Abstract
The UNUSUAL FLORAL ORGANS (UFO) gene is required for multiple processes in the developing Arabidopsis flower, including the proper patterning and identity of both petals and stamens. The gene encodes an F-box-containing protein, UFO, which interacts physically and genetically with the Skp1 homolog, ASK1. In this report, we describe four ufo alleles characterized by the absence of petals, which uncover another role for UFO in promoting second whorl development. This UFO-dependent pathway is required regardless of the second whorl organ to be formed, arguing that it affects a basic process acting in parallel with those establishing organ identity. However, the pathway is dispensable in the absence of AGAMOUS (AG), a known inhibitor of petal development. In situ hybridization results argue that AG is not transcribed in the petal region, suggesting that it acts non-cell-autonomously to inhibit second whorl development in ufo mutants. These results are combined into a genetic model explaining early second whorl initiation/proliferation, in which UFO functions to inhibit an AG-dependent activity.
Organ formation involves the coordinated execution of several processes that result in primordia capable of generating a given structure. These include the proper allocation of progenitor cells, establishment of appropriate gene expression patterns, and regulated control of cell division. In higher plants, flower formation serves as a model for understanding how these developmental strategies are controlled and integrated. The Arabidopsis flower consists of four organ types arranged in a whorled phyllotaxis; these whorls (w1–w4) contain four sepals (w1), four petals (w2), six stamen (w3), and two fused carpels forming the gynoecium (w4). Genes required for proper development of the floral meristem (meristem identity genes) and the individual organs themselves (organ identity genes) have been isolated and genetic interactions among many of these genes determined. A model in which combinatorial interactions between three classes (A, B, and C) of organ identity genes result in the four organ types observed has been proposed (1, 2). Subsequent investigation has largely validated this model as well as identifying new genes that function in concert with these key regulators (3).
The UNUSUAL FLORAL ORGANS gene (UFO) is required for proper floral development, and strong loss-of-function mutations result in defects in all whorls of the flower. These abnormalities are most severe in w2 and w3, where, in addition to aberrant organ number and phyllotaxis, homeotic transformations are prevalent (4, 5). Specifically, petals are commonly replaced by sepals and stamen by carpels, although mosaic organs are also frequently found in both whorls. These transformations imply that B class function is compromised. In fact, UFO is necessary for proper transcriptional control of these genes. In ufo flowers, both AP3 and PI transcript levels are reduced (4, 6), and ectopic expression of UFO, in the presence of LEAFY (LFY), leads to the ectopic activation of AP3 and PI, both in flowers and vegetative tissue (6, 7). Further, coexpression of AP3 and PI from heterologous promoters partially rescues a strong ufo mutant (8).
UFO is expressed in the shoot apical meristem (SAM) throughout development (6, 9), although loss of UFO activity does not detectably affect vegetative development. Like its Antirrhinum homolog, FIMBRIATA (FIM), UFO is expressed in a complex temporal and spatial pattern during floral development (6, 10). In floral meristems, transcription is initially repressed during stage 1, then activated during stage 2 in the central region. By stage 3, expression is lost in the central meristem, but it expands laterally in a cone-shaped pattern. Expression becomes restricted to the petal primordia region by stage 5 and is maintained there through most of the floral organ development. This pattern is consistent with the proposed role of UFO in promoting B class function, because both AP3 and PI patterns are established during stage 3 (11, 12), before w2/w3 organ initiation during stage 5 (13, 14).
UFO encodes a 442-aa protein that, like FIM, contains an F-box motif that mediates interaction with an evolutionarily conserved protein, SKP1 (15). Despite the large number of F-box-containing proteins in Arabidopsis (16), UFO and FIM currently show extensive homology only to the Impatiens Stp and Lotus Pfo proteins (17, 18). F-box protein-SKP1 complexes most commonly function together with a third subunit, CULLIN/Cdc53, to form the SKP-Cullin-F-box (SCF) class of E3 ubiquitin protein ligases. SCF, in concert with an E2 ubiquitin-conjugating enzyme, targets substrates bound to the F-box protein for polyubiquitination and then degradation by the 26S proteasome (19). Both UFO and FIM associate with plant SKP1 proteins in vitro (20, 21), and UFO also interacts genetically with one SKP1 homolog, ASK1 (22, 23). However, neither the corresponding Cullin nor a target protein has been isolated.
