Abstract
Important progress has been achieved in the knowledge about the pathogenesis of cancer. However, despite these advances, the therapeutic strategies are still limited. Leukemias are often characterized by specific balanced translocations, with the t(8;21) balanced translocation being the most frequent chromosomal aberration in acute myeloid leukemia (AML). This translocation produces the AML1-ETO fusion protein, which binds to AML1 target promoter sequences. Transcriptional repression of AML1-dependent genes by AML1-ETO and associated corepressors represents the pathogenetic mechanisms of t(8;21). Here, we show that targeting of AML1-ETO to essential, MYB-dependent gene promoters induces t(8;21)-restricted cell death. We constructed a chimeric protein that contained the MYB DNA-binding domain and the AML1-binding domain of myeloid Elf-1-like factor (MEF). This protein associated with AML1-ETO and directed the complex to MYB-responsive promoters in vitro and in vivo. In the presence of AML1-ETO, the chimeric protein repressed the activity of MYB-responsive promoters, rapidly induced apoptosis, and specifically inhibited colony growth. All these effects occurred only in AML1-ETO-positive cells, whereas no adverse effects were observed in cells not expressing AML1-ETO. Taken together, this study demonstrates that redirection of oncogenic proteins can be used as a strategy to dramatically influence their cellular effects, with the ultimate goal to design highly specific therapies for cancer.
Leukemias are often characterized by specific balanced translocations (1). Most of the chromosomal translocations in acute myeloid leukemia (AML) result in chimeric proteins involving transcription factors, often fusing a DNA-binding domain of a transcriptional activator to a transcriptional repressor. Thus, the transcriptional repressor is dislocated to target genes of the transcriptional activator, which are thought to play an important role in the differentiation process of hematopoietic cells.
The most frequent chromosomal translocation in AML is the t(8;21) translocation, found in 10–15% of adult patients with this disease (2). Due to this translocation, the C terminus of the transcriptional activator AML1 is replaced by the transcriptional repressor ETO and results in the fusion protein AML1-ETO (3, 4). ETO recruits corepressors and histone deacetylases (HDACs), and by this mechanism AML1-ETO represses AML1 target genes. This mechanism is thought to be responsible for the differentiation block that is characteristic of AML. In addition, we have recently shown that AML1-ETO inhibits expression of the p14ARF tumor suppressor (5). AML1-ETO also interacts with various transcription factors including myeloid elf-like factor (MEF), CCAAT/enhancer binding protein A (C/EBPA), other E26-transformation-specific (ETS) family members, and activating-protein 1 (AP-1). In fact it is the physical interaction between AML1-ETO and C/EBPA that is thought to be responsible for the repression of C/EBPA expression, which may contribute to the block in differentiation (6–9).
The goal of our study was to analyze whether redirection of an oncogenic transcriptional repressor protein could be used to specifically induce cell death in malignant cells. We chose t(8;21)-positive leukemia as an example. Here, we show evidence that AML1-ETO can be targeted to a different set of promoters by using a specifically designed chimeric protein. This redirection induced rapid cell death in AML1-ETO-expressing cells. Importantly, no adverse effects were noted for AML1-ETO-negative cells.
Materials and Methods
Plasmids. The GFP-M&M expression plasmid in pcDNA3.1 was constructed by PFU-PCR (Pyrococcus furiosus DNA polymerase) using a murine Myb expression plasmid and cDNA from KCL22 cells as templates by using specific primers for the DNA-binding domain of Myb and the AML1-binding domain of MEF. PCR products were cloned in frame into GFP-pcDNA3.1. GFP and GFP-M&M were also cloned into the retroviral vector pMSCV 2.2 (a gift from J. Duyster, Technical University of Munich, Munich). A deletion mutant, GFP-ΔM&M, was cloned accordingly by using a PCR fragment missing the first 159 bp of the DNA-binding domain of Myb (primer for the AML1-binding domain of MEF: MEF-BamHI for: 5′-ATA GGA TCC GCC ACC TCG CAC ACC ATG TCA-3′; MEF-EcoRI rev: 5′-CAG AAT TCG CCT TTG CCA TCC TTT GAT TTC-3′; primer for the DNA-binding domain of Myb: Myb-KpnI for: 5′-CAG AGA GGT ACC GTC ATT GCC AAT TAT CTG-3′; Myb-BamHI rev: 5′-CAG AGA GGA TCC GTA GCC TTC CTG TTC CAC-3′). The MYB-TK (thymidine kinase) luciferase construct was a gift of K. H. Klempnauer (University of Münster, Münster, Germany).
Cell Lines and Transfection. The human myeloid cell lines KCl22 and Kasumi-1, the monkey kidney cell line Cos7, the murine myeloid cell line 32Dcl3, and the Phoenix-E packaging cell line were cultured and transfected as described (10).
