Abstract
Type 2 diabetes mellitus (DM) is characterized by insulin resistance and pancreatic β cell dysfunction. In high-risk subjects, the earliest detectable abnormality is insulin resistance in skeletal muscle. Impaired insulin-mediated signaling, gene expression, glycogen synthesis, and accumulation of intramyocellular triglycerides have all been linked with insulin resistance, but no specific defect responsible for insulin resistance and DM has been identified in humans. To identify genes potentially important in the pathogenesis of DM, we analyzed gene expression in skeletal muscle from healthy metabolically characterized nondiabetic (family history negative and positive for DM) and diabetic Mexican–American subjects. We demonstrate that insulin resistance and DM associate with reduced expression of multiple nuclear respiratory factor-1 (NRF-1)-dependent genes encoding key enzymes in oxidative metabolism and mitochondrial function. Although NRF-1 expression is decreased only in diabetic subjects, expression of both PPARγ coactivator 1-α and-β (PGC1-α/PPARGC1 and PGC1-β/PERC), coactivators of NRF-1 and PPARγ-dependent transcription, is decreased in both diabetic subjects and family history-positive nondiabetic subjects. Decreased PGC1 expression may be responsible for decreased expression of NRF-dependent genes, leading to the metabolic disturbances characteristic of insulin resistance and DM.
Insulin resistance precedes and predicts the development of type 2 diabetes mellitus (DM) (1, 2). Defects in insulin signal transduction, gene expression, and muscle glycogen synthesis, and accumulation of intramyocellular triglycerides have all been identified as potential mediators of insulin resistance in high-risk individuals (1, 3–7). However, the molecular pathogenesis of DM remains unknown. Mouse data highlight the importance of glucose uptake into muscle but suggest a role for novel mechanisms, distinct from insulin signaling pathways (8). The importance of genetic risk factors is exemplified by the high concordance of DM in identical twins, the strong influence of family history and ethnicity on risk, and the identification of DNA sequence alterations in both rare and common forms of DM (9). Environmental factors, including obesity, inactivity, and aging, also play critical roles in DM risk. Because both genotype and environment converge to influence cellular function via gene and protein expression, we hypothesize that alterations in expression define a phenotype that parallels the metabolic evolution of DM and provides potential clues to pathogenesis. We used high-density oligonucleotide arrays to identify genes differentially expressed in skeletal muscle from nondiabetic and type 2 diabetic subjects. Because hyperglycemia per se can modulate expression, we also evaluated gene expression in insulin-resistant subjects at high risk for DM (“prediabetes”) on the basis of family history of DM and Mexican–American ethnicity (10). We demonstrate that prediabetic and diabetic muscle is characterized by decreased expression of oxidative phosphorylation genes, many of which are regulated by nuclear respiratory factor (NRF)-dependent transcription. Furthermore, expression of peroxisomal proliferator activator receptor γ coactivator (PGC1)α and -β (PPARGC1 and PERC), coactivators of both PPARG and NRF-dependent transcription, is significantly reduced in both prediabetic and diabetic subjects. Taken together, these data indicate that decreased PGC1 expression may be responsible for decreased expression of NRF-dependent metabolic and mitochondrial genes and may contribute to the metabolic disturbances characteristic of insulin resistance and DM.
Methods
Subject Recruitment and Characterization. The clinical protocol was approved by the University of Texas Health Science Center Institutional Review Board, and written informed consent was obtained. Subjects had no significant medical problems except DM, were not taking medications affecting glucose metabolism, and were sedentary. Subjects maintained their usual diet and refrained from vigorous exercise for 2 days before the study. Subjects were stratified by family history (FH) of DM (first-degree relative). For nondiabetics, normal glucose tolerance was confirmed with a 75-g glucose load. Diabetic subjects were treated with lifestyle or sulfonylureas (discontinued 48 h prestudy).
After an overnight fast, a biopsy was taken from the vastus lateralis muscle and frozen in liquid nitrogen. A 2 h, 40 milliunits/m2/min hyperinsulinemic euglycemic clamp was performed to assess glucose disposal (11).
RNA Isolation, cRNA Preparation, and Array Hybridization. Muscle biopsies from 10 nondiabetics (six FH- and four FH+) and five diabetics, and five muscle aliquots from a single subject, were homogenized in RNA Stat (Tel-Test, Friendswood, TX). Total RNA was purified with RNeasy (Qiagen, Chatsworth, CA), and used for cRNA synthesis (12). Fifteen micrograms of adjusted cRNA were hybridized to Affymetrix (Santa Clara, CA) HuGeneFL arrays. Intensity values were quantitated by using mas 4.0 software (Affymetrix). Average correlation between replicates (five independent preparations of RNA and cRNA) was 0.96 ± 0.01.
Regression Normalization. All 20 arrays were normalized for overall intensity using linear regression. Each array was normalized against the microarray most similar to all others by using the central 95% of expression values (13).
Identification of Differentially Expressed Genes and Pathways. Differences between groups were evaluated by using the t test with unequal variances (14). Annotations were compiled by using genespring (Silicon Genetics, Redwood City, CA), unchip (Children's Hospital Informatics Program, Boston), and onto express, Ver. 2 (15). genmapp, mappfinder, and onto express were used to integrate expression data with known pathways (16, 17) and to determine confidence levels for differential expression within ontology groups (15).
Quantitative PCR. Muscle expression of selected genes was determined by using two-step real-time quantitative PCR (Applied Biosystems PRISM 7700) in an independent cohort of 15 FH- controls, 12 FH+ controls, and 15 diabetic subjects (Table 1 Right). cDNA was synthesized from DNAse I-treated total RNA by using random hexamer primers (Advantage, BD Biosciences, San Jose, CA). Primer and probe sequences were selected by using PRIMER EXPRESS (Applied Biosystems). Target gene and endogenous control amplicons were labeled with FAM and VIC, respectively.