Here, we report the isolation of four additional ufo alleles that specifically block w2 development. This UFO-dependent pathway is activated after B class organ identity gene expression is established, just before w2 primordia initiation and/or proliferation. The pathway appears to counteract the inhibitory effects of an AG-dependent pathway, allowing w2 organogenesis to occur. These results are incorporated with existing data to offer a modified framework of how A, B, and C class genes contribute to second whorl development.
Methods
Plant Material. Plants were grown in Sunshine Mix (Fisons Horticultural, Bellevue, WA) soil under long day conditions (18 h light) in chambers maintained at 22°C. ufo-11 and ufo-12 were isolated from T-DNA mutagenized seeds of the Wassilewskija (WsO) ecotype (24). ufo-13 and ufo-14 were identified from an ethyl methane sulfonate (EMS)-treated population of Columbia seeds (the generous gift of S. Steinberg and L. Feldman, University of California, Berkeley). All other mutants were obtained from the laboratory of Elliot Meyerowitz (California Institute of Technology, Pasadena).
Identification of the ufo Lesions. ufo11 and ufo-12 mutations were first mapped to the distal end of the UFO promoter by Southern blot analysis by using probes to different regions of UFO and the T-DNA borders. Precise endpoints of the ufo-12 lesion were determined by PCR amplification (see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org).
ORFs from each ufo allele were amplified by PCR and subcloned. These inserts were sequenced completely. ufo-11 and ufo-12 were found to contain no mutations, whereas ufo-13 and ufo-14 were found to each contain a single base-pair substitution.
Scanning Electron Microscopy. Fixation, drying, and viewing were done as described (25).
In Situ Hybridization. In situ hybridization was performed by using digoxigenin (DIG)-labeled RNA probes and visualized with an anti-DIG antibody conjugated to alkaline phosphatase according to Drews (26) and the manufacturer (Boehringer Mannheim). Probes were generated by in vitro transcription by using T7 RNA polymerase and DIG-labeled ribonucleotides. For detection of UFO transcripts, the plasmid pG11/UfoFL (see Supporting Methods) was linearized with NcoI before in vitro transcription. AP3 antisense probes were generated from pIa (27) digested with BglII. AG transcripts were detected with probes from CIT565 (28) cleaved with HindIII.
Generation and Identification of Double Mutants. To obtain double mutants, ufo plants were crossed with either homozygous (ap2–1, pi-1) or heterozygous lines (lfy-1, ag-1), and F2 families with both single mutant phenotypes identified. From these families, seeds from individual petalless plants and other mutants (when possible) were sown and the F3 generation analyzed for any new phenotypes. No background effects on the petalless phenotype were observed
Two-Hybrid Analysis. Y153 (29) was the recipient for all transformations. For β-galactosidase activity, cells were prepared and assayed as described (29) by using chlorophenol-red-β-d-galactopyranoside (Boehringer Mannheim) as substrate. Gal4-DNA-binding domain fusions with UFO were constructed as described in Supporting Methods.
Results
Isolation of ufo Alleles Specifically Affecting Early Petal Development. During two independent screens for mutants affecting floral organ number, we identified four lines, dubbed petalless, with severe defects in petal formation (Fig. 1). Subsequent analysis indicated that the phenotype segregated as a single recessive Mendelian trait, and that all lines were allelic. Crosses between these mutants and the strong ufo-2 allele (4) resulted in flowers with the petalless phenotype; thus, these four mutants contain partial loss-of-function mutations in UFO.