Immunoblotting. Protein lysates were prepared from Cos cells transfected with the expression vectors for GFP, GFP-M&M, or GFP-ΔM&M. The three proteins were detected with a mouse monoclonal GFP antibody (CLONTECH), followed by an IgG-horseradish peroxidase-conjugated secondary antibody against mouse IgG (Jackson ImmunoResearch). Immunoblotting of the protein lysates from the chromatin immunoprecipitation (IP) (see below) were performed with the monoclonal GFP antibody, a murine monoclonal actin antibody (Sigma), and a murine monoclonal FLAG antibody (Sigma).
Electrophoretic Mobility-Shift Assay. Cos cells were transfected with a total amount of 5 μg of the expression vectors for Myb, AML1-ETO, GFP, and GFP-M&M in different combinations. Preparation of nuclear extracts from transfected Cos cells, binding reaction, and the oligonucleotide containing the MYB consensus-binding site have been described (10). For competition experiments, 100 ng of double-stranded oligonucleotide containing either the MYB-consensus site or nonspecific binding site were used.
Chromatin IP. KCL22 cells were transfected with FLAG-AML1-ETO and GFP or GFP-M&M. DNA–protein complexes in the transfected cells were crosslinked with 1% formaldehyde for 10 min. Crosslinking was quenched with 0.125 M glycine before cell lysis in 1 ml of RIPA lysis buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 8.0/1% NP-40/0.5% sodium deoxycholate/0.1% SDS) with protease inhibitors, 200 μM sodium orthovanadate, and 50 μM NaF. Chromatin was sheared by sonication, and debris was removed by centrifugation. The lysates were precleared with protein A/G agarose and 5 μg of rabbit and mouse IgG. The precleared lysates were divided into two samples each, and IP was carried out either with 3 μg of anti-FLAG or control mouse IgG with 40 μl of protein A/G agarose overnight. Immunocomplexes were washed eight times with a low salt wash buffer (0.1% SDS/150 μM NaCl/1% Triton X-100/2 μM EDTA, pH 8.0/20 μM Tris·HCl, pH 8.1). Crosslinks were reversed, and DNA was phenol:chloroform extracted. Specific promoter sequences were detected with PCR for the KIT promoter region and the p14ARF promoter region (p14ARF promoter region: forward primer 5′-AGT GGC TAC GTA AGA GTG ATC GC-3′, reverse primer 5′-CTT ACA GAT CAG ACG TCA AGC CC-3′; KIT promoter region: forward primer 5′-ACT GTT GTT GCT TTC CGT TCA A-3′, reverse primer 5′-TTA AGC CCG ATT TCA CTG CC-3′). PCR was performed with Taq Polymerase (Promega) on a Mastercycler (Eppendorf) with the cycling parameters of 95°C for 3 min, 37 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min. PCR products were run on a 2% agarose gel and stained with ethidium bromide.
Luciferase Reporter Assays. Reporter assays were carried out as described (11). In brief, a total amount of 15.5 μg of plasmid was electroporated. The mixture consisted of 5 μg of MYB-TK luciferase construct, 0.5 μg of PRL-null plasmid (Promega), and 5 μg of the expression vectors for AML-1, AML1-ETO, GFP-ΔM&M, and GFP-M&M in different combinations. Empty expression vector was used to equalize the amount of DNA. The Dual Luciferase Assay system (Promega) was used for these analyses. Values present the mean ± SE of three independent experiments.
Retroviral Transduction of Primary Bone Marrow Cells. Bone marrow cells were harvested from the femurs of 6-mo-old BALB/c mice and cultured in RPMI medium 1640 in the presence of murine IL-3. Phoenix cells were transiently transfected with GFP or GFP-M&M in MSCV2.2 by Lipofectamine Plus (Invitrogen). Medium was changed after 24 h. Forty-eight hours after transfection, the supernatants were harvested, passed through a 0.45-μM filter, and added to the bone marrow cells in the presence of 4 μg/ml polybrene. After spinning at 1,000 × g for 45 min and 2 h incubation at 37°C, a second round of transduction was performed. Another two rounds were performed the next day.
Twenty-four hours after transduction, the bone marrow cells were analyzed for the expression of GFP and KIT (anti CD117-PE from PharMingen) and for apoptosis with annex-inV-PE (PharMingen) by flow cytometry according to the instructions of the manufacturer.
Clonal Growth in Methylcellulose. 32Dcl3 cells and Kasumi cells were transiently transfected with a total amount of 15 μg of the expression vectors for AML1, AML1-ETO, GFP, GFP-M&M, and GFP-ΔM&M in different combinations. To analyze clonal growth, cells were separated by gradient-centrifugation on the day after electroporation and seeded at a concentration of 1 × 105 viable cells per 35-mm dish in 1 ml of a culture mixture containing Iscove's modified Dulbecco's medium (IMDM; Life Technologies, Grand Island, NY), 1% methylcellulose, 20% FCS, IL-3 (1 ng/ml), and 0.6 mg/dl G418. All assays were plated as triplicates, and colonies were counted on day 10. A clone of >50 cells was defined as a colony for 32D cells, and >40 Kasumi cells were regarded as a colony. The indicated numbers show the results of three independent experiments (two independent experiments for Kasumi-1 cells and 32D cells with GFP-ΔM&M) per transfection.
