system with a limited number of mutants (Fig. 4d). There was no correlation between the transactivity for a specific promoter and the mutation frequency in both yeast and human cells (data not shown). From these observations, we predict that a wide range of activity against various types of downstream genes, rather than a limited range of activity against a specific downstream gene(s), is necessary for the full tumor-suppressive function of p53; this could be the major reason why TP53 mutations are frequently found in the p53 pathway, yet mutations in p53 downstream genes are rarely found in tumors (see Fig. 9, which is published as supporting information on the PNAS web site). These results also support the hypothesis that p53 acts at a critical nodal point between upstream modulators and downstream effectors (25). During the study, we have mutagenized each of the known phosphorylation sites at Ser and Thr residues, acetylation sites at Lys residues as well as prolyl isomerase sites at Ser-Pro and at Thr-Pro motifs in the NH2 or COOH terminus. However any of these residues were basically not sensitive by single amino acid substitutions because almost all of the mutants did not lead to significant reduction of the p53 transactivities. This result is not surprising because the previous studies have shown that multiple mutations are necessary to inactivate p53 transactivity (8–10, 26).
Conclusion
A key finding of our study is that high-resolution mutation analysis over the full length of a protein provides a precise map of functionally susceptible as well as mutation-susceptible residues. In the future, additions to the map of clinical information such as response to chemotherapy and prognostic factors may permit mutation-specific prediction of clinical outcomes on a functional basis. Furthermore, the high-resolution p53 mutant library allows the isolation of a number of temperature-sensitive mutants and intragenic suppressor mutants (data not shown) that reactivate the function of p53 by permissive temperatures or second-site mutations, and may also provide mutants with enhanced ability to induce apoptosis and/or cell-cycle arrest compared with wild-type p53. Such specific mutants may be useful in investigation of p53-targeted molecular therapeutics (27).
A second key finding is that this high-resolution missense mutation analysis has advanced our understanding of the interrelation among p53 structure, function, and tumor-derived mutations. Our results support the view that, among the endogenous and exogenous mechanisms of mutation of the TP53 gene (28), inactivation of p53 function is the most important factor in the spectrum of TP53 mutation and that sequence-specific transactivation is the critical function in p53-dependent tumor suppression. Therefore, we propose that the strategy described here can provide insights into the function–structure and function–mutation relationships of a disease-associated protein, as well as a better understanding of the role of the gene product in structural and functional genetics and in molecular pathogenesis and epidemiology.
Supplementary Material
Acknowledgments
We thank Ms. Yuka Fujimaki for technical assistance. This study was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture (to C.I. and S.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: p53BS, p53 binding sequence; IARC, International Agency for Research on Cancer; EGFP, enhanced GFP.
References
- 1.el-Deiry, W. S., Kern, S. E., Pietenpol, J. A., Kinzler, K. W. & Vogelstein, B. (1992) Nat. Genet. 1 45-49. [DOI] [PubMed] [Google Scholar]
- 2.el-Deiry, W. S. (1998) Semin. Cancer Biol. 8 345-357. [DOI] [PubMed] [Google Scholar]
