Abstract
NAPVSIPQ (NAP), an active fragment of the glial-derived activity-dependent neuroprotective protein, is protective at femtomolar concentrations against a wide array of neural insults and prevents ethanol-induced fetal wastage and growth retardation in mice. NAP also antagonizes ethanol inhibition of L1-mediated cell adhesion (ethanol antagonism). We performed an Ala scanning substitution of NAP to determine the role of ethanol antagonism and neuroprotection in NAP prevention of ethanol embryotoxicity. The Ser-Ile-Pro region of NAP was crucial for both ethanol antagonism and protection of cortical neurons from tetrodotoxin toxicity (neuroprotection). Ala replacement of either Ser-5 or Pro-7 (P7A-NAP) abolished NAP neuroprotection but minimally changed the efficacy of NAP ethanol antagonism. In contrast, Ala replacement of Ile-6 (I6A-NAP) caused a decrease in potency (>2 logarithmic orders) with only a small reduction (<10%) in the efficacy of NAP neuroprotection but markedly reduced the efficacy (50%) and the potency (5 logarithmic orders) of NAP ethanol antagonism. Ethanol significantly reduced the number of paired somites in mouse whole-embryo culture; this effect was prevented significantly by 100 pM NAP or by 100 pM P7A-NAP, but not by 100 pM I6A-NAP. The structure–activity relation for NAP prevention of ethanol embryotoxicity was similar to that for NAP ethanol antagonism and different from that for NAP neuroprotection. These findings support the hypothesis that NAP antagonism of ethanol inhibition of L1 adhesion plays a central role in NAP prevention of ethanol embryotoxicity and highlight the potential importance of ethanol effects on L1 in the pathophysiology of fetal alcohol syndrome.
Fetal alcohol syndrome (FAS) is the most common preventable cause of mental retardation (1). Ethanol has multiple cellular targets in the nervous system (2); hence, it is not surprising that it damages the fetus through a variety of mechanisms: oxidative injury, induction of apoptosis, suppression of neurogenesis, disruption of cell–cell interactions, and alterations in the release and signaling of growth factors, morphogens, and chemical messengers (3–10). Several drugs that block specific molecular actions of ethanol have been shown to prevent or mitigate ethanol's teratogenesis in animal models (11, 12), an unexpected finding, given the complex pathophysiology of FAS. Delineating the mechanism of action of these drugs would help to identify the most critical mechanisms that underlie ethanol's teratogenesis.
Brain lesions in children with FAS resemble those of children with mutations in the gene for the L1 cell adhesion molecule, suggesting that ethanol might perturb fetal development in part by disrupting the actions of L1 (7). Interestingly, ethanol potently inhibits L1-mediated cell–cell adhesion (7, 13, 14) and L1-mediated neurite extension (15). A series of straight, cyclic, and branched alcohols shows unexpectedly strict structural requirements for alcohol inhibition of cell adhesion, consistent with a ligand-receptor interaction (16). Several alcohols proved to be competitive or noncompetitive antagonists of ethanol inhibition of L1 adhesion (17), and one such molecule, 1-octanol, also prevented ethanol-induced dysmorphology and apoptosis in mouse whole-embryo culture (11). These findings highlight the potential importance of ethanol's actions on L1 in the pathophysiology of FAS and raise the possibility of identifying safe ethanol antagonists.
A second class of compounds also prevents ethanol teratogenesis (12). NAPVSIPQ (NAP) and SALLRSIPA (SAL) are small peptide fragments of the glial-derived activity-dependent neuroprotective protein (ADNP) and activity-dependent neurotrophic factor (ADNF), respectively (18, 19). Both NAP and SAL are protective at femtomolar concentrations in vitro against the neural toxicity of a wide range of compounds and cellular insults (18, 20, 21). NAP and SAL are also neuroprotective in vivo against diverse neural insults, including excitotoxicity (22), closed head injury (23), ischemic brain injury (24), apoplipoprotein E deficiency (19), exposure to the cholinotoxin ethylcholine aziridium (25), and prenatal ethanol exposure (12). The precise mechanism by which NAP and SAL produce neuroprotection is not clear, although a variety of biochemical actions may contribute. NAP, SAL, or their parent compounds increase levels of cGMP and nitric oxide (26), promote the release of neurotrophic factor-3 (27), induce the expression of heat shock protein 60 (20), activate protein kinase C and mitogen-associated protein kinase kinase (28), increase NF-κB DNA-binding activity (29), protect against oxidative injury, and reduce neuronal apoptosis (21, 24, 30, 31). Protection against oxidative injury may partly explain NAP prevention of ethanol teratogenesis, because NAP decreases ethanol-induced depletion of reduced glutathione (12).
We recently showed (32) that NAP and SAL potently antagonize ethanol inhibition of L1 adhesion. This observation raises the question of whether NAP and SAL, like 1-octanol, prevent ethanol embryotoxicity by antagonizing a specific action of ethanol, rather than through broad neuroprotective actions. To answer this question, we identified derivatives of NAP that differentially affect ethanol inhibition of L1 adhesion and protection against tetrodotoxin (TTX) neurotoxicity and tested these for prevention of ethanol embryotoxicity. Electrical blockade with TTX was chosen as a model system to study neuroprotection because of its relevance to activity-dependent mechanisms of neuronal development as well as its first use in characterizing ADNP, the parent protein of NAP (19).
Methods
Materials. Ethanol was purchased from Fisher Scientific; all other chemicals were purchased from Sigma-Aldrich (St. Louis) or as indicated. Peptides were purchased from Peptide Technologies (Washington, DC), Sigma Genosys (The Woodlands, TX), and New England Peptides (Fitchburg, MA). Purity (>95%) and identity were assessed by these companies by using HPLC and MS analyses. The peptides were dissolved in 10% DMSO in PBS (0.13 M NaCl/0.003 M KCl/0.01 M Na2HPO4/0.002 M KH2PO4) and stored as 1-mM aliquots. NAP was stable in solution and could be aliquoted and frozen for later use without loss of activity.