Apoptosis Assay. 32Dcl3 cells were transiently transfected as described above. After 24 h, cells were sorted by flow cytometry for GFP-positive cells. Within the positively sorted cells, the percentage of apoptotic cells was detected in a terminal deoxynucleotidyltransferase-mediated UT P end labeling (TUNEL) assay (APO-BRDU Kit from PharMingen). The results of one of two independent experiments with similar results are shown.
Results
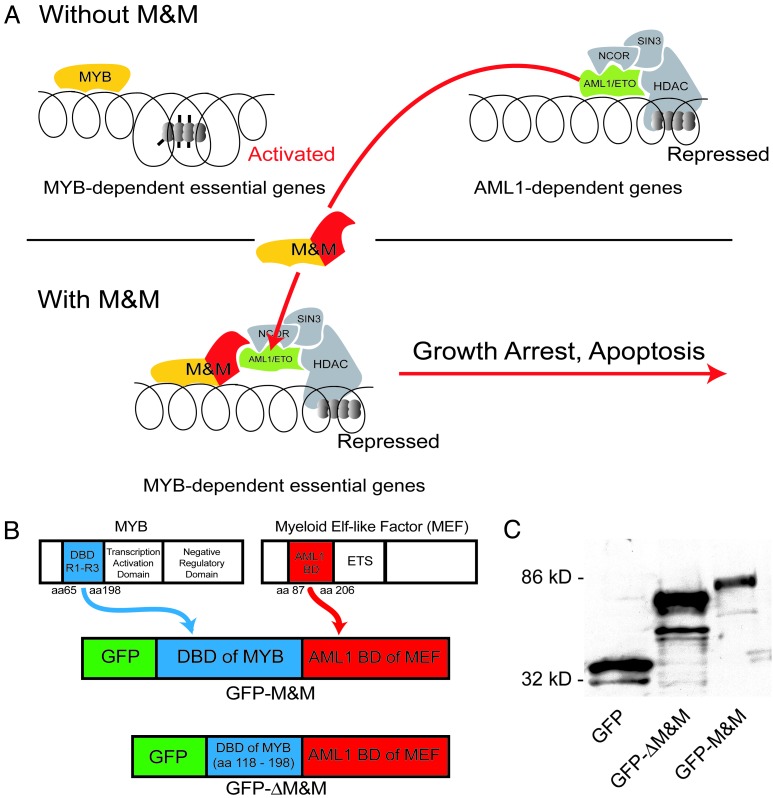
Cloning of GFP-M&M and Expression in Cos Cells. We hypothesized that targeting of AML1-ETO to promoters essential for myeloid cell survival could induce leukemia-specific cell death and growth inhibition. Binding sites of MYB were chosen as target sequences, because MYB is essential for myeloid cell survival and proliferation (refs. 12–16 and Fig. 1A). We constructed a recombinant protein containing the DNA-binding domain of murine Myb (amino acids 65–198 of the murine Myb; ref. 17) and the AML1-binding domain of human Myeloid Elf-1 factor, MEF (amino acids 87–206; ref. 18). The amino acids 87–206 of MEF bind to AML1 and AML1-ETO in vivo and in vitro (18). The protein was tagged with enhanced GFP for visualization purposes. This expression construct was named GFP-M&M (Fig. 1B). For control purposes, we cloned a deletion mutant that lacked the first 53 aa of the DNA-binding domain of Myb. The deletion mutant was called GFP-ΔM&M.
Fig. 1.
Construction of an AML1-ETO-redirecting chimeric protein. (A) Model of antileukemic activity of a chimeric protein (M&M) consisting of the MYB DNA-binding domain and the MEF AML1-binding domain. (B) Construction of an expression vector for the chimeric M&M protein and a deletion mutant lacking DNA-binding activity. (C) Expression analysis of the proteins encoded by the various constructs. Immunoblotting of Cos cell lysates after transfection with GFP, GFP-ΔM&M, and GFP-M&M. Western blot analysis was performed by using an anti-GFP antibody.
Expression of the recombinant proteins was verified after transient transfection into Cos cells and immunoblotting by using an anti-GFP antibody (GFP alone, 35 kDa; GFP-ΔM&M, 80 kDa; GFP-M&M, 85 kDa; Fig. 1C).