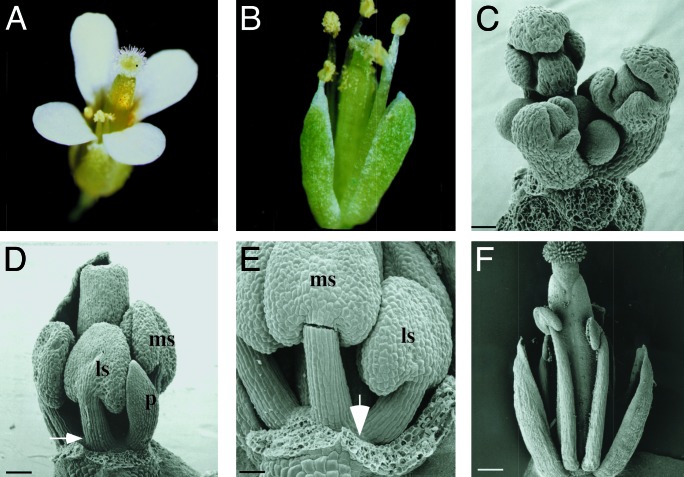
Fig. 1.
Phenotype of weak ufo mutants. (A) Wild-type flower. (B) Typical flower from ufo-12. Petals are completely absent in almost all flowers. (C) Scanning electron micrograph of ufo-11 inflorescence. (D) Stage-9 flower showing one petal (p) arising between the lateral (ls) and medial (ms) stamen but no organ in adjacent w2 position (arrow). Three sepals were removed. (E) Higher magnification of stage-9 flower showing the lateral (ls) and medial (ms) stamen and position where a petal would normally arise (arrow). Sepals were removed. (F) Scanning electron microscopy image of a mature ufo-11 flower. One medial sepal removed. [Bars, 52 μm(C), 81 μm(D), 55 μm(E), and 227 μm (F).]
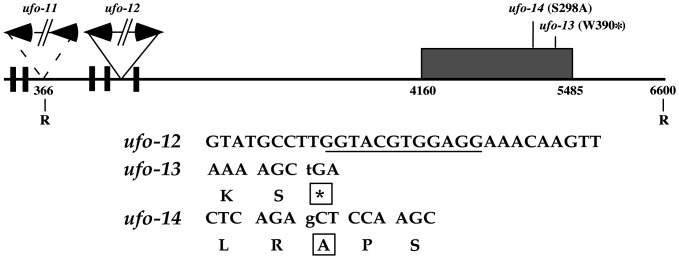
Two alleles, ufo-11 and ufo-12, are from a T-DNA mutagenized collection (24). Sequencing of the UFO ORF from these lines detected no mutations. Instead, both T-DNA insertion sites localize to the 5′ distal end of the UFO promoter (Fig. 2), suggesting that transcriptional regulation of the gene is compromised. The ufo-12 lesion contains multiple T-DNAs inserted 764 bp downstream of the 5′ EcoRI site with a concomitant loss of 12 bp of wild-type sequence. The ufo-11 insertion site lies just 5′ to an EcoRI site, although rearrangement in this area has confounded efforts to precisely locate the lesion. This region of the UFO promoter contains five CArG box-like sequences that are recognition elements for MADS box DNA-binding proteins (Fig. 2), implying that one or more of these factors may activate w2 expression of UFO.
Fig. 2.
Location of weak ufo lesions. (Upper) Schematic of UFO gene with sites of four new mutations marked. UFO ORF is indicated by hatched box. Numbering begins at an arbitrary point 5′ of the putative w2 enhancer element. Potential CArG boxes are denoted with black boxes. T-DNAs are indicated by the broken solid line and are not drawn to scale. Dashed line marking the ufo-11 insertion indicates that the exact location is unknown. R, EcoRI site. (Lower) Locations of ufo-12, -13, and -14 lesions. For ufo-12, the underlined nucleotides (1129–1140) are replaced by T-DNA sequences. For ufo-13 and -14, nucleotide changes are lowercase letters, and amino acid substitutions are boxed. The asterisk denotes a stop codon.
ufo-13 and ufo-14 alleles were isolated from EMS mutagenized plants, and sequence analysis of these alleles revealed that each contains a single point mutation in the UFO ORF (Fig. 2). ufo-13 contains a nonsense mutation leading to a stop codon at position 390, whereas a missense mutation in ufo-14 results in a serine-to-alanine substitution at residue 298.
Phenotypic quantification of these lines (Table 1, which is published as supporting information on the PNAS web site) indicates that mutants produce less than one petal per flower on average, although most basal flowers can have more. Basal flowers also can have petal/stamen mosaic organs in either w2 or w3, although these generally contain only a small amount of transformed tissue. Sepal and carpel whorls develop normally, and flowers are fully fertile.