Culture of L1-Expressing NIH 3T3 Cells. NIH 3T3 cells were cultured in DMEM supplemented with 10% normal calf serum and 400 μg/ml G418 (all from Life Technologies, Rockville, MD). Two subclones were used in these studies: 2A2-L1 and Vec-1A5. The 2A2-L1 cell line is an ethanol-sensitive subclone derived from a stable transfection of NIH 3T3 cells with the human L1 cDNA, and Vec-1A5 is a subclone from a transfection with the empty expression vector (14). Both cell lines were cultured at 37°C, in an atmosphere of 90% air and 10% CO2.
Cell Adhesion Assay. Cell–cell adhesion was measured by using a short-term aggregation assay of subconfluent cells (32). Cells were detached by gentle agitation with calcium-free and magnesium-free PBS supplemented with 2 mM EDTA and 0.1 mg/ml DNase, mechanically dissociated to obtain a single-cell suspension, and diluted to 330,000 cells/ml. One ml of the cell suspension was added per well (4.5 cm2) to a 12-well plate. Peptides and ethanol were mixed together before their addition to the cells, which were then mixed gently for 30 min on ice. Cells were viewed at a final magnification of ×200 and each well was scored for single and adherent cells in five or six microscopic fields of view. We counted ≈150–200 cells per field of view and 750–1,200 cells per well. The percentage of adherent cells was calculated for each microscopic field of view and averaged. To ensure the reliability of the cell adhesion assays, most assays were scored without knowledge of the experimental conditions.
We define L1-mediated cell–cell adhesion (L1 adhesion) as the difference in the percent of adherent cells between an L1-expressing cell line (2A2-L1) and a vector-transfected cell line (Vec-1A5). In L1-transfected cells, this component of cell adhesion is fully inhibited by Fab fragments of an anti-L1 polyclonal antibody (7, 14, 33). Ethanol inhibition of cell adhesion was calculated as 100 × (1 - the ratio of L1 adhesion in the presence and absence of ethanol). We define antagonists as compounds that alone have no effect on L1 adhesion, but block the ethanol inhibition of L1 adhesion. Antagonist activity was calculated as 100 × 1 - [(percent inhibition of cell adhesion by ethanol plus peptide)/(percent inhibition of cell adhesion by ethanol alone)].
Assay of TTX Toxicity in Neuronal Cells. Dissociated cerebral cortical tissue from newborn rats was seeded on a confluent layer of astroglial cultures derived from rat cerebral cortex (34). We plated 250,000 cells into a 35-mm dish in a volume of 1.5 ml. The mixed cultures were maintained in medium (35) consisting of 5% horse serum in MEM supplemented with defined medium components (36) and 9-fluoro-2-deoxyuridine (15 μg/ml) plus uridine (3 μg/ml). The mixed cultures were >95% astrocytes. Four days after adding the cerebral cortical suspension to astrocyte feeder cultures, the culture preparations were given a complete change of medium (before peptide treatment). Peptides and 1 μM TTX were added once and cultures were assayed for neuronal survival after a 4-day incubation period. Neuronal cell counts were conducted after fixation with glutaraldehyde (35). Neuronal identity was established with sister cultures immunocytochemically stained with antiserum against neuron-specific enolase (37). Neurons were counted in 20 fields in each culture dish without knowledge of the treatment group. Previous studies demonstrated that 1 μM TTX blocked synaptic activity in CNS cultures (38) and produced decrements as measured with many neuronal parameters, including choline acetyltransferase, tetanus toxin fixation, saxitoxin binding (39) and neuronal survival (40).
Whole-Embryo Culture. On gestational day 8, C57BL/6J embryos were explanted under a dissecting microscope with removal of the maternal decidua, trophoblast, parietal yolk sac, and Reichert's membranes, whereas the visceral yolk sac, ectoplacental cone, and amnion remained intact (4). Those embryos having three to five somite pairs were used for culture. Each embryo was placed into a 30-ml vial containing 2.5 ml of medium (75% heat-inactivated rat serum, 25% Tyrode's solution). The vials were flushed with a mixture of 5% O2, 5% CO2, and 90% N2 and attached to a rotating wheel in an incubator maintained at 37°C. Explanted embryos were exposed to experimental agents for 6 h only, followed by culture for an additional 20 h in control medium. Extraembryonic membranes were removed and the embryos were examined under a dissecting microscope, without knowledge of treatment condition, for morphological assessment, and to allow determination of the number of somite pairs. Each embryo was cultured separately, constituting an independent experiment.
Operational Definitions. The ability of NAP to antagonize ethanol inhibition of L1-mediated cell adhesion is referred to as NAP antagonism. NAP protection of cortical neurons from TTX toxicity is referred to as NAP neuroprotection. NAP prevention of ethanol-induced reductions in somite number is referred to as NAP prevention of ethanol embryotoxicity.
Statistical Analysis. By using STATVIEW software, the differences among means of groups were analyzed by ANOVA. Multiple comparison post-tests between groups were conducted by using Bonferroni/Dunn comparisons.