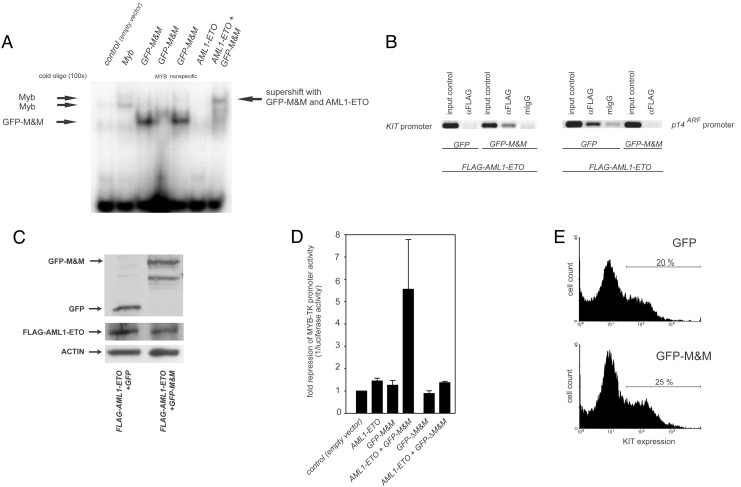
GFP-M&M Specifically Binds to MYB-Binding Sites and Recruits AML1-ETO in Vitro. To analyze the interaction of GFP-M&M with MYB DNA-binding sites, we performed electrophoretic mobility shift assays using nuclear extracts from transiently transfected Cos cells (Fig. 2A). A MYB-consensus oligonucleotide served as target DNA. These experiments indicated, that GFP-M&M, similar to Myb, could bind specifically to MYB DNA-binding sites. AML1-ETO alone did not bind to the MYB sites, but GFP-M&M recruited AML1-ETO to DNA. This interaction resulted in a supershift of the complex consisting of DNA, GFP-M&M, and AML1-ETO (Fig. 2 A).
Fig. 2.
Specific binding of GFP-M&M to MYB-binding sites, recruitment of AML1-ETO, and repression of MYB-responsive promoters by AML1-ETO in the presence of GFP-M&M. (A) In vitro recruitment of AML1-ETO to MYB-binding sites. Nuclear extracts from Cos cells transfected with Myb, GFP-M&M, and AML1-ETO as indicated were analyzed by an electrophoretic mobility-shift assay (EMSA). Competition experiments with specific MYB and nonspecific oligonucleotides demonstrated the specificity of M&M binding. The supershift of M&M cotransfected with AML1-ETO evidenced the recruitment of AML1-ETO to MYB-binding sites. (B) Recruitment of AML1-ETO in vivo. KCL22 cells transfected with FLAG-AML1-ETO and GFP or GFP-M&M were crosslinked with formaldehyde, lysed, sonicated, and immunoprecipitated with anti-Flag or with unspecific mouse IgG. Immunoprecipitated chromatin was analyzed by PCR for the KIT promoter region and the p14ARF promoter region. One representative of two experiments with similar results is shown. (C) Expression of FLAG-AML1-ETO and GFP/GFP-M&M in the transfected cells. The expression level of FLAG-AML1-ETO and GFP/GFP-M&M in transfected cells was detected by Western blot. (D) KCL22 cells were transiently transfected with a MYB-responsive luciferase construct and AML1, AML1-ETO, GFP-ΔM&M, and GFP-M&M (as indicated). Firefly luciferase values were standardized to expression of a cotransfected Renilla luciferase vector construct. Results are shown as means and SEM of three independent experiments. (E) Primary murine bone marrow cells were transduced with GFP or GFP-M&M and analyzed for the expression of KIT. The results of one of two independent experiments are shown.
GFP-M&M Recruits AML1-ETO to the Endogenous KIT Promoter. We next tested whether GFP-M&M was able to redirect AML1-ETO to endogenous MYB target promoters in vivo using chromatin IP assay (ChIP). We have recently demonstrated that AML1-ETO inhibits expression of the p14ARF tumor suppressor by binding and suppressing the endogenous p14ARF promoter (5). As an MYB-dependent endogenous promoter, we chose the KIT promoter (19). A FLAG-tagged form of AML1-ETO was ex-pressed in the myeloid cell line KCL22 in the presence or absence of GFP-M&M. After transfection, expression levels of FLAG-AML1-ETO and GFP or GFP-M&M were similar as detected by Western blot analysis (Fig. 2C). Transcription factors were crosslinked to DNA, and, subsequent to cell lysis and DNA fragmentation, DNA/AML1-ETO complexes were immunoprecipitated by using anti-FLAG or, for control purposes, unspecific antibodies. The crosslinks were reversed, and the presence of KIT and p14ARF promoter DNA was analyzed by PCR. On expression of GFP-M&M, AML1-ETO was no longer bound to the p14ARF promoter, but was now immunoprecipitated with the promoter of the MYB target gene, KIT (Fig. 2B). Thus, GFP-M&M could redirect AML1-ETO to the endogenous MYB-dependent KIT promoter in vivo.