To investigate at what stage petal primordia arrest in the ufo mutants, ufo-11 flowers were analyzed by scanning electron microscopy during various stages of development. As expected, the gross overall morphology of ufo-11 apices closely resembled that of wild type (compare Fig. 1C with ref. 14). In wild-type flowers, petal primordia are first visible as bumps on the floral meristem during stage 5 and then undergo a lag phase before beginning rapid expansion (14). In contrast, no evidence of petal primordia is visible in ufo-11 flowers (Fig. 1 D–F) except in rare cases (two petals in 45 ufo-11 flowers) where an organ will develop fully (Fig. 1D). Note that when petals do form, they do so in the proper temporal and spatial manner (Fig. 1D). Thus, affected petal primordia arrest at an early stage in the mutants before the primordia are clearly visible.
The proper positioning of the stamens and the rare petals that form argue that the primordia patterning function of UFO is intact in these mutants. That these organs generally have the correct identity further suggests that the UFO role in activating AP3/PI transcription also is retained. Indeed, AP3 transcript expression in developing ufo-12 flowers appears normal (Fig. 8, which is published as supporting information on the PNAS web site). Given that petal primordia are absent in these ufo alleles and that wild-type petal primordia are comprised of few cells, these data imply that AP3 expression mainly identifies w3 stamen primordia cells in wild-type tissues at these early stages. Nonetheless, these results indicate that the weak ufo mutations likely affect an additional UFO activity required for controlling petal primordia initiation and/or proliferation.
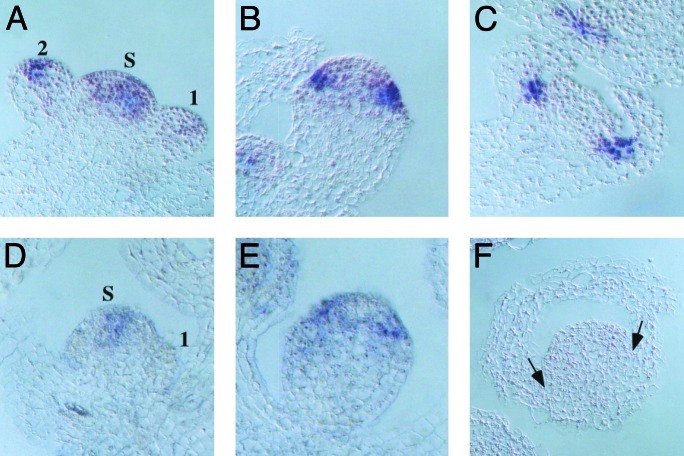
ufo-11 and -12 Mutations Disrupt Petal Primordia Expression. Localization of ufo-11 and -12 T-DNA insertions to the promoter suggests the UFO expression pattern might be altered. As reported (6, 9), UFO transcripts are present in the SAM of wild-type plants but absent in stage 1 floral meristems. Transcripts are readily detected in the central portion of the developing flower by stage 2 (Fig. 3A). During stage 3, they are excluded from the presumptive fourth whorl but remain expressed in regions that will give rise to second and third whorl organs (Fig. 3B). Finally, by stage 5, expression is limited to the region that will give rise to the petals (Fig. 3C), where it is maintained throughout floral organ development.
Fig. 3.
Distribution of UFO transcripts in wild-type and ufo-12 flowers. Longitudinal sections through wild type (A–C) and ufo-12 (D–F) were subjected to in situ hybridization with digoxigenin-labeled UFO anti-sense RNA probes. Signal is dark blue. (A) Inflorescence apex with SAM (S) and stage-1 and -2 floral primordia. UFO is expressed in the SAM and stage-2 flowers but down-regulated during stage 1. (B) Stage-3 flower showing the cone-shaped expression in regions that will give rise to petals and stamens. (C) Stage-5 wild-type flower. Expression is limited to regions of developing petal primordia. (D) SAM (S) and stage-1 floral meristem; expression appears lower than wild type. (E) Stage-3 ufo-12 flower showing weak but detectable levels of transcripts in the w2/w3 region. (F) UFO transcripts are not detected in stage-5 ufo-12 flowers in the region that would form petals in wild type (arrows).