Results
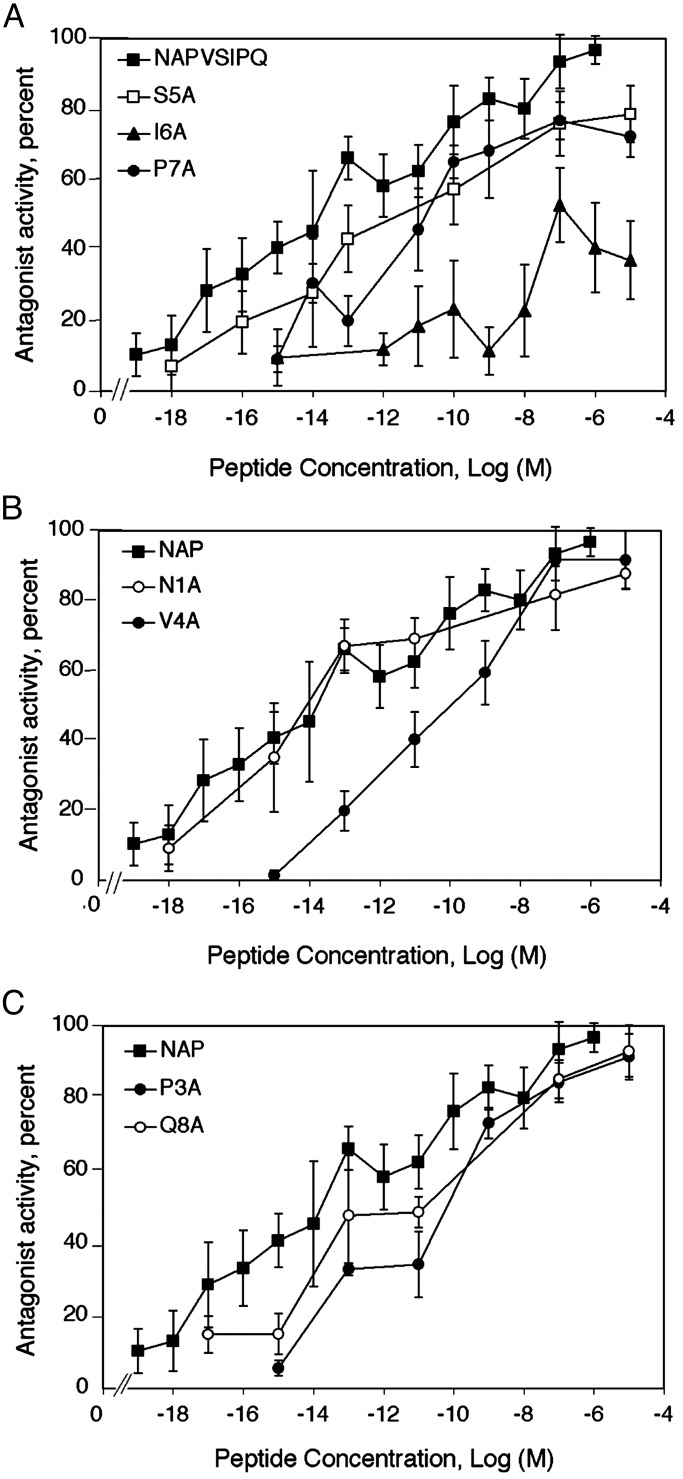
Structure–Activity Relation for NAP Antagonism. L1-mediated cell adhesion was measured in NIH 3T3 cells stably transfected with human L1. Ethanol (100 mM) inhibited L1-mediated cell adhesion and NAP completely antagonized this action of ethanol, as reported (32). The dose–response curve for NAP revealed a monophasic curve of antagonist activity over many logarithmic orders, with half-maximal effects at 36 fM (Fig. 1).
Fig. 1.
Structure–activity relation for NAP antagonism of ethanol inhibition of L1 adhesion. Adhesion assays were carried out in the absence and presence of 100 mM ethanol and the indicated peptides, by using NIH 3T3 cells stably transfected with human L1 cDNA (2A2-L1). Antagonist activity was calculated as described (see Methods), based on the ability of each peptide to decrease ethanol inhibition of L1-mediated cell adhesion. Alanine-scanning substitution produced seven NAP-derived peptides, which are named for the single-letter amino acid that is replaced with Ala and its position with respect to the N terminus (e.g., S5A, Ala replaces Ser-5 in NAP). Dose–response curves are shown as follows: NAP, S5A, I6A, and P7A (A); NAP, N1A, and V4A (B); and NAP, P3A, and Q8A (C). The NAP dose–response curve is reproduced in each figure to facilitate comparison with the NAP mutants. Shown are the mean ± SEM values for antagonist activity derived from 3–15 independent experiments. Mean values for adhesion were 39.1 ± 0.8% in the absence of ethanol and 25.7 ± 0.8% in the presence of ethanol. Some of the data points in the NAP dose–response curve were taken from a previous publication (32) but were derived contemporaneously with the Ala-substituted NAP derivatives.
Alanine scanning substitution produced seven NAP-derived peptides, which are named for the single-letter amino acid that is replaced by Ala and its position with respect to the N terminus (e.g., S5A-NAP, Ala replaces Ser-5 in NAP). Dose–response curves were determined for this series of NAP-derived peptides. The Ala substitutions had strikingly different effects on NAP antagonism (Fig. 1 and Table 1). Ala substitutions at the Ser-5 (S5A), Ile-6 (I6A), and Pro-7 (P7A) positions reduced both the efficacy and potency of NAP. Of these three amino acids, Ile-6 was the most sensitive to Ala substitution. Compared with NAP, the potency of I6A-NAP was reduced by 19,000-fold and its efficacy was reduced by half. The efficacies of S5A-NAP (79%) and P7A-NAP (77%) were reduced slightly compared with NAP, but their potencies were decreased 500-fold and 1,400-fold, respectively. In contrast, Ala substitution for the Asn residue at the N-terminal position of NAP (N1A-NAP) had the least effect, producing only a slight reduction in antagonist potency and efficacy. Likewise, Ala substitutions at the Pro-3, Val-4, and Gln-8 positions affected antagonist efficacy minimally, whereas the dose–response curves for all three peptides showed marked rightward shifts. These data define a structure-activity relation in which the Ser-Ile-Pro (SIP) region of NAP is important for antagonizing ethanol inhibition of L1 adhesion, with Ile being a critical site.