Table 1. Metabolic characteristics of array and PCR subject cohorts, including FH- and FH+ control and diabetic (DM2) subjects.
|
Array subject cohort
|
PCR subject cohort
|
|||||
|---|---|---|---|---|---|---|
| Control FH- | Control FH+ | DM 2 | Control FH- | Control FH+ | DM 2 | |
| Subject number | 6 | 4 | 5 | 15 | 12 | 15 |
| Sex | 4 F, 2 M | 3 F, 1 M | 4 F, 1 M | 5 F, 10 M | 6 F, 6 M | 5 F, 10 M |
| Age | 38.5 ± 3.7 | 40.8 ± 2.6 | 43.8 ± 2.1 | 31.1 ± 2.0 | 40.3 ± 3.0* | 47.3 ± 1.9*** |
| BMI, kg/m2 | 31.2 ± 0.8 | 28.9 ± 1.4 | 37.4 ± 5.8 | 27.0 ± 1.1 | 26.9 ± 0.9 | 33.4 ± 1.2*† |
| Fasting glucose, mg/dl | 99 ± 3 | 92 ± 2 | 201 ± 33*† | 95 ± 2 | 93 ± 3 | 176 ± 12***† |
| Two-hour glucose | 100 ± 11 | 115 ± 10 | 263 ± 23**† | 100 ± 5 | 85 ± 10 | 227 ± 14**† |
| Hemoglobin A1c, % | 4.9 ± 0.3 | 5.7 ± 0.1* | 9.1 ± 0.5***† | 4.8 ± 0.1 | 5.0 ± 0.1 | 7.4 ± 0.4***† |
| Fasting insulin, microunit/ml | 8.3 ± 0.8 | 14.8 ± 1.9* | 13.1 ± 1.3* | 7.2 ± 0.8 | 13.5 ± 2.6* | 17.0 ± 2.5** |
| Insulin-stimulated glucose disposal, mg/kg per min | 4.1 ± 0.3 | 3.1 ± 0.5 | 2.4 ± 0.7 | 5.1 ± 0.4 | 2.7 ± 0.2*** | 2.6 ± 0.4*** |
All P values relative to FH- control unless specified
, P < 0.05
, P < 0.01
, P < 0.001
, P < 0.05 vs. FH+
Results
We used high-density oligonucleotide arrays and quantitative real-time PCR to identify genes differentially expressed in skeletal muscle from type 2 diabetic subjects and in nondiabetic insulin-resistant subjects at high risk for DM, based on family history of DM (FH+) and Mexican–American ethnicity. Clinical characteristics of the initial array subject cohort and independent cohort used for PCR are provided in Table 1. Diabetic subjects had increased fasting and 2-h glucose levels and hemoglobin A1c (all P < 0.01). Fasting insulin was significantly increased in FH+ and DM, whereas insulin-stimulated glucose disposal, a measure of insulin sensitivity, decreased progressively from FH- to FH+ to DM (P < 0.001). cRNA was prepared from muscle biopsies and hybridized to oligonucleotide microarrays. Primary data are available at www.diabetesgenome.org.
Of 7,129 sequences represented on the array, 187 were differentially expressed (P < 0.05) between control (FH-) and diabetic subjects (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). No single gene remained differentially expressed after controlling for multiple comparison false discovery by using the Benjamini–Hochberg method (18, 19). Therefore, to identify groups of genes with similar regulation between FH- and DM subjects, we ranked genes by P values and analyzed ontology for genes with uncorrected P < 0.05. By using mapp finder (17), the top-ranked cellular component terms were mitochondrion, mitochondrial membrane, mitochondrial inner membrane, and ribosome (Z scores 7.8, 7.2, 7.8, and 6.8, respectively, indicating overrepresentation in FH- vs. DM comparison). Similarly, the top-ranked process term was ATP biosynthesis (Z 5.5). Similar results were obtained by using ontoexpress (15) with multiple correction testing, which indicated that energy generation (P = 0.0029), protein biosynthesis/ribosomal proteins (P = 0.013), RNA binding (P = 0.007), ribosomal structural protein (P = 0.0007), and ATP synthase complex (P = 2 × 10-5) ontology groups were represented to a significantly greater extent in FH- vs. DM than expected if ontology groups were randomly distributed within the list of top-ranked genes.
Genes differentially expressed between control and diabetic subjects may reflect either the pathophysiology of insulin resistance (primary alterations) or secondary effects of hyperglycemia, hyperlipidemia, and other metabolic factors. To identify potentially primary expression changes associated with insulin resistance, we compared gene expression in FH+ (nondiabetic but insulin resistant) and FH- controls. One hundred sixty-six genes were differentially expressed between FH+ and FH- (P < 0.05) (Table 3, which is published as supporting information on the PNAS web site); 55 were common to both [FH- vs. DM] and [FH- vs. FH+] comparisons. No single gene remained differentially expressed after Benjamini–Hochberg multiple comparison testing. However, ontology classification analysis (17) revealed that 20S and 26S proteasome complexes were the top-ranked cellular component terms (Z 7.7 and 7.3); mitochondrion-linked genes were also overrepresented (Z 3.2). Cell structure (P = 0.004), protein degradation (P = 3.7 × 10-4), and energy generation (P = 0.003) groups were represented to a greater extent than expected for random distribution; with multiple comparison testing, the protein degradation/26S proteasome (P = 1 × 10-5) group remained significant.
To evaluate the effects of hyperglycemia or other metabolic consequences of DM per se on expression, we identified 12 genes altered in DM as compared with both nondiabetic groups but not as a function of family history (Table 4, which is published as supporting information on the PNAS web site). This included a 70-kDa heat-shock protein (HSP701A), which was decreased by 42% in DM and whose expression correlated inversely with fasting glucose for all subjects (r = -0.77). Expression of a related HSP70 gene was previously found to be reduced in Caucasian diabetic subjects (20).
By using quantitative real-time PCR, we measured expression of 10 genes selected on the basis of biological interest or P value ranking in muscle from an independent cohort of control (FH-) and DM (Table 1 Right). Expression of three genes was concordant with array data, and four genes were both concordant and statistically different between FH- and DM (P < 0.05, PCR) (Table 5, which is published as supporting information on the PNAS web site).