- 3.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265 346-355. [DOI] [PubMed] [Google Scholar]
- 4.Clore, G. M., Omichinski, J. G., Sakaguchi, K., Zambrano, N., Sakamoto, H., Appella, E. & Gronenborn, A. M. (1994) Science 265 386-391. [DOI] [PubMed] [Google Scholar]
- 5.Jeffrey, P. D., Gorina, S. & Pavletich, N. P. (1995) Science 267 1498-1502. [DOI] [PubMed] [Google Scholar]
- 6.Prives, C. & Hall, P. A. (1999) J. Pathol. 187 112-126. [DOI] [PubMed] [Google Scholar]
- 7.Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., Nishimori, H., Tamai, K., Tokino, T., Nakamura, Y. & Taya, Y. (2000) Cell 102 849-862. [DOI] [PubMed] [Google Scholar]
- 8.Barlev, N. A., Liu, L., Chehab, N. H., Mansfield, K., Harris, K. G., Halazonetis, T. O. & Berger, S. L. (2001) Mol. Cell 8 1243-1254. [DOI] [PubMed] [Google Scholar]
- 9.Zheng, H., You, H., Zhou, X. Z., Murray, S. A., Uchida, T., Wulf, G., Gu, L., Tang, X., Lu, K. P. & Xiao, Z. X. (2002) Nature 419 849-853. [DOI] [PubMed] [Google Scholar]
- 10.Zacchi, P., Gostissa, M., Uchida, T., Salvagno, C., Avolio, F., Volinia, S., Ronai, Z., Blandino, G., Schneider, C. & Del, S. G. (2002) Nature 419 853-857. [DOI] [PubMed] [Google Scholar]
- 11.Samuels-Lev, Y., O'Connor, D. J., Bergamaschi, D., Trigiante, G., Hsieh, J. K., Zhong, S., Campargue, I., Naumovski, L., Crook, T. & Lu, X. (2001) Mol. Cell 8 781-794. [DOI] [PubMed] [Google Scholar]
- 12.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253 49-53. [DOI] [PubMed] [Google Scholar]
- 13.Soussi, T., Dehouche, K. & Beroud, C. (2000) Hum. Mutat. 15 105-113. [DOI] [PubMed] [Google Scholar]
- 14.Olivier, M., Eeles, R., Hollstein, M. A., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19 607-614. [DOI] [PubMed] [Google Scholar]
- 15.Adzhubei, I. A., Adzhubei, A. A. & Neidle, S. (1998) Nucleic Acids Res. 26 327-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishioka, C., Frebourg, T., Yan, Y. X., Vidal, M., Friend, S. H., Schmidt, S. & Iggo, R. (1993) Nat. Genet. 5 124-129. [DOI] [PubMed] [Google Scholar]
- 17.Shimada, A., Kato, S., Enjo, K., Osada, M., Ikawa, Y., Kohno, K., Obinata, M., Kanamaru, R., Ikawa, S. & Ishioka, C. (1999) Cancer Res. 59 2781-2786. [PubMed] [Google Scholar]
- 18.Christianson, T. W., Sikorski, R. S., Dante, M., Shero, J. H. & Hieter, P. (1992) Gene 110 119-122. [DOI] [PubMed] [Google Scholar]
- 19.Wang, Y., Geer, L. Y., Chappey, C., Kans, J. A. & Bryant, S. H. (2000) Trends Biochem. Sci. 25 300-302. [DOI] [PubMed] [Google Scholar]
- 20.Guex, N. & Peitsch, M. C. (1997) Electrophoresis 18 2714-2723. [DOI] [PubMed] [Google Scholar]
- 21.Bargonetti, J., Manfredi, J. J., Chen, X., Marshak, D. R. & Prives, C. (1993) Genes Dev. 7 2565-2574. [DOI] [PubMed] [Google Scholar]
- 22.Pavletich, N. P., Chambers, K. A. & Pabo, C. O. (1993) Genes Dev. 7 2556-2564. [DOI] [PubMed] [Google Scholar]
- 23.Clore, G. M., Ernst, J., Clubb, R., Omichinski, J. G., Kennedy, W. M., Sakaguchi, K., Appella, E. & Gronenborn, A. M. (1995) Nat. Struct. Biol. 2 321-333. [DOI] [PubMed] [Google Scholar]
- 24.Saller, E., Tom, E., Brunori, M., Otter, M., Estreicher, A., Mack, D. H. & Iggo, R. (1999) EMBO J. 18 4424-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, P. A., Meek, D. & Lane, D. P. (1996) J. Pathol. 180 1-5. [DOI] [PubMed] [Google Scholar]
- 26.Ashcroft, M., Kubbutat, M. H. & Vousden, K. H. (1999) Mol. Cell. Biol. 19 1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane, D. P. & Lain, S. (2002) Trends Mol. Med. 8 S38-S42. [DOI] [PubMed] [Google Scholar]
- 28.Harris, C. C. & Hollstein, M. (1993) N. Engl. J. Med. 329 1318-1327. [DOI] [PubMed] [Google Scholar]
- 29.Soussi, T., Caron de Fromentel, C. & May, P. (1990) Oncogene 5 945-952. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.