Table 1. Comparison of structure-activity relation for NAP ethanol antagonism and NAP neuroprotection.
|
Maximal effect, %
|
||||||
|---|---|---|---|---|---|---|
|
TTX
|
TTX
|
|||||
| NAP-derived peptides | L1 | P1 | P2 | L1 EC50, fM | P1 EC50, fM | P2 EC50, pM |
| NAPVSIPQ (NAP) | 97±4 (14) | 100±2 | 97±4 | 36 | 0.003 | 3 |
| AAPVSIPQ (N1A) | 88±4 (5)* | 89±3 | 101±3 | 69 | 0.1 | 100 |
| NAAVSIPQ (P3A) | 92±6 (3) | 95±2 | 89±2* | 46,000 | 100 | 300 |
| NAPASIPQ (V4A) | 92±2 (7)** | 95±2 | 96±4 | 90,000 | 3 | 3 |
| NAPVAIPQ (S5A) | 79±8 (10)* | 0*** | 0*** | 19,000 | — | — |
| NAPVSAPQ (I6A) | 53±10 (15)*** | 91±1* | 91±2* | 700,000 | 3 | 3,000 |
| NAPVSIAQ (P7A) | 77±5 (15)*** | 0*** | 0*** | 50,000 | — | — |
| NAPVSIPA (Q8A) | 93±7 (5) | 97±1 | 98±2 | 4,400 | 0.003 | 3 |
NAP potency and efficacy were estimated from dose-response curves for NAP antagonism of ethanol inhibition of L1 adhesion (Fig. 1) and NAP protection of neuronal cultures from TTX toxicity (Fig. 2). NAP potency for ethanol antagonism was calculated from linear regression analysis of the average values in the dose-response curves (n = 3-15). NAP potency for neuroprotection was estimated from the two peaks (P1 and P2) of the dose-response curves shown in Fig. 2 (n = 3). L1, NAP antagonism of ethanol inhibition of L1 adhesion. TTX, NAP protection of cortical neurons from TTX toxicity
, P < 0.05
, P < 0.01
, P < 0.001 on paired t tests comparing efficacy of NAP derivatives to NAP
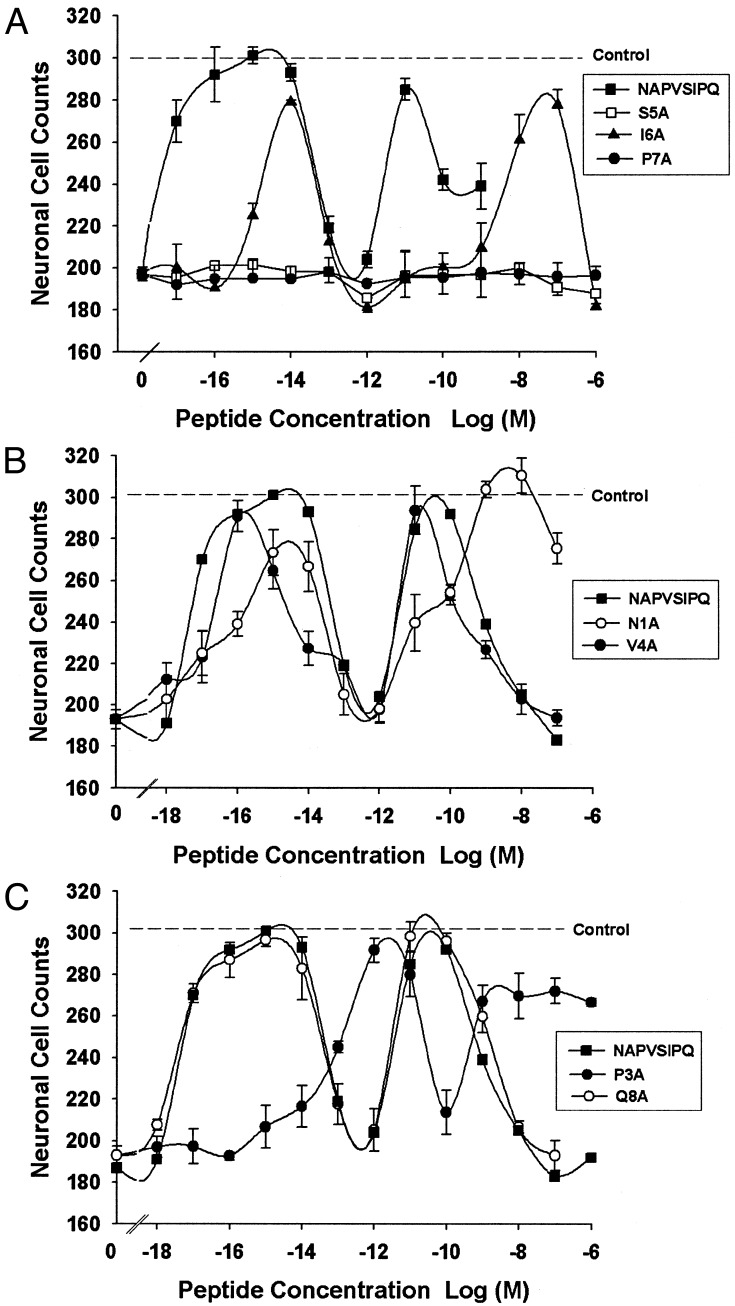
Structure–Activity Relation for NAP Neuroprotection. A structure– activity-relation analysis was performed for the neuroprotective action of NAP on cortical neurons exposed to TTX (Fig. 2 and Table 1). Four days of treatment with 1 μM TTX reduced neuronal survival by 37 ± 2%. NAP potently and completely prevented TTX neurotoxicity. The dose–response curve for NAP neuroprotection showed two major peaks of activity with potencies (EC50) of ≈0.003 fM and 3 pM. Ala substitution at the Ser-5 and Pro-7 positions abolished the neuroprotective activity of NAP. In contrast, Ala substitution at the Ile-6 position caused a rightward shift in both peaks (>2 logarithmic orders) of the dose–response curve and only a small reduction (<10%) in efficacy for the higher affinity peak. Ala substitution at Asn-1, Val-4, and Glu-8 did not reduce neuroprotective efficacy, and among these three mutants, only N1A-NAP showed a modest (33-fold) reduction in potency. Ala substitution at the Pro-3 position reduced the neuroprotective potency of NAP by 4.5 logarithmic orders, without reducing efficacy at the high-affinity peak. The low-affinity peak of P3A-NAP showed a 2-logarithmic-orders decrease in potency and an 11% decrease in efficacy compared with that of NAP. These data demonstrate that the SIP region of NAP is also critical for neuroprotection; however, in this action, the Ser-5 and Pro-7 sites are much more sensitive than the Ile-6 site.