Taken together, our data indicate that differences in muscle gene expression between healthy diabetic and control subjects at steady-state are modest. Although expression differences for individual genes did not meet statistical significance after rigorous multiple testing correction, pattern analysis demonstrated that genes encoding proteins related to mitochondrial oxidative metabolism were overrepresented in both [control vs. DM] and [FH- vs. FH+] comparisons. Because altered expression of genes regulating oxidative metabolism might be a major DM-associated phenotype, we performed more detailed analysis of gene expression patterns within lipid and carbohydrate metabolism groups. (Data refer to uncorrected P values.)
Lipid Transport and Metabolism. Expression of several genes involved in fatty acid oxidation were significantly decreased in DM relative to FH- controls, including 3-hydroxyacyl CoA dehydrogenase (also decreased in FH+ vs. FH-), mitochondrial 3,2-transenoyl-CoA isomerase, and 2,4-dienoyl CoA reductase 1. Monoglyceride lipase, a key enzyme in triglyceride hydrolysis, was significantly decreased in both FH+ and DM. Lipoprotein lipase expression was increased in FH+. These changes could contribute to muscle triglyceride accumulation, as in mice over-expressing LPL (21) and insulin-resistant or diabetic humans (6).
Oxidative Metabolism. Expression of multiple glycolysis and tricarboxylic acid cycle genes was significantly decreased in DM, including glucose phosphate isomerase, fructose-1,6 bisphosphatase 2, pyruvate kinase, pyruvate dehydrogenase A1, α-ketoglutarate dehydrogenase, and succinate dehydrogenase B. Expression of many of these was also reduced in FH+, reaching significance for pyruvate kinase and α-ketoglutarate dehydrogenase.
Multiple components of the mitochondrial respiratory chain were reduced in FH+ and significantly decreased in DM, including two complex I, one complex II, two complex III, and three complex IV subunits, and multiple subunits of ATP synthase (four of five Fo subunits and the ATP5O subunit of F1); ATP5D was significantly increased in both FH+ and DM. Uncoupling proteins 1–3 did not differ significantly. Expression of ANT1, which determines ADP/ATP flux between cytosol and mitochondria, and the voltage-dependent anion channel porin was reduced in FH+ and significantly decreased in DM.
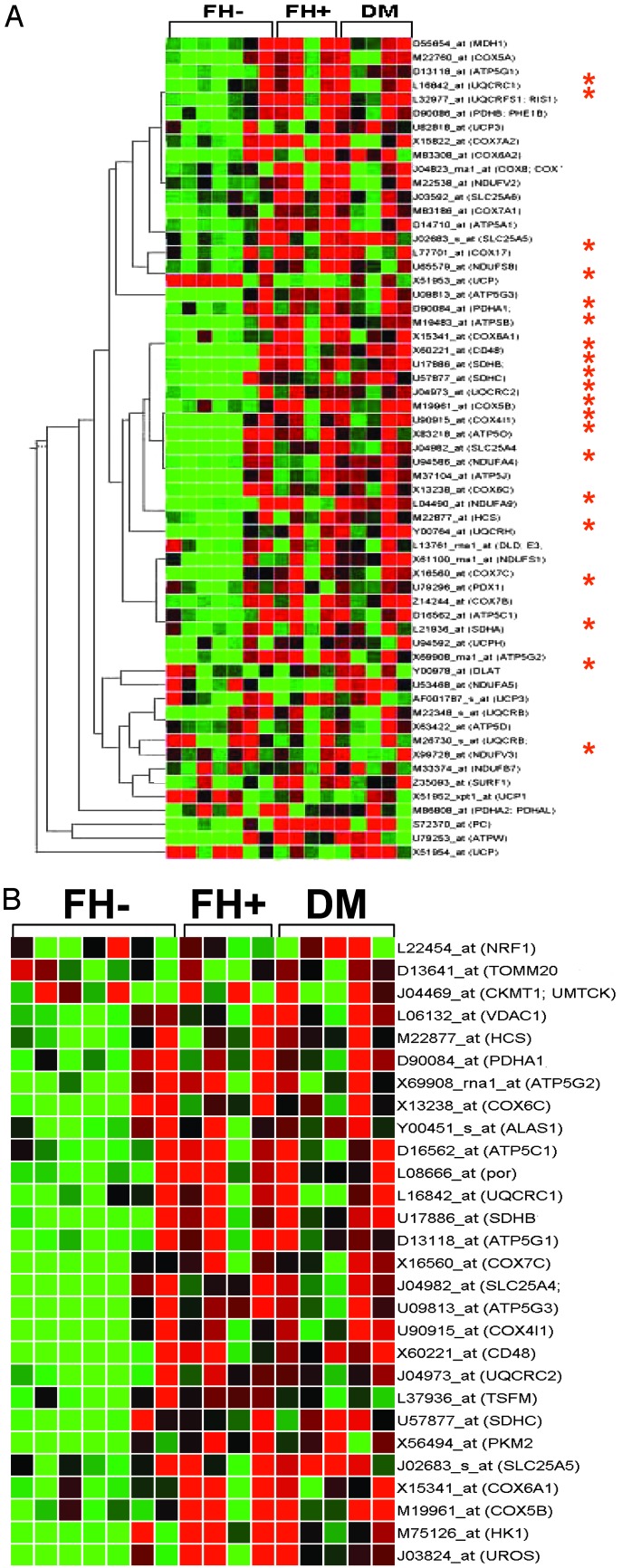
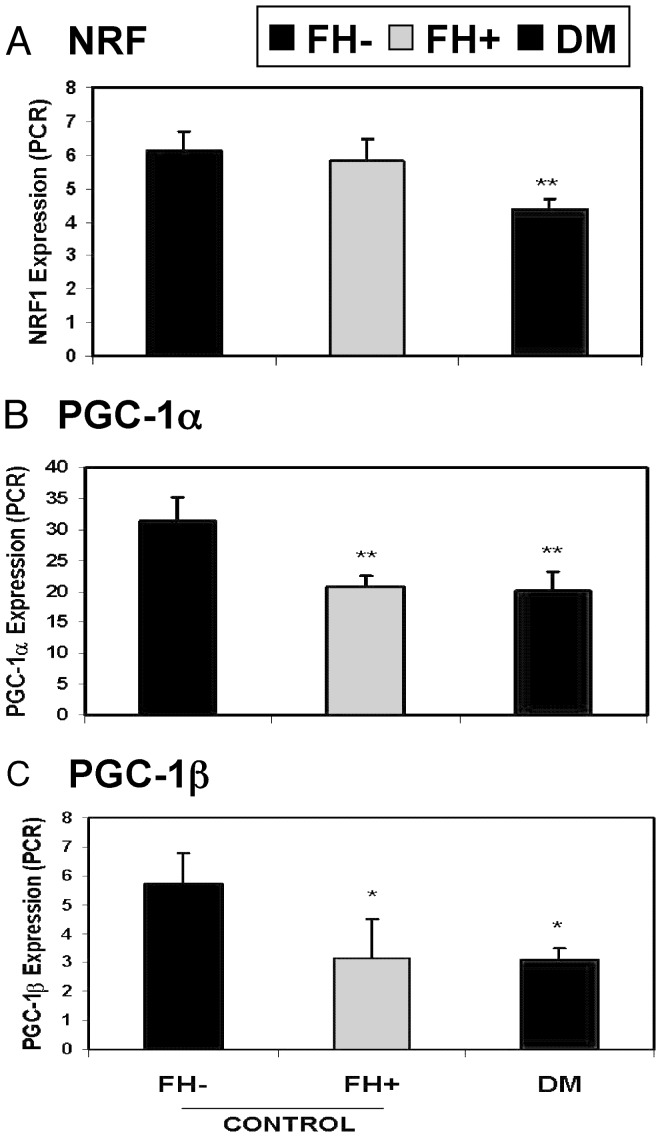
Thus, these data demonstrate progressive decreases in expression of genes encoding key proteins in oxidative metabolism in insulin-resistant and DM subjects (Fig. 1A). Because many of these genes are regulated by the NRF transcription factor family (22) (indicated by asterisk), we hypothesized that this pattern might result from coordinated reductions in NRF transcriptional activation. In addition, other genes known to be regulated by NRF were progressively