In ufo-12 flowers, the proper expression pattern is maintained through stage 3, although RNA levels appear to be reduced (Fig. 3 D and E). However, transcripts were absent in the subsequent stages when they are normally found in developing w2 cells (compare Fig. 3 C to F). Identical results were obtained with ufo-11 flowers (data not shown). The specific loss of UFO transcripts in the petal region argues that both T-DNA insertions have disrupted an enhancer element essential for w2 expression. This result argues that the w2-specific UFO expression commencing during stage 4/5 is necessary for petal initiation/proliferation.
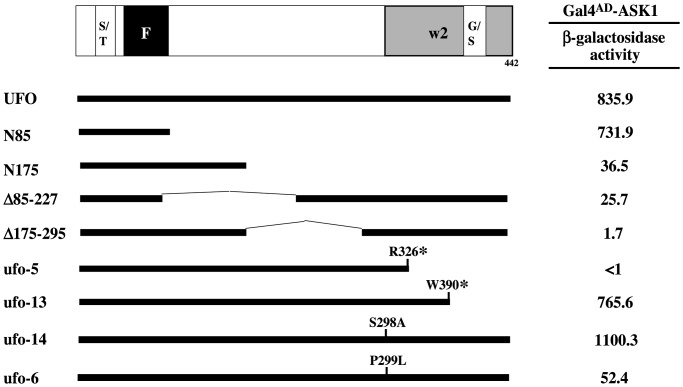
ufo-13 and ufo-14 Mutations Do Not Affect Interaction with ASK1. Genetic experiments argue that UFO interaction with the Arabidopsis SKP1 homolog, ASK1, is important for its function (22, 23). To determine the region(s) of UFO necessary for binding ASK1, a deletion series was constructed in the yeast expression vector, pAS1 (29), creating fusions with the Gal4 DNA-binding domain. These hybrids were then analyzed for the ability to interact with ASK1 (21) via the two-hybrid assay. Full length UFO/ASK1 interaction results in a large increase in β-galactosidase activity, suggesting a strong interaction between the two proteins (Fig. 4). However, only the N-terminal 85-aa fragment, which terminates at the end of the F-box, interacts with ASK1 with similar apparent affinity as full length UFO. All other engineered deletions show dramatically reduced affinity for ASK1 despite containing an intact F-box. These results suggest that, whereas the F-box is apparently sufficient for high-affinity interaction with ASK1, the overall structure of the protein may be important for allowing ASK1 access to the motif.
Fig. 4.
Two-hybrid interactions between a UFO mutation series and ASK1. UFO fusions to the Gal4 DNA-binding domain were constructed in pAS1 and introduced into yeast strain Y153 with a plasmid expressing full-length ASK1 fused to the Gal4 activation domain. Nn contain N-terminal residues 1-n. Δn-m are internal deletions of amino acid n-m. Amino acid substitutions in ufo mutations are indicated in single-letter code or an asterisk denoting a stop codon. β-Galactosidase activity (Miller units) was quantified with chlorophenol red-β-d-galactopyranoside (CPRG), representing the average from three independent transformants. Locations of F-box (F), Ser/Thr-rich (S/T), and Gly/Ser-rich (G/S) domains in UFO are indicated. The region inferred to be necessary for second whorl specific function is indicated in the light shaded box marked w2.
To ascertain how various ufo mutations affect UFO interaction with ASK1, DNA-binding domain fusions were made with both strong and weak mutants and the association with ASK1 tested (Fig. 4). The strong ufo-5 allele contains a nonsense mutation at codon 326 (6) and was chosen because it results in the smallest deletion of the strong mutants characterized to date. This mutant UFO protein is unable to associate with ASK1 in this assay, consistent with the hypothesis that interaction with ASK1 is essential for UFO function.