Fig. 2.
Structure–activity relation for NAP protection of cortical cultures from TTX toxicity. Cortical neuronal cultures were incubated for 4 days in the absence (control) and presence of 1 μM TTX. Peptides were added with TTX as indicated. Shown is the mean ± SEM number of neurons from three to four determinations. The results shown were derived from two independent experiments. The dotted horizontal line indicates the number of surviving neurons in control cultures (305 ± 10, n = 5) that were not treated with TTX. Dose–response curves are shown as follows: NAP, S5A, I6A, and P7A (A); NAP, N1A, and V4A (B); and NAP, P3A, and Q8A (C).
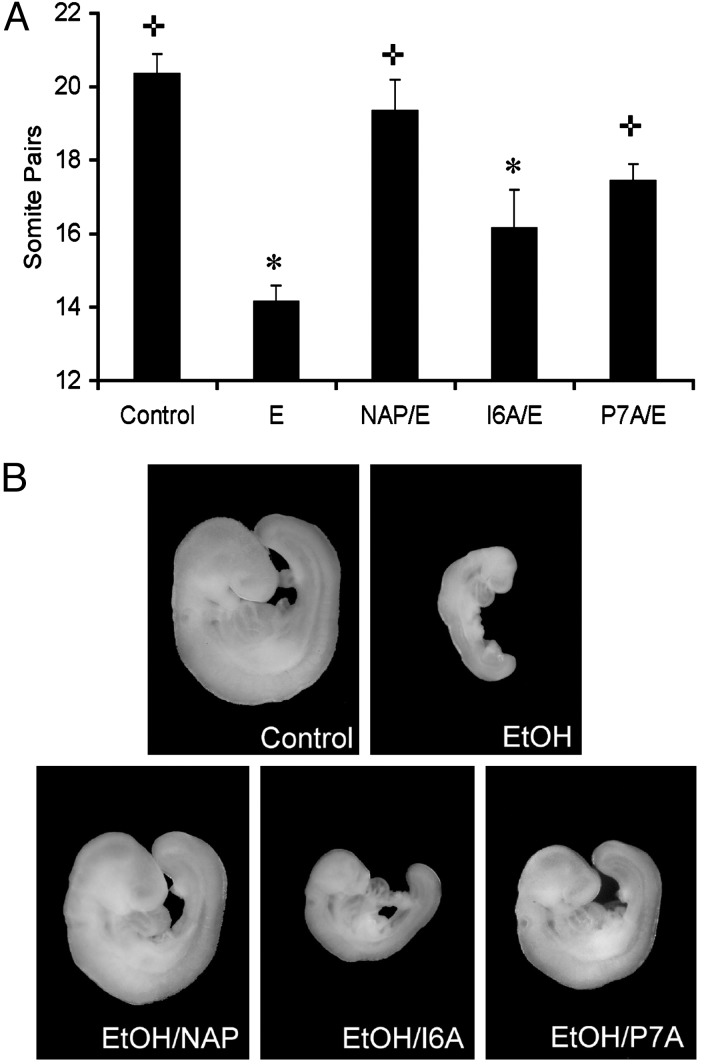
NAP Prevention of Ethanol Embryotoxicity. Whole-embryo culture allows the controlled administration of drugs and ethanol in the absence of maternal influences, including nutritional variables. At these early stages of embryogenesis, exposure to teratogenic concentrations of ethanol in vivo, as well as in vitro (whole-embryo culture), yields comparable results, including excessive apoptotic cell death, neural tube defects, and craniofacial abnormalities (4, 11, 41, 42). We exploited the differences in structure–activity relation for NAP antagonism and NAP neuroprotection to ask which action contributes to NAP prevention of ethanol embryotoxicity. A single concentration (100 pM) of NAP, I6A-NAP, and P7A-NAP was tested for prevention of ethanol-induced growth retardation in mouse whole-embryo culture. This concentration was chosen for its ability to produce nearly maximal effects with NAP and markedly different effects for the NAP derivatives. Gestational day 8 mouse embryos (3–5 somites) were cultured for 6 h in the absence or presence of 100 mM ethanol or peptide plus ethanol and then transferred to control medium for an additional 20 h. Somite pairs were counted after a total of 26 h in culture. Embryos cultured for 6 h with 100 mM ethanol showed markedly delayed in vitro development as compared with control embryos (Fig. 3). The coin-cubation of cultured embryos with NAP or P7A-NAP significantly reduced ethanol-induced growth retardation. In contrast, treatment of cultured embryos with I6A-NAP did not significantly reduce ethanol-induced growth retardation.
Fig. 3.
Effect of ethanol and NAP peptides on cultured C57BL/6J gestational day 8
mouse embryos. (A) Mean ± SEM number of somite pairs in
individual embryos treated for 6 h under control conditions (n = 13),
in the presence of 100 mM ethanol alone (E, n = 44), or with 100 mM
ethanol combined with 100 pM NAP (NAP/E, n = 17), I6A-NAP (I6A/E,
n = 23), or P7A-NAP (P7A/E, n = 28) and after an additional
20 h of culture.  , P < 0.0001, compared with ethanol alone.
*, P < 0.0001, compared with control. (B) Shown is
a representative embryo (median number of somites) from each of the five
experimental groups: control, 21 somites; ethanol (EtOH), 14 somites; ethanol
plus NAP (EtOH/NAP), 20 somites; ethanol plus I6A-NAP (EtOH/I6A), 16 somites;
and ethanol plus P7A-NAP (EtOH/P7A), 18 somites.