reduced from FH- to FH+ to DM (Fig. 1B). To test the potential contribution of NRF, we assessed NRF-1 expression by real-time quantitative RT-PCR. NRF-1 expression was decreased by 29% in DM (P = 0.01) (Fig. 2A) and inversely correlated with fasting glucose (r = -0.46, P = 0.03); expression of NRF-2 (GABPA) did not differ. Array expression levels for other transcription factors implicated in nuclear-encoded mitochondrial gene transcription, including TFAM, MEF-2 isoforms, YY1, CREM, CREB1, and CREB3, did not differ but were near background levels. Sp1 (2-fold increase, P = 0.003) and CREB-RP (61% increase, P = 0.004) were increased in FH+ but not in DM.
Fig. 1.
(A) Expression of many oxidative metabolism genes is reduced in FH+ insulin-resistant nondiabetic and type 2 DM subjects. Hierarchical clustering was performed (genespring, algorithm similar to that of Eisen et al. (51) by using glycolysis, tricarboxylic acid cycle, and electron transport gene groups (genmapp). Genes known to be regulated by NRF transcription in humans or rodents are indicated by an asterisk. Colors represent gene expression values in individual subject expression changes relative to the mean (normalized to 1 for each gene), with red and green representing decreases or increases in expression, respectively by >50%. (B) Expression of genes regulated by NRF transcription is decreased in FH+ and DM2. The gene tree was created by compiling a list of NRF-regulated genes (52) as in A.
Fig. 2.
Decreases in transcription factor NRF-1 and coactivator PGC1 expression contribute to reductions in oxidative gene expression. (A) Expression of NRF-1 (quantitative PCR), is decreased in DM (*, P = 0.01 vs. FH- controls). (B) PGC1α expression (PCR) is reduced in prediabetic FH+ (34% reduction, P = 0.001 vs. FH-) and DM (36% decrease, P = 0.0009 vs. FH-). (C) PGC1β expression (PCR) is reduced in prediabetic FH+ (45% reduction, P = 0.045 vs. FH-) and DM (46% decrease, P = 0.01 vs. FH-).
PPARγ coactivator 1 (PPARGC1, PGC1α) also regulates NRF-dependent transcription (22), increases expression of both nuclear and mitochondrial-encoded genes of oxidative metabolism (e.g., tricarboxylic acid cycle, lipid oxidation, and electron transport complexes), and induces mitochondrial biogenesis (23). To test the potential contribution of PGC1α (PPARGC1) and related PGC1β (PERC) (not on array) to the DM expression phenotype, we performed quantitative real-time RT-PCR. Both PGC1α and -β expression was significantly reduced in DM subjects (PGC1α: 36% decrease, P = 0.0009 vs. FH-; PGC1β: 46% decrease, P = 0.01) (Fig. 2 B and C). Expression of PRC, a related NRF coactivator, was not altered. Even more striking was the significant reduction in expression of both PGC1α and -β in nondiabetic FH+ subjects (PGC1α: 34% decrease as compared with FH- controls, P = 0.007, PGC1β: 45% decrease, P = 0.047) (Fig. 2 B and C).