GFP-M&M Represses MYB-Responsive Promoters in the Presence of AML1-ETO. According to our model, GFP-M&M binds to MYB-dependent promoters, recruits AML1-ETO (if present) to these promoters, and thereby represses gene expression. To test this hypothesis, we performed luciferase assays with an MYB-responsive promoter construct (20). Neither AML1-ETO, GFP, nor GFP-M&M alone significantly influenced luciferase activity. However, cells expressing GFP-M&M and AML1-ETO together suppressed the promoter activity >5-fold (Fig. 2D). We also analyzed whether a functional interaction between GFP-M&M- and MYB DNA-binding sites was necessary for the repression of luciferase activity by GFP-M&M in AML1-ETO-positive cells. The mutation of the DNA-binding domain in GFP-ΔM&M inhibits DNA binding of the recombinant protein, whereas expression of the protein is not altered (Fig. 1B). Experiments using GFP-ΔM&M demonstrated that promoter activity was not regulated by the mutant deficient in DNA binding. Thus, DNA binding was necessary for the repression of MYB-responsive genes in the presence of AML1-ETO (Fig. 2D). To demonstrate that GFP-M&M has no repressing effect on Myb-dependent promoters without AML1-ETO in vivo, we retroviraly transduced primary bone marrow cells with GFP or GFP-M&M. No significant difference in the expression of KIT (percentage and geometric mean, Fig. 2E) and in the rate of apoptosis (data not shown) was found in the GFP-positive cells.
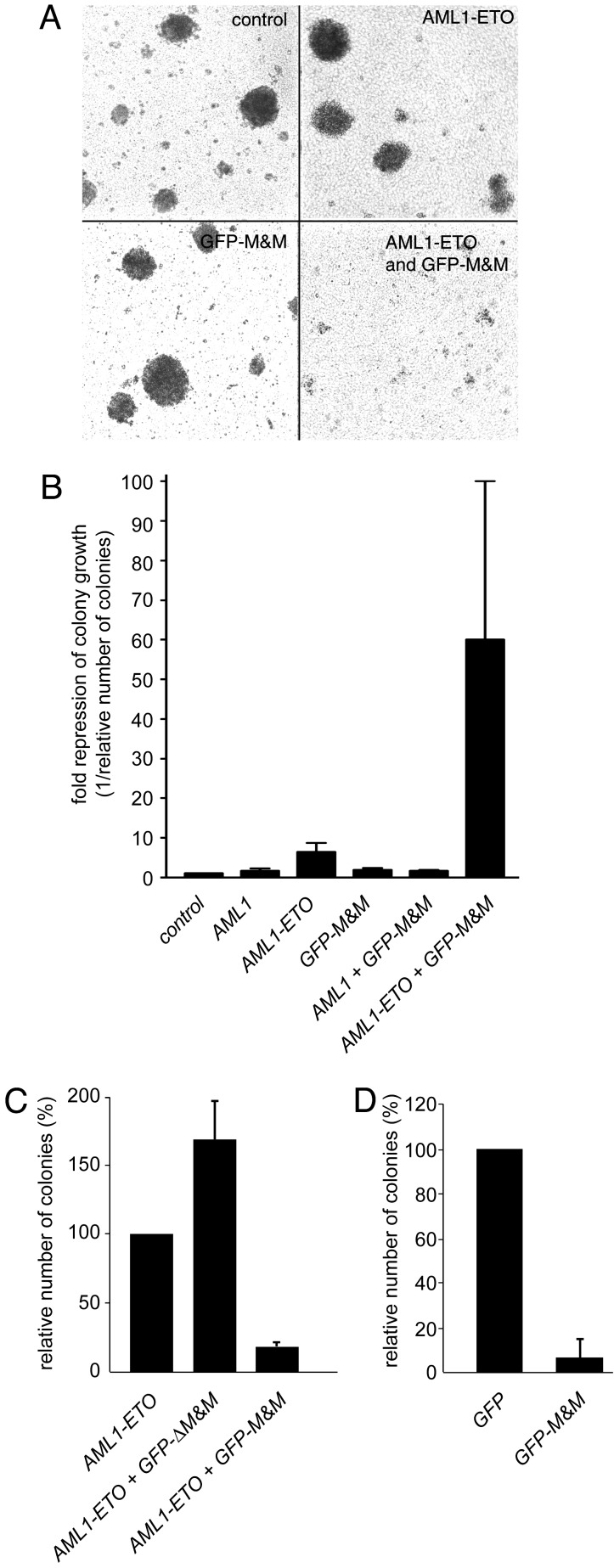
GFP-M&M Represses Colony Growth of AML1-ETO-Expressing Cells. Next, we analyzed the effects of GFP-M&M on proliferation and survival of AML1-ETO-expressing cells. First, we examined the ability of hematopoietic 32D cells to form colonies. Colony growth was not repressed in cells transfected with AML1, GFP-M&M, or AML1 together with GFP-M&M. Cells transfected with AML1-ETO alone showed a 6-fold repression of colony growth, most likely due to the toxic effect of AML1-ETO, itself (11). However, transfection of 32D cells with GFP-M&M in the presence of AML1-ETO decreased colony growth almost 60-fold (Fig. 3 A and B). A direct comparison of GFP-M&M and GFP-ΔM&M in colony assays indicated that the DNA-binding activity of the molecule was necessary to mediate inhibition of colony growth (Fig. 3C).