, P < 0.0001, compared with ethanol alone.
*, P < 0.0001, compared with control. (B) Shown is
a representative embryo (median number of somites) from each of the five
experimental groups: control, 21 somites; ethanol (EtOH), 14 somites; ethanol
plus NAP (EtOH/NAP), 20 somites; ethanol plus I6A-NAP (EtOH/I6A), 16 somites;
and ethanol plus P7A-NAP (EtOH/P7A), 18 somites.
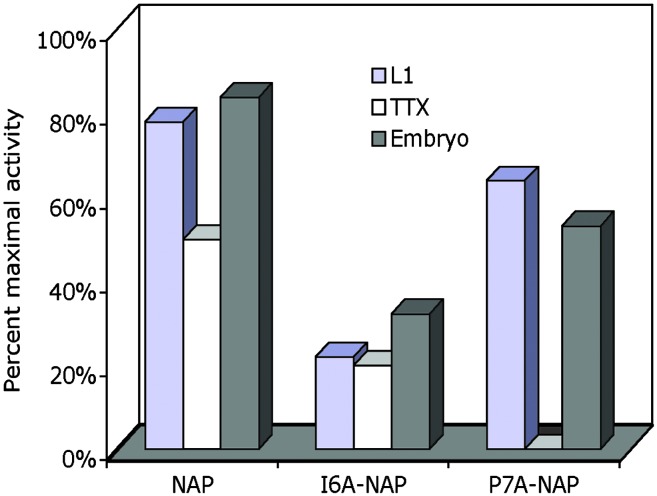
Comparison of Structure–Activity Relation for NAP Antagonism, Neuroprotection, and Prevention of Ethanol Embryotoxicity. The laborious and costly nature of whole-embryo culture permitted the evaluation of only a single concentration of selected NAP mutants. To compare the efficacy of NAP mutants among three different assay systems, we expressed the data for 100 pM peptide as a percent of maximal NAP activity for ethanol antagonism and neuroprotection, and as a percent of maximal possible prevention of ethanol embryotoxicity (Fig. 4). The activities of NAP, P7A-NAP, and I6A-NAP were remarkably similar for ethanol antagonism and prevention of ethanol embryotoxicity. In contrast, P7A-NAP had no neuroprotective activity, but showed 56% of maximal NAP ethanol antagonism and 53% of maximal prevention of ethanol embryotoxicity.
Fig. 4.
Relative effect of NAP mutants on ethanol antagonism, neuroprotection, and prevention of ethanol embryotoxicity. Data for 100 pM of the indicated peptides were expressed as a percentage of maximal NAP effect (ethanol antagonism, ethanol neuroprotection) or as a percentage of maximal possible prevention of ethanol embryotoxicity. L1, percentage of maximal NAP antagonism of ethanol inhibition of L1 adhesion (see Fig. 1). TTX, percentage of maximal NAP protection of TTX neurotoxicity (see Fig. 2). Embryo, percentage of peptide protection from the full reduction in somite number caused by ethanol (see Fig. 3). A different measure was used in the whole-embryo experiments, because it was not feasible to establish a maximal NAP effect through a dose–response curve.
Discussion
NAP is a multifunctional peptide that protects neural cells against a wide array of toxins and insults (19, 24, 25, 30, 31, 43). This broad neuroprotective profile suggests that NAP acts downstream of convergent cellular pathways that trigger cell death. Therefore, NAP might prevent ethanol teratogenesis by a general protective effect against ethanol-induced oxidative injury or apoptosis. NAP also blocks ethanol inhibition of L1-mediated cell–cell adhesion (32). We have speculated that ethanol inhibition of L1 adhesion might cause anoikis, the induction of apoptosis through loss of cell–cell contact (11). This hypothesis suggests a second possible mechanism for NAP prevention of ethanol teratogenesis: the upstream antagonism of a specific action of ethanol that triggers cell death.
We undertook the present study to learn which of two actions of NAP correlated best with NAP prevention of ethanol embryotoxicity: antagonism of ethanol inhibition of L1 adhesion or NAP's broad neuroprotective actions. We chose to examine NAP protection against TTX toxicity in cortical cultures, because this was the first and best established model of NAP's broad neuroprotective actions (18, 44). TTX blocks electrical activity in cortical neurons, activating a process that eliminates neurons that fail to make or sustain functional connections (40). We used 100 mM ethanol in our experiments, because this high concentration is observed in alcoholics (45) and reliably produces teratogenic effects in several models of FAS (4, 11, 46). Moreover, a high concentration of ethanol provides a stringent test of any putative ethanol antagonists.
Structure–activity analysis revealed both similarities and differences for NAP antagonism and NAP neuroprotection. The SIP region of NAP was crucial for both actions of NAP. This finding is not surprising, given the conservation of the SIP region in NAP and SAL. NAP and SAL have a similar spectrum of neuroprotective activity, and both antagonize ethanol inhibition of L1 adhesion (32). The N-terminal Asn of NAP was relatively insensitive to Ala replacement, as was the C-terminal Gln. In this respect, the structure–activity relation of NAP neuroprotection differs from that of SAL, which was sensitive to changes in both the N-terminal and C-terminal regions (18). The relative insensitivity of NAP to N-terminal and C-terminal replacement suggests the feasibility of adding reporter molecules to either end of the NAP peptide to further study its mechanism of action.
The most striking difference in the structure–activity relation between NAP antagonism and NAP neuroprotection was within the SIP region. Ala replacement of either Ser-5 or Pro-7 abolished NAP neuroprotection but had relatively minor effects on the efficacy of NAP antagonism. In contrast, Ala replacement of Ile-6 caused only a small reduction in the efficacy of NAP neuroprotection but reduced by half the efficacy of NAP antagonism. The potency of NAP for ethanol antagonism was reduced by Ala substitution at all sites except the N-terminal Asn; however, the Ile-6 site was by far the most sensitive. The potency of NAP for neuroprotection was reduced primarily by Ala replacement at Asn-1, Pro-3, and within the SIP region. Thus, the efficacy and potency of NAP for neuroprotection depended most on Ser-5 and Pro-7, whereas the efficacy and potency of NAP for ethanol antagonism depended most on Ile-6.