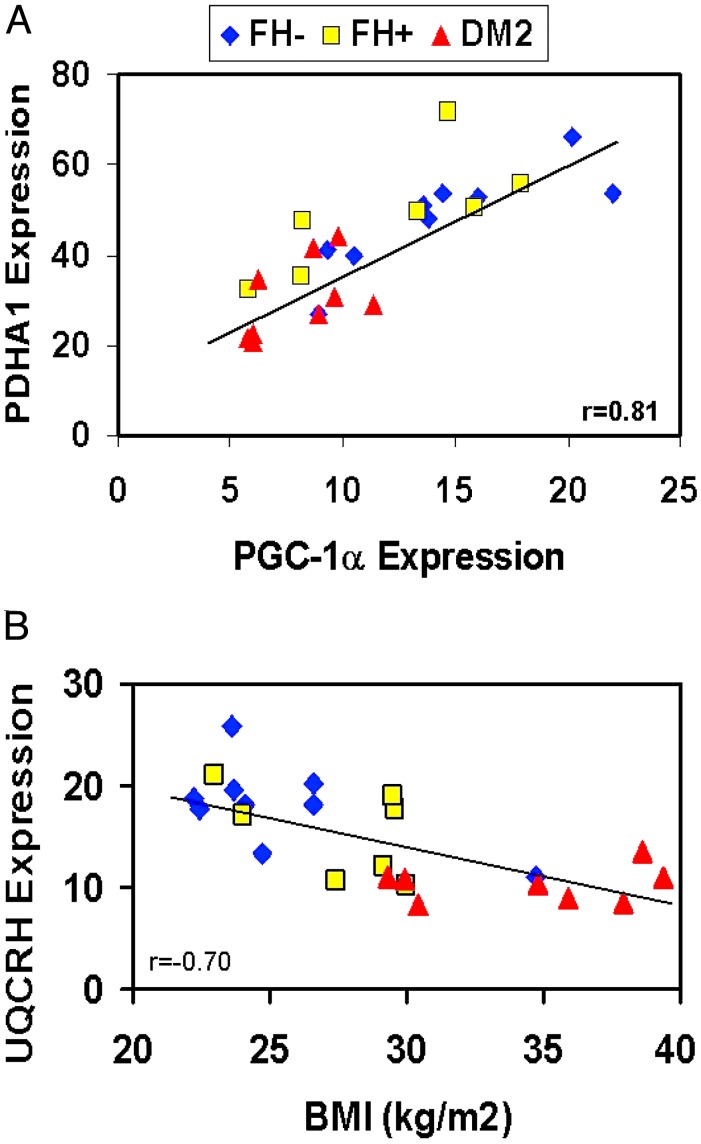
To identify potential relationships between clinical data and PCR gene expression, we performed regression analysis incorporating family history and diabetes status. PGC1α expression was highly correlated with that of two representative oxidative genes, pyruvate dehydrogenase A1 (r = 0.81, P < 0.0001, Fig. 3A), and the complex III subunit UQCRH (r = 0.75, P < 0.0001), supporting a link between PGC1 and the array expression phenotype. In the univariate model, expression of PGC1α was inversely related to FH status (r = -0.49, P = 0.001) and modestly to insulin-stimulated glucose disposal (r = 0.31, P = 0.07), but not to age or body mass index (BMI). In a multivariate model incorporating age, BMI, and family history (r = 0.50, P = 0.01), family history status remained the primary correlate with PGC1α expression (P = 0.0037). Although expression of PGC1α and PGC1β were significantly correlated (r = 0.43, P = 0.005), expression of PGC1β did decrease modestly with age (r = -0.32, P = 0.05). In the multivariate model (r = 0.58, P = 0.008), FH, but not age, remained a significant covariate for PGC1β expression (P = 0.017), as for PGC1α.
Fig. 3.
Representative metabolic and expression correlates. (A) PGC1α expression correlates with that of pyruvate dehydrogenase (PDHA) (r = 0.81, P < 0.0001). (B) The complex III subunit UQCRH correlates inversely with BMI (r = -0.70, P = 0.0002).
Correlation patterns for the representative complex III UQCRH differed somewhat. In univariate analysis, UQCRH expression was positively correlated with insulin-stimulated glucose disposal (r = 0.48, P = 0.05) and negatively with fasting plasma insulin (r = -0.64, P = 0.003), BMI (r = -0.70, P = 0.0002, Fig. 3B), fasting glucose (r = -0.44, P = 0.03), and family history of DM (r = 0.56, P = 0.005). In the best fit model, incorporating family history status, age, BMI, and fasting insulin (r = 0.88, P = 0.0002), both BMI and fasting insulin, but not FH, remained significant covariates (P = 0.0063 and 0.0025).
Discussion
Our array data demonstrate that expression of many genes of oxidative metabolism is reduced in DM. We recognize that microarray approaches are limited by multiple comparison caveats and false positives. Furthermore, many alterations in gene expression are of relatively low magnitude, and the small subject number in our study limits our power to detect differences between groups. Despite these limitations, expression differences for representative genes were validated by PCR, and many interesting patterns of regulation in oxidative pathways were detected in both prediabetic and diabetic subjects.
A potential role for dysregulation of oxidative metabolism gene expression in DM may be inferred from other studies. In streptozotocin-induced DM mice (12), expression of oxidative phosphorylation genes is decreased. Similarly, expression of multiple energy metabolism genes is altered in poorly controlled type 2 DM humans (24). In both studies, some differences were partially normalized by insulin, suggesting that differential regulation in DM may partly reflect secondary changes, perhaps due to decreases in NRF-1 expression or transcriptional activity. However, we also observed similar, although less pronounced, alterations in oxidative phosphorylation genes in insulin-resistant nondiabetics. In agreement, expression of two electron chain subunits (NADH dehydrogenase 1 and ATP5C1) is reduced in insulin-resistant nondiabetic Pima Indians (25), and ATP synthase subunit F expression is reduced in the insulin-resistant normoglycemic ob/ob mouse (26). Mootha et al. (27) found similar reductions in expression of oxidative phosphorylation genes in Caucasians with impaired glucose tolerance and type 2 DM. Our data extend these findings to insulin-resistant but completely glucose-tolerant individuals and suggest that this pattern of expression may be a primary feature of prediabetic pathophysiology, related to insulin resistance rather than hyperglycemia. Interestingly, expression patterns in our subjects do not mirror closely those in the severely insulin-resistant muscle insulin receptor knockout mouse,∥ suggesting that the molecular mechanism underlying insulin resistance is critical for determining patterns of gene expression.