Fig. 3.
GFP-M&M represses colony growth in cells expressing AML1-ETO. (A) 32D cells were transfected with GFP-pcDNA3.1 as a control or AML1-ETO and GFP-M&M as indicated, and a total of 1 × 105 cells were seeded in colony assays. Pictures of representative colonies were taken on day 10. (B) Transfected 32D cells (as in A) were seeded in colony assays. The repression of colony growth compared with the control transfection with GFP-pcDNA3.1 (set as 1) is shown. (C) Analysis of the importance of DNA binding of GFP-M&M for colony growth. AML1-ETO alone or in combination with either GFP-M&M or GFP-ΔM&M was transfected into 32D cells, which were subsequently plated in colony assays. (D) Human leukemic Kasumi-1 cells that naturally express AML1-ETO were transfected with GFP or GFP-M&M. Cells were subsequently plated in colony assays.
In addition to the effects of GFP-M&M in AML1-ETO-transfected 32D cells, we also analyzed the activity of GFP-M&M in t(8;21)-positive Kasumi-1 leukemia cells. Expression of AML1-ETO in these cells was verified by Western blot analysis (data not shown). The colony-forming ability was reduced 12-fold when Kasumi-1 cells were transfected with GFP-M&M, compared with cells transfected with GFP-pcDNA3.1 alone (Fig. 3D). Thus, GFP-M&M was effective in inhibiting transfected as well as naturally occurring AML1-ETO.
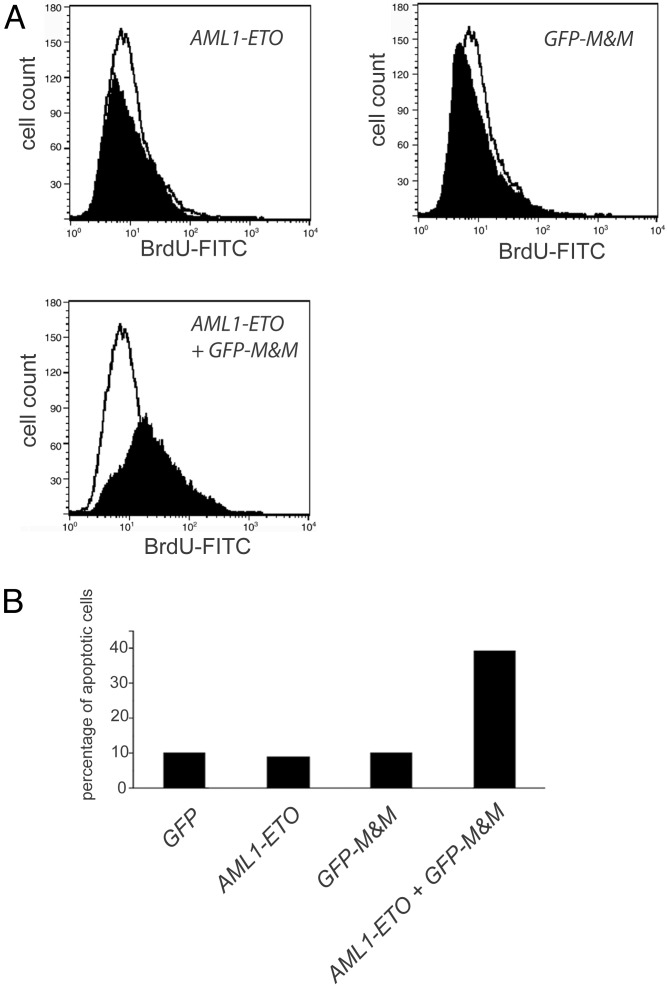
Induction of Apoptosis by GFP-M&M in AML1-ETO-Expressing Cells. Hematopoietic cells devoid of MYB activity are known to undergo apoptosis (21). To elucidate the role of apoptosis for the effect of GFP-M&M in AML1-ETO-positive cells, we analyzed the presence of DNA strand breaks by TUNEL assay. 32D cells were transfected with GFP, AML1-ETO, or GFP-M&M, or with the combination of AML1-ETO and GFP-M&M. After 24 h, ≈10% of the cells expressing GFP, AML1-ETO, or GFP-M&M alone underwent apoptosis. In contrast, in 32D cells that expressed both AML1-ETO and GFP-M&M, the percentage of apoptotic cells increased almost 4-fold to 39% (Fig. 4). In addition, the number of AML1-ETO- and GFP-M&M-positive cells that could be acquired by flow cytometry was always much lower than the number of cells in the other samples (data not shown), probably due to rapid cell death induced by the presence of the combination of both proteins. These data provided evidence that GFP-M&M specifically induced apoptotic cell death in AML1-ETO-positive cells.