Dose–response curves also differed for NAP antagonism and NAP neuroprotection. NAP antagonism demonstrated a broad dose–response curve with half-maximal effects in the femtomolar range (32). In contrast, NAP neuroprotection showed two peaks of activity with half-maximal effects in the subfemtomolar and picomolar range. The two peaks of NAP neuroprotective activity could result from the interaction of NAP with two different molecular targets or different states of the same target. However, neither target appears to be the same as that which mediates NAP antagonism, because Ala substitution for Ser-5 or Pro-7 abolished both peaks in the neuroprotection assay, but did not eliminate NAP antagonism. These data imply that NAP interacts with at least two, and possibly three, distinct molecular targets to produce neuroprotection and ethanol antagonism.
Picomolar concentrations of NAP significantly prevented ethanol-induced growth retardation in mouse whole-embryo culture. This finding confirms observations in a different animal model of FAS, where fetal wastage and growth retardation at birth were the primary outcome measures (12). Most importantly, P7A-NAP significantly reduced ethanol embryotoxicity, whereas I6A-NAP did not. This profile of activity strongly resembles that for NAP antagonism but differs strikingly from that for NAP neuroprotection. Taken together, these data indicate that NAP prevention of ethanol's embryotoxicity correlates better with NAP antagonism than with NAP neuroprotection.
It is noteworthy that despite the multiplicity of mechanisms by which ethanol disrupts fetal development, antagonists for a single action of ethanol have such robust effects in preventing ethanol teratogenesis. This finding suggests that ethanol disruption of L1 plays an important role in the pathogenesis of FAS. It remains possible that as-yet-unknown actions of NAP share the same structure–activity relation as NAP antagonism and are responsible for NAP prevention of ethanol embryotoxicity. However, this seems unlikely, given that two structurally unrelated molecules, NAP and 1-octanol, antagonize ethanol inhibition of L1 adhesion and prevent ethanol teratogenesis (11, 12, 16, 32). At least in mouse whole-embryo culture, antagonism of ethanol inhibition of L1 adhesion is sufficient for preventing ethanol-induced embryotoxicity.
Acknowledgments
This work was supported in part by Public Health Service Grants AA12974 (to M.E.C.) and AA11605 (to K.K.S.), and the Medical Research Service, Department of Veterans Affairs.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FAS, fetal alcohol syndrome; TTX, tetrodotoxin; NAP, peptide fragment NAPVSIPQ; SAL, peptide fragment SALLRSIPA.
See commentary on page 8043.
References
- 1.Abel, E. L. & Sokol, R. J. (1987) Drug Alcohol Depend. 19 51-70. [DOI] [PubMed] [Google Scholar]
- 2.Diamond, I. & Gordon, A. S. (1997) Physiol. Rev. 77 1-20. [DOI] [PubMed] [Google Scholar]
- 3.Gressens, P., Lammens, M., Picard, J. J. & Evrard, P. (1992) Alcohol Alcohol. 27 219-226. [PubMed] [Google Scholar]
- 4.Kotch, L. E., Chen, S. Y. & Sulik, K. K. (1995) Teratology 52 128-136. [DOI] [PubMed] [Google Scholar]
- 5.Guerri, C., Pascual, M. & Renau-Piqueras, J. (2001) Neurotoxicology 22 593-599. [DOI] [PubMed] [Google Scholar]
- 6.Climent, E., Pascual, M., Renau-Piqueras, J. & Guerri, C. (2002) J. Neurosci. Res. 68 213-225. [DOI] [PubMed] [Google Scholar]
- 7.Ramanathan, R., Wilkemeyer, M. F., Mittal, B., Perides, G. & Charness, M. E. (1996) J. Cell Biol. 133 381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikonomidou, C., Bittigau, P., Ishimaru, M. J., Wozniak, D. F., Koch, C., Genz, K., Price, M. T., Stefovska, V., Horster, F., Tenkova, T., et al. (2000) Science 287 1056-1060. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell, J. J., Paiva, M. & Heaton, M. B. (1999) Neurosci. Lett. 263 189-192. [DOI] [PubMed] [Google Scholar]
- 10.Goodlett, C. R. & Horn, K. H. (2001) Alcohol Res. Health 25 175-184. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S.-Y., Wilkemeyer, M. F., Sulik, K. K. & Charness, M. E. (2001) FASEB J. 15 1649-1651. [DOI] [PubMed] [Google Scholar]
- 12.Spong, C. Y., Abebe, D. T., Gozes, I., Brenneman, D. E. & Hill, J. M. (2001) J. Pharmacol. Exp. Ther. 297 774-779. [PubMed] [Google Scholar]
- 13.Charness, M. E., Safran, R. M. & Perides, G. (1994) J. Biol. Chem. 269 9304-9309. [PubMed] [Google Scholar]
- 14.Wilkemeyer, M. F. & Charness, M. E. (1998) J. Neurochem. 71 2382-2391. [DOI] [PubMed] [Google Scholar]