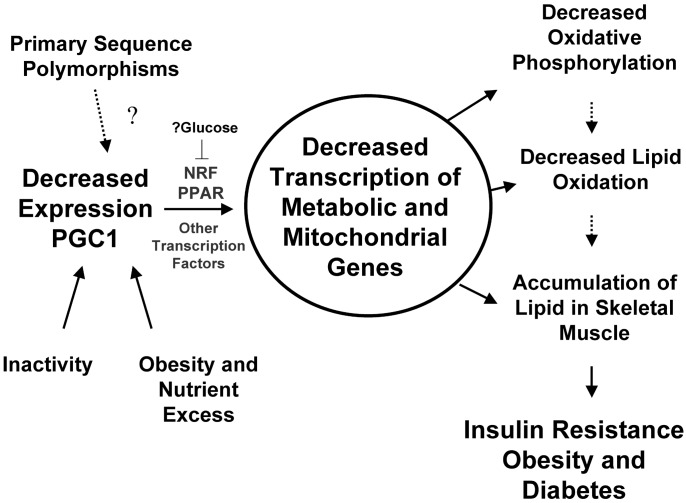
Our expression phenotype is also consistent with studies implicating mitochondrial dysfunction in DM pathogenesis. Mutations in mitochondrial DNA result in altered expression of oxidative genes, myopathy, and “mitochondrial” DM (28). In both obesity and common forms of type 2 DM, glucose oxidation and storage are reduced, in parallel with reduced activity of tricarboxylic acid cycle, β-oxidation, and electron transport enzymes (29). Moreover, reductions in mitochondrial area (30), number (31, 32), and complex I activity correlate with decreased insulin-stimulated glucose disposal (30). Our data provide a potential molecular mechanism for these results (Fig. 4). Impaired mitochondrial function may result from decreased expression of critical nuclear-encoded mitochondrial genes in “prediabetic” and diabetic subjects, related to alterations in PGC1-mediated coactivation of PPAR- and NRF-dependent transcription. These transcriptional changes may also contribute to altered glucose and fatty acid metabolism characteristic of the evolution to type 2 DM, including decreased fat oxidation (lack of decrease in respiratory quotient during fasting) (33), increased lipid esterification and accumulation in skeletal muscle (6, 34), and insulin resistance, perhaps via effects of lipids or reduced lipid oxidation on glucose metabolism (35, 36). In DM, expression of oxidative phosphorylation genes is further decreased, perhaps as a result of decreased expression or transcriptional activity of NRF-1, and might further impair oxidative metabolism, thus establishing a vicious metabolic cycle with the onset of DM.
Fig. 4.
Proposed contribution of PGC1 and NRF-1 to expression and metabolic phenotype of insulin resistance and type 2 DM.
Both primary sequence alterations and environmental risk factors for DM may contribute to decreased PGC1α and -β expression and/or function and thus NRF-dependent transcription. The 4p15 locus has been linked to fasting insulin levels in Pima Indians (37) and to obesity in both Caucasians and Mexican–Americans (38, 39), whereas polymorphisms in PGC1α have been associated with obesity** and increased risk of DM in Danish and Japanese populations (40, 41). Environmental risk factors for DM may also contribute to decreased expression or function of PGC1. We cannot completely exclude a role for age or BMI in differential expression of PGC1α and -β, because FH+ and DM subjects in the PCR cohort were older, and DM subjects were more obese. However, multiple regression demonstrated no significant contribution of age or BMI, and oxidative metabolic defects have been previously demonstrated in diabetic subjects even when compared with age-matched controls (42). Similarly, we cannot exclude insulin resistance itself in contributing to differential expression of PGC1 isoforms and oxidative phosphorylation genes. However, isolated insulin resistance in transgenic mice does not alter expression of oxidative metabolism genes or PGC1α (27) (V. Yechoor, personal communication), and family history of DM, rather than insulin resistance, was the predominant covariate for both PGC1α and -β expression in our human cohort.
Inactivity could also contribute to the expression phenotype in our sedentary subjects. Exercise training and activation of both AMP kinase and calcium/calmodulin-dependent protein kinase increase mitochondrial gene expression and oxidative capacity, perhaps via increases in PGC1 (43–45). Because PGC1 is preferentially expressed in type 1 fibers (46), fiber type composition might contribute to differences in PGC1 expression. However, relative expression of slow and fast myosin or troponin isoforms did not differ between our groups, and oxidative enzyme activity and lipid content have been shown to be independent of fiber type in obese and diabetic subjects (47). Moreover, decreased PGC1 expression may be a primary contributor to increased type IIb fiber content in nondiabetic FH+ subjects (48).
The inverse relationship among UQCRH expression, obesity, and fasting insulin raises the possibility that an adipocyte product, nutrient excess, or insulin resistance itself may further contribute to down-regulation of oxidative phosphorylation gene expression in susceptible individuals. Activation of the hexosamine nutrient signaling pathway decreases expression of nuclear-encoded mitochondrial genes in rats (49). Although caloric restriction increases PGC1α expression in obese subjects (50), we cannot exclude an additive role for other transcription factors and coactivators in mediating the oxidative metabolism expression phenotype.
In summary, we demonstrate that expression of PGC1α and -β and multiple genes of oxidative metabolism is reduced in DM and in high-risk nondiabetic subjects with a family history of DM. We postulate that in genetically susceptible individuals, inactivity, over-nutrition, and the development of insulin resistance may further reduce expression of NRF-regulated oxidative metabolism genes, aggravating the metabolic phenotype and increasing DM risk. Our data illustrate the utility of assessing coordinated changes in gene expression to identify pathways important in pathogenesis of insulin resistance and DM. Future studies focused on understanding the upstream molecular mechanisms by which complex oxidative metabolic pathways are dysregulated in insulin-resistant “prediabetics” may ultimately help to develop novel methods for detecting and interrupting the vicious cycle of metabolic derangements and to prevent the onset of overt diabetes.