Next, three weak mutants, ufo-6, -13, and -14, were tested. Interestingly, proteins encoded by both ufo-13 and -14 still interact with ASK1, and these associations lead to comparable increases in β-galactosidase activity as wild type. Although the ufo-6- encoded polypeptide still binds to ASK1, its affinity is dramatically compromised, potentially explaining its slightly more severe phenotype (4). Intriguingly, the ufo-6 mutation maps directly adjacent to ufo-14, changing the proline at position 299 to leucine (6).
Thus, although ASK1-UFO association may be necessary, it is not sufficient for full UFO function. Because ufo-13 and ufo-14 retain primordia patterning and B class gene activation functions, the results imply that the C-terminal region (amino acids 298–442) is required specifically for w2 initiation/proliferation.
Genetic Interactions with Key Floral Development Genes. To identify additional genetic components in this UFO-dependent pathway, double mutants were generated carrying either ufo-11 or ufo-12 in combination with mutations in other known floral genes. In all cases, either ufo allele resulted in an indistinguishable double mutant phenotype. Four combinations led to informative results.
Strong mutations in LFY, such as lfy-1, result in plants with increased numbers of secondary inflorescences and severe floral defects (30–32). lfy-1 flowers are composed of two outer whorls of sepal-like structures and an aberrantly formed central gynoecium. Organs formed in outer whorls are arranged in a more spiral phyllotaxis than wild type, consistent with their arising from a shoot-like floral meristem. lfy-1 ufo-11 flowers are phenotypically identical to lfy-1 (Fig. 5 A and B), formally placing LFY upstream of UFO in this pathway, consistent with earlier observations that LFY activity is generally required for UFO function (4–6). This result also indicates that the “w2” structures in lfy-1 are not under identical regulatory mechanisms as similarly positioned organs arising from wild-type floral meristems.
Fig. 5.
Double mutant combinations with weak ufo alleles and four key floral development genes. Indistinguishable results were obtained with either ufo-11 or ufo-12.(A)A lfy-1 flower showing sepal-like structures produced in “w1” and “w2”; these structures are arranged in a more spiral phyllotaxis than wild type. (B) lfy-1 ufo-11 flowers are phenotypically identical to lfy-1. (C) ap2-1 flowers contain leaf-like structures in w2 and petals and stamenoid petals in w2 positions. (D) ap2-1 ufo-11 flowers have first whorl leaf-like organs as in ap2-1 but are missing w2 organs. (E) pi-1 flowers have four sepals in both w1 and w2. w3 stamens are transformed into filamentous structures and are often fused to the w4 carpels. (F) pi-1 ufo-11 flowers have an additive phenotype where w2 pi-1 sepals are absent. (G) Typical ag-1 flower with w3 stamens converted to petals and a new flower initiated in w4. This pattern is repeated multiple times. (H) ag-1 ufo-11 double mutants are phenotypically identical to ag-1. (I)An ag-5 flower with petaloid stamens in w3 and fused w4 carpelloid sepals encasing a new flower. w1 and w2 develop normally. (J) ag-5 ufo-11 flowers display the same phenotype as the ag-5 single mutant with four petals forming in w2. Note that w4 organs are unfused.
Weak mutations in the A class gene, AP2, lead to floral defects in both outer two whorls with leaf-like organs replacing sepals and stamenoid petals forming in w2 (33) (Fig. 5C). ap2-1 ufo-11 plants are missing w2 stamenoid petals found in ap2-1 (Fig. 5D). Either ap2-1 and ufo have an additive affect, or the ap2 w2 phenotype is enhanced by the weak ufo alleles, because strong ap2 mutants also lack w2 organs (33).
Strong mutations in the B class gene, PI, cause homeotic transformations in w2 and w3, with sepals replacing petals and carpels or filaments replacing stamens, respectively (13, 33) (Fig. 5E). In addition, w3 organs commonly fuse with w4 carpels. In contrast to lfy-1 ufo-11, pi-1 ufo-11 flowers showed an additive phenotype in that they lacked w2 organs, but w3 filaments were maintained and often were fused to the gynoecium (Fig. 5F). Thus, in the presence of LFY activity, the w2 function of UFO is required regardless of the organ to be formed and therefore is affecting a process independent of organ identity.