Fig. 4.
GFP-M&M induces apoptosis in AML1-ETO-containing cells. 32D cells were transfected with AML1-ETO, GFP-M&M, or both and subsequently transfected cells were sorted by flow cytometry. The transfected cell population was analyzed in a TUNEL assay. (A) Fluorescence-activated cell sorter (FACS) analysis for BrdUrd-positive apoptotic cells is shown. Apoptosis in pcDNA3.1-transfected control cells is indicated in the open histograms. (B) The percentages of apoptotic cells within the transfected 32D cell populations.
Discussion
The limited availability of agents that specifically target and kill cancer cells is a major reason for the still dismal prognosis of many types of cancer. So far, there exist only a limited number of strategies for cancer therapy (22, 23).
By far the most resources are allocated to efforts to find small molecule inhibitors of oncogenic proteins, for example, specific inhibitors of tyrosine kinases. Imatinib mesylate, an inhibitor of several tyrosine kinases including BCR-ABL, has been shown to be effective in t(9;22)-containing leukemias (22). However, despite the effectiveness of imatinib mesylate in inhibiting its molecular targets in BCR-ABL-associated diseases, it seems to be most effective in chronic myelogenous leukemia (CML) patients with early (chronic phase), not yet fully transformed disease. In contrast, most patients with BCR-ABL-positive acute lymphoblastic leukemia and CML blast crisis do relapse. Besides mutational events leading to specific drug resistance, the likely reason for this primary resistance is that fully malignant diseases are the result of a series of genetic events. Reversal of one such oncogenic event by a drug is probably not sufficient to cure the disease. This result might also account for the numerous reports of primary resistance of solid tumors to targeted therapies, despite the well-advanced knowledge about the targeted oncogenes in these diseases.
McCormick and colleagues (23) followed a different principle. They constructed a deficient adenovirus that could replicate only in tumors with genetic alterations affecting the p53 pathway. As a consequence, tumor cells harboring p53 mutations were killed, whereas normal cells should not be affected. Currently, clinical trials are underway to define the practical value of this therapy (24).
We chose an approach according to the same principle: the oncogenic function is turned against the tumor to specifically eliminate oncogene-containing cells. The approach is distinctive, because our experiments were designed to redirect rather than inhibit an oncogenic protein.
The main activity of AML1-ETO is its repressive activity on AML1-dependent genes. We constructed a recombinant fusion protein to redirect this repressive activity to promoters that are essential for myeloid cell survival and proliferation. A high degree of specificity was obtained in our approach. We used MYB-binding sites as the target sites for the GFP-M&M and AML1-ETO repressor complex. The MYB protein is essential for hematopoietic cells but not for development of other organs (12). Targeting the recombinant fusion protein to MYB DNA-binding sites limits its effects outside of the hematopoietic system. The essential role of MYB in leukemia cell proliferation is well known, and inhibition of MYB-dependent genes represents a valuable target for leukemia therapy (12, 13, 15, 16).
Although these experiments evidence that redirection of fusion proteins derived from balanced translocation provides a novel and specific therapeutic approach, delivery problems need to be solved. One possibility would be to deliver the recombinant protein through gene therapy approaches. Novel developments in this field such as the targeting of specific cell types by pseudotyped viral vectors, and the more and more efficient transduction of target cells will increase the feasibility of this therapeutic approach. Because much of the required tumor specificity is included in the principle of the redirection therapy, specific delivery is not necessarily needed and transduction by replication-competent systems might be feasible. Another possibility would be to deliver recombinant protein itself into the cells, or to construct small molecules that simultaneously bind to AML1-ETO and MYB. Because no specific action besides the binding to MYB and AML1-ETO is necessary to be effective, such components could be identified in regular high-throughput screens.
Taken together, this study demonstrates that knowledge about the molecular pathogenesis of cancers can be used to design highly effective and specific therapies that are based on intracellular protein redirection.
Acknowledgments
This work is dedicated to the memory of Kerstin Meister. We thank Maria Möller for excellent technical assistance. This work was supported by grants from the José-Carreras Leukemia Foundation, Deutsche Forschungsgemeinschaft (DFG:SE 600/2; MU 1328/2-3), the Innovative Medizinische Forschung, and the Interdisziplinäres Zentrum für Klinische Forschung (H2) at the University of Münster.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AML, acute myeloid leukemia; MEF, myeloid Elf-1-like factor; IP, immunoprecipitation; TUNEL, terminal deoxynucleotidyltransferase-mediated UTP end labeling.