Supplementary Material
Acknowledgments
We appreciate data sharing by Vamsi Mootha, Whitehead Institute/ Massachusetts Institute of Technology Center for Genome Research, and helpful suggestions from John Rogus. We gratefully acknowledge support from National Institutes of Health (NIH) Grant DK02526, the Iacocca Foundation, and Harvard Medical School 50th Anniversary Scholars in Medicine (to M.E.P.); Endocrine Fellows Foundation, Lawson Wilkins Pediatric Endocrinology Society, Harvard Center for Neurodegeneration and Repair, NIH Grants HL066582 and DK063696 (to A.J.B.); American Diabetes Association (to R.B.); NIH Grant DK24092 (to R.D.); NIH Grant DK060837 (to C.R.K.); NIH Grants DK47936 and GCRC RR-01346 (to L.J.M.); and NIH Grant U24 DK058739 (National Institute of Diabetes and Digestive and Kidney Diseases Biotechnology Center). Studies were facilitated by the Affymetrix Academic User Center/NIH Grant P01HG01323.
Abbreviations: DM, type 2 diabetes mellitus; PGC1α and -β, peroxisomal proliferator activator receptor γ coactivator α and β (PPARGC1 and PERC); NRF-1, nuclear respiratory factor 1; FH, family history; BMI, body mass index.
Footnotes
Yechoor, V. K., Patti, M. E., Saccone, R. & Kahn, C. R. (2002) Diabetes, 51, p. A258 (abstr.).
Arya, R., Blangero, J., Almasy, L., O'Connell, P. & Stern, M. P. (2001) Obes. Res. 9, 70S (abstr.).
References
- 1.Eriksson, J., Franssila-Kallunki, A., Ekstrand, A., Saloranta, C., Widen, E., Schalin, C. & Groop, L. (1989) N. Engl. J. Med. 321 337-343. [DOI] [PubMed] [Google Scholar]
- 2.Martin, B. C., Warram, J. H., Krolewski, A. S., Bergman, R. N., Soeldner, J. S. & Kahn, C. R. (1992) Lancet 340 925-929. [DOI] [PubMed] [Google Scholar]
- 3.Vaag, A., Henriksen, J. E. & Beck-Nielsen, H. (1992) J. Clin. Invest. 89 782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cusi, K., Maezono, K., Osman, A., Pendergrass, M., Patti, M. E., Mandel, G., DeFronzo, R. A., Kahn, C. R. & Mandarino, L. J. (2000) J. Clin. Invest. 105 311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducluzeau, P. H., Perretti, N., Laville, M., Andreelli, F., Vega, N., Riou, J. P. & Vidal, H. (2001) Diabetes 50 1134-1142. [DOI] [PubMed] [Google Scholar]
- 6.Jacob, S., Machann, J., Rett, K., Brechtel, K., Volk, A., Renn, W., Maerker, E., Matthaei, S., Schick, F., Claussen, C. D., et al. (1999) Diabetes 48 1113-1119. [DOI] [PubMed] [Google Scholar]
- 7.Perseghin, G., Scifo, P., De Cobelli, F., Pagliato, E., Battezzati, A., Arcelloni, C., Vanzulli, A., Testolin, G., Pozza, G., Del Maschio, A., et al. (1999) Diabetes 48 1600-1606. [DOI] [PubMed] [Google Scholar]
- 8.Saltiel, A. R. & Kahn, C. R. (2001) Nature 414 799-806. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy, M. I. & Froguel, P. (2002) Am. J. Physiol Endocrinol. Metab 283 E217-E225. [DOI] [PubMed] [Google Scholar]
- 10.Burke, J. P., Williams, K., Haffner, S. M., Villalpando, C. G. & Stern, M. P. (2001) Diabetes Care 24 1573-1578. [DOI] [PubMed] [Google Scholar]
- 11.Pratipanawatr, W., Pratipanawatr, T., Cusi, K., Berria, R., Adams, J. M., Jenkinson, C. P., Maezono, K., DeFronzo, R. A. & Mandarino, L. J. (2001) Diabetes 50 2572-2578. [DOI] [PubMed] [Google Scholar]
- 12.Yechoor, V. K., Patti, M. E., Saccone, R. & Kahn, C. R. (2002) Proc. Natl. Acad. Sci. USA 99 10587-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadt, E. E., Li, C., Su, C. & Wong, W. H. (2000) J. Cell Biochem. 80 192-202. [PubMed] [Google Scholar]
- 14.Press, W. H., Teukolsky, S. A., Vetterlin, W. T. & Flannery, B. P. (1993) Numerical Recipes in C: The Art of Scientific Computing (Cambridge Univ. Press, Cambridge, U.K.).