- 15.Bearer, C. F., Swick, A. R., O'Riordan, M. A. & Cheng, G. (1999) J. Biol. Chem. 274 13264-13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkemeyer, M. F., Sebastian, A. B., Smith, S. A. & Charness, M. E. (2000) Proc. Natl. Acad. Sci. USA 97 3690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkemeyer, M. F., Menkari, C. E. & Charness, M. E. (2002) Mol. Pharmacol. 62 1053-1060. [DOI] [PubMed] [Google Scholar]
- 18.Brenneman, D. E., Hauser, J., Neale, E., Rubinraut, S., Fridkin, M., Davidson, A. & Gozes, I. (1998) J. Pharmacol. Exp. Ther. 285 619-627. [PubMed] [Google Scholar]
- 19.Bassan, M., Zamostiano, R., Davidson, A., Pinhasov, A., Giladi, E., Perl, O., Bassan, H., Blat, C., Gibney, G., Glazner, G., et al. (1999) J. Neurochem. 72 1283-1293. [DOI] [PubMed] [Google Scholar]
- 20.Zamostiano, R., Pinhasov, A., Bassan, M., Perl, O., Steingart, R. A., Atlas, R., Brenneman, D. E. & Gozes, I. (1999) Neurosci. Lett. 264 9-12. [DOI] [PubMed] [Google Scholar]
- 21.Glazner, G. W., Boland, A., Dresse, A. E., Brenneman, D. E., Gozes, I. & Mattson, M. P. (1999) J. Neurochem. 73 2341-2347. [DOI] [PubMed] [Google Scholar]
- 22.Gressens, P., Marret, S., Hill, J. M., Brenneman, D. E., Gozes, I., Fridkin, M. & Evrard, P. (1997) J. Clin. Invest. 100 390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beni-Adani, L., Gozes, I., Cohen, Y., Assaf, Y., Steingart, R. A., Brenneman, D. E., Eizenberg, O., Trembolver, V. & Shohami, E. (2001) J. Pharmacol. Exp. Ther. 296 57-63. [PubMed] [Google Scholar]
- 24.Leker, R. R., Teichner, A., Grigoriadis, N., Ovadia, H., Brenneman, D. E., Fridkin, M., Giladi, E., Romano, J. & Gozes, I. (2002) Stroke (Dallas) 33 1085-1092. [DOI] [PubMed] [Google Scholar]
- 25.Gozes, I., Giladi, E., Pinhasov, A., Bardea, A. & Brenneman, D. E. (2000) J. Pharmacol. Exp. Ther. 293 1091-1098. [PubMed] [Google Scholar]
- 26.Ashur-Fabian, O., Giladi, E., Furman, S., Steingart, R. A., Wollman, Y., Fridkin, M., Brenneman, D. E. & Gozes, I. (2001) Neurosci. Lett. 307 167-170. [DOI] [PubMed] [Google Scholar]
- 27.Blondel, O., Collin, C., McCarran, W. J., Zhu, S., Zamostiano, R., Gozes, I., Brenneman, D. E. & McKay, R. D. (2000) J. Neurosci. 20 8012-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gressens, P., Marret, S., Bodenant, C., Schwendimann, L. & Evrard, P. (1999) J. Mol. Neurosci. 13 199-210. [DOI] [PubMed] [Google Scholar]
- 29.Glazner, G. W., Camandola, S. & Mattson, M. P. (2000) J. Neurochem. 75 101-108. [DOI] [PubMed] [Google Scholar]
- 30.Offen, D., Sherki, Y., Melamed, E., Fridkin, M., Brenneman, D. E. & Gozes, I. (2000) Brain Res. 854 257-262. [DOI] [PubMed] [Google Scholar]
- 31.Steingart, R. A., Solomon, B., Brenneman, D. E., Fridkin, M. & Gozes, I. (2000) J. Mol. Neurosci. 15 137-145. [DOI] [PubMed] [Google Scholar]
- 32.Wilkemeyer, M. F., Menkari, C., Spong, C. Y. & Charness, M. E. (2002) J. Pharmacol. Exp. Ther. 303 110-116. [DOI] [PubMed] [Google Scholar]
- 33.Wilkemeyer, M. F., Pajerski, M. & Charness, M. E. (1999) Alcohol. Clin. Exp. Res. 23 1711-1720. [PubMed] [Google Scholar]
- 34.McCarthy, K. D. & de Vellis, J. (1980) J. Cell Biol. 85 890-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenneman, D. E. & Foster, G. A. (1987) Peptides (Tarrytown, NY) 8 687-694. [DOI] [PubMed] [Google Scholar]
- 36.Romijn, H. J., van Huizen, F. & Wolters, P. S. (1984) Neurosci. Biobehav. Rev. 8 301-334. [DOI] [PubMed] [Google Scholar]
- 37.Schmechel, D., Marangos, P. J., Zis, A. P., Brightman, M. & Goodwin, F. K. (1978) Science 199 313-315. [DOI] [PubMed] [Google Scholar]
- 38.Ransom, B. R. & Holz, R. W. (1977) Brain Res. 136 445-453. [DOI] [PubMed] [Google Scholar]
- 39.Brenneman, D. E. (1986) Can. J. Physiol. Pharmacol. 64 356-362. [DOI] [PubMed] [Google Scholar]
- 40.Brenneman, D. E., Neale, E. A., Habig, W. H., Bowers, L. M. & Nelson, P. G. (1983) Brain Res. 285 13-27. [DOI] [PubMed] [Google Scholar]
- 41.Sulik, K. K., Johnston, M. C. & Webb, M. A. (1981) Science 214 936-938. [DOI] [PubMed] [Google Scholar]
- 42.Kotch, L. E. & Sulik, K. K. (1992) Int. J. Dev. Neurosci. 10 273-279. [DOI] [PubMed] [Google Scholar]
- 43.Zemlyak, I., Furman, S., Brenneman, D. E. & Gozes, I. (2000) Regul. Pept. 96 39-43. [DOI] [PubMed] [Google Scholar]
- 44.Brenneman, D. E. & Gozes, I. (1996) J. Clin. Invest. 97 2299-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charness, M. E., Simon, R. P. & Greenberg, D. A. (1989) N. Engl. J. Med. 321 442-454. [DOI] [PubMed] [Google Scholar]
- 46.Kotch, L. E., Dehart, D. B., Alles, A. J., Chernoff, N. & Sulik, K. K. (1992) Teratology 46 323-332. [DOI] [PubMed] [Google Scholar]