References
- 1.Rowley, J. D. (1973) N. Engl. J. Med. 289 220-221. [DOI] [PubMed] [Google Scholar]
- 2.Downing, J. R. (1999) Br. J. Haematol. 106 296-308. [DOI] [PubMed] [Google Scholar]
- 3.Meyers, S., Lenny, N. & Hiebert, S. W. (1995) Mol. Cell. Biol. 15 1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenny, N., Meyers, S. & Hiebert, S. W. (1995) Oncogene 11 1761-1769. [PubMed] [Google Scholar]
- 5.Linggi, B., Müller-Tidow, C., van de Locht, L., Hu, M., Nip, J., Serve, H., Berdel, W. E., van der Reijden, B., Quelle, D. E., Rowley, J. D., et al. (2002) Nat. Med. 8 743-750. [DOI] [PubMed] [Google Scholar]
- 6.Britos-Bray, M. & Friedman, A. D. (1997) Mol. Cell. Biol. 17 5127-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, R., Zhang, J., Uchida, H., Meyers, S., Hiebert, S. W. & Nimer, S. D. (1995) Oncogene 11 2667-2674. [PubMed] [Google Scholar]
- 8.Pabst, T., Mueller, B. U., Harakawa, N., Schoch, C., Haferlach, T., Behre, G., Hiddemann, W., Zhang, D. E. & Tenen, D. G. (2001) Nat. Med. 7 444-451. [DOI] [PubMed] [Google Scholar]
- 9.Oelgeschlager, M., Nuchprayoon, I., Luscher, B. & Friedman, A. D. (1996) Mol. Cell. Biol. 16 4717-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller, C., Yang, R., Idos, G., Tidow, N., Diederichs, S., Koch, O. M., Verbeek, W., Bender, T. P. & Koeffler, H. P. (1999) Blood 94 4255-4262. [PubMed] [Google Scholar]
- 11.Müller, C., Readhead, C., Diederichs, S., Idos, G., Yang, R., Tidow, N., Serve, H., Berdel, W. E. & Koeffler, H. P. (2000) Mol. Cell. Biol. 20 3316-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mucenski, M. L., McLain, K., Kier, A. B., Swerdlow, S. H., Schreiner, C. M., Miller, T. A., Pietryga, D. W., Scott, W. J., Jr., & Potter, S. S. (1991) Cell 65 677-689. [DOI] [PubMed] [Google Scholar]
- 13.Ratajczak, M. Z., Kant, J. A., Luger, S. M., Hijiya, N., Zhang, J., Zon, G. & Gewirtz, A. M. (1992) Proc. Natl. Acad. Sci. USA 89 11823-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White, J. R. & Weston, K. (2000) Oncogene 19 1196-1205. [DOI] [PubMed] [Google Scholar]
- 15.Gewirtz, A. M. & Calabretta, B. (1988) Science 242 1303-1306. [DOI] [PubMed] [Google Scholar]
- 16.Gewirtz, A. M. (1999) Oncogene 18 3056-3062. [DOI] [PubMed] [Google Scholar]
- 17.Sakura, H., Kanei-Ishii, C., Nagase, T., Nakagoshi, H., Gonda, T. J. & Ishii, S. (1989) Proc. Natl. Acad. Sci. USA 86 5758-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao, S., Frank, R. C., Zhang, J., Miyazaki, Y. & Nimer, S. D. (1999) Mol. Cell. Biol. 19 3635-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratajczak, M. Z., Perrotti, D., Melotti, P., Powzaniuk, M., Calabretta, B., Onodera, K., Kregenow, D. A., Machalinski, B. & Gewirtz, A. M. (1998) Blood 91 1934-1946. [PubMed] [Google Scholar]
- 20.Ziebold, U., Bartsch, O., Marais, R., Ferrari, S. & Klempnauer, K. H. (1997) Curr. Biol. 7 253-260. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, D., Badiani, P. & Weston, K. (1996) Genes Dev. 10 2732-2744. [DOI] [PubMed] [Google Scholar]
- 22.Vigneri, P. & Wang, J. Y. (2001) Nat. Med. 7 228-234. [DOI] [PubMed] [Google Scholar]
- 23.Bischoff, J. R., Kirn, D. H., Williams, A., Heise, C., Horn, S., Muna, M., Ng, L., Nye, J. A., Sampson-Johannes, A., Fattaey, A. & McCormick, F. (1996) Science 274 373-376. [DOI] [PubMed] [Google Scholar]
- 24.McCormick, F. (2000) Semin. Cancer Biol. 10 453-459. [DOI] [PubMed] [Google Scholar]