- 15.Draghici, S., Khatri, P., Martins, R., Ostermeier, G. & Krawetz, S. (2003) Genomics 81 1-7. [DOI] [PubMed] [Google Scholar]
- 16.Dahlquist, K. D., Salomonis, N., Vranizan, K., Lawlor, S. C. & Conklin, B. R. (2002) Nat. Genet. 31 19-20. [DOI] [PubMed] [Google Scholar]
- 17.Doniger, S. W., Salomonis, N., Dahlquist, K. D., Vranizan, K., Lawlor, S. C. & Conklin, B. R. (2003) Genome Biol. 4 R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. Ser. B 57 289-300. [Google Scholar]
- 19.Dudait, S., Yang, Y. H. & Speed, T. P. (2002) Stat. Sin. 12 111-139. [Google Scholar]
- 20.Kurucz, I., Morva, A., Vaag, A., Eriksson, K. F., Huang, X., Groop, L. & Koranyi, L. (2002) Diabetes 51 1102-1109. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. K., Fillmore, J. J., Chen, Y., Yu, C., Moore, I. K., Pypaert, M., Lutz, E. P., Kako, Y., Velez-Carrasco, W., Goldberg, I. J., et al. (2001) Proc. Natl. Acad. Sci. USA 98 7522-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarpulla, R. C. (2002) Gene 286 81-89. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., et al. (1999) Cell 98 115-124. [DOI] [PubMed] [Google Scholar]
- 24.Sreekumar, R., Halvatsiotis, P., Schimke, J. C. & Nair, K. S. (2002) Diabetes 51 1913-1920. [DOI] [PubMed] [Google Scholar]
- 25.Yang, X., Pratley, R. E., Tokraks, S., Bogardus, C. & Permana, P. A. (2002) Diabetologia 45 1584-1593. [DOI] [PubMed] [Google Scholar]
- 26.Vicent, D., Piper, M., Gammeltoft, S., Maratos-Flier, E. & Kahn, C. R. (1998) Diabetes 47 1451-1458. [DOI] [PubMed] [Google Scholar]
- 27.Mootha, V., Lindgren, C., Eriksson, K., Subramian, A., Sihaq, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet., in press. [DOI] [PubMed]
- 28.Heddi, A., Stepien, G., Benke, P. J. & Wallace, D. C. (1999) J. Biol. Chem. 274 22968-22976. [DOI] [PubMed] [Google Scholar]
- 29.Simoneau, J. A., Veerkamp, J. H., Turcotte, L. P. & Kelley, D. E. (1999) FASEB J. 13 2051-2060. [DOI] [PubMed] [Google Scholar]
- 30.Kelley, D. E., He, J., Menshikova, E. V. & Ritov, V. B. (2002) Diabetes 51 2944-2950. [DOI] [PubMed] [Google Scholar]
- 31.Song, J., Oh, J. Y., Sung, Y. A., Pak, Y. K., Park, K. S. & Lee, H. K. (2001) Diabetes Care 24 865-869. [DOI] [PubMed] [Google Scholar]
- 32.Antonetti, D. A., Reynet, C. & Kahn, C. R. (1995) J. Clin. Invest. 95 1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley, D. E. & Simoneau, J. A. (1994) J. Clin. Invest. 94 2349-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin, K., Daa, S. H., Alford, F. P. & Beck-Nielsen, H. (2001) Diabetologia 44 824-833. [DOI] [PubMed] [Google Scholar]
- 35.Boden, G. & Shulman, G. I. (2002) Eur. J. Clin. Invest. 32 Suppl. 3, 14-23. [DOI] [PubMed] [Google Scholar]
- 36.Perseghin, G., Scifo, P., Danna, M., Battezzati, A., Benedini, S., Meneghini, E., Del Maschio, A. & Luzi, L. (2002) Am. J. Physiol Endocrinol. Metab 283 E556-E564. [DOI] [PubMed] [Google Scholar]
- 37.Pratley, R. E., Thompson, D. B., Prochazka, M., Baier, L., Mott, D., Ravussin, E., Sakul, H., Ehm, M. G., Burns, D. K., Foroud, T., et al. (1998) J. Clin. Invest. 101 1757-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone, S., Abkevich, V., Hunt, S. C., Gutin, A., Russell, D. L., Neff, C. D., Riley, R., Frech, G. C., Hensel, C. H., Jammulapati, S., et al. (2002) Am. J. Hum. Genet. 70 1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esterbauer, H., Oberkofler, H., Linnemayr, V., Iglseder, B., Hedegger, M., Wolfsgruber, P., Paulweber, B., Fastner, G., Krempler, F. & Patsch, W. (2002) Diabetes 51 1281-1286. [DOI] [PubMed] [Google Scholar]
- 40.Ek, J., Andersen, G., Urhammer, S. A., Gaede, P. H., Drivsholm, T., Borch-Johnsen, K., Hansen, T. & Pedersen, O. (2001) Diabetologia 44 2220-2226. [DOI] [PubMed] [Google Scholar]
- 41.Hara, K., Tobe, K., Okada, T., Kadowaki, H., Akanuma, Y., Ito, C., Kimura, S. & Kadowaki, T. (2002) Diabetologia 45 740-743. [DOI] [PubMed] [Google Scholar]
- 42.Kelley, D. E., Mokan, M. & Mandarino, L. J. (1992) Diabetes 41 698-706. [DOI] [PubMed] [Google Scholar]
- 43.Zong, H., Ren, J. M., Young, L. H., Pypaert, M., Mu, J., Birnbaum, M. J. & Shulman, G. I. (2002) Proc. Natl. Acad. Sci. USA 99 15983-15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terada, S., Goto, M., Kato, M., Kawanaka, K., Shimokawa, T. & Tabata, I. (2002) Biochem. Biophys. Res. Commun. 296 350-354. [DOI] [PubMed] [Google Scholar]
- 45.Wu, H., Kanatous, S. B., Thurmond, F. A., Gallardo, T., Isotani, E., Bassel-Duby, R. & Williams, R. S. (2002) Science 296 349-352. [DOI] [PubMed] [Google Scholar]
- 46.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418 797-801. [DOI] [PubMed] [Google Scholar]
- 47.He, J., Watkins, S. & Kelley, D. E. (2001) Diabetes 50 817-823. [DOI] [PubMed] [Google Scholar]
- 48.Nyholm, B., Qu, Z., Kaal, A., Pedersen, S. B., Gravholt, C. H., Andersen, J. L., Saltin, B. & Schmitz, O. (1997) Diabetes 46 1822-1828. [DOI] [PubMed] [Google Scholar]
- 49.Obici, S., Wang, J., Chowdury, R., Feng, Z., Siddhanta, U., Morgan, K. & Rossetti, L. (2002) J. Clin. Invest. 109 1599-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larrouy, D., Vidal, H., Andreelli, F., Laville, M. & Langin, D. (1999) Int. J. Obes. Relat Metab Disord. 23 1327-1332. [DOI] [PubMed] [Google Scholar]
- 51.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scarpulla, R. C. (2002) Biochim. Biophys. Acta 1576 1-14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.