The strong ag mutation, ag-1, leads to flowers in which stamens are converted to petals, and w4 is replaced by another flower reiterating the same pattern of whorled organs (ref. 33; Fig. 5G). ag-1 ufo-11 flowers are phenotypically indistinguishable from the ag-1 single mutant with w2 petals restored (Fig. 5H), indicating that ag is epistatic to ufo.
To confirm this epistasis, ufo-11 was crossed to the weak ag allele, ag-5 (34). ag-5 flowers are characterized by petaloid stamens in w3 and the replacement of w4 carpels with carpelloid sepals surrounding a new flower (Fig. 5I). The w4 organs are often fused. Consistent with the ag-1 ufo-11 result, ag-5 ufo-11 flowers have the same phenotype as the ag-5 single mutant (Fig. 5J).
These results provide two important insights into the w2 function of UFO. First, the existence of petals in both ag ufo double mutants further confirms that these “petalless” ufo alleles are not affecting petal formation per se. Second, the reappearance of petals in the double mutants indicates AG is essential for the loss of petals in the ufo mutants. The epistasis of ag over ufo is consistent with previous results indicating that AG can inhibit petal development (35, 36) and argues that UFO normally acts to antagonize AG activity in w2.
AG Expression in ufo-12 Flowers. The requirement of AG to block petal formation in ufo plants suggests that AG may be ectopically expressed in mutant w2 cells. To address this issue, AG expression in ufo-12 flowers was monitored by in situ hybridization using a probe lacking the MADS box encoding sequences and compared with wild type (Fig. 6). In wild-type flowers, AG transcripts are present in the region interior to the sepal primordia starting late in stage 2 and become clearly confined to the inner two whorls starting from approximately stage 6 (28). As petal initiation occurs during stage 5 (14), it was anticipated that ectopic AG expression might be detected in ufo w2 cells at that time. However, serial sections of either wild-type or ufo-12 flowers showed no clear evidence of AG transcripts in presumptive w2 cells at any point in development, including stage-5 flowers (Fig. 6). By stage 6, expression was clearly confined to the developing w3 and w4 organs (data not shown). These data argue that AG acts non-cell-autonomously to inhibit petal development in ufo mutants. Confirmation awaits tools to precisely identify and track both AG mRNA and/or protein in the two-celled petal anlagen (37).
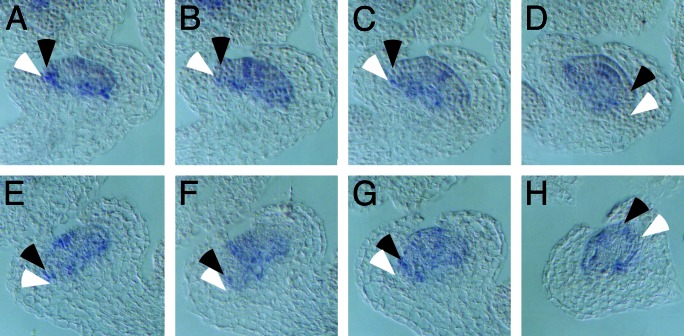
Fig. 6.
AG expression patterns in wild-type and ufo-12 flowers. Adjacent longitudinal sections from wild type (A–D) and ufo-12 (E–H) stage-5 flowers were analyzed by in situ hybridization with AG anti-sense RNA probes. Signal is dark blue. Black arrows indicate emerging stamen primordia, and white arrowheads denote the approximate region where petal primordia will arise. No AG transcripts are clearly visible in the w2 region of either wild-type or ufo-12 flowers. Transcripts are readily detected in both the developing third and fourth whorls.
Discussion
This work describes four new ufo alleles that define a role for UFO in promoting initiation and/or early proliferation of w2 organ primordia. The phenotypes of these mutants are distinct from those observed in strong ufo mutants where both w2 and w3 are severely affected, indicating that at least some UFO functions are separable. These “weak” alleles are unusual in resulting in the loss of w2 organs, whereas “strong” alleles appear to form some “w2” organs albeit with frequent homeotic transformations and aberrant positioning (4, 5).
A comparison of the ufo mutant analyses with its expression pattern suggests that UFO acts in at least three pathways at different times during floral development, consistent with recent studies using inducible UFO expression (38). Early, UFO likely functions in two pathways essential for w2 and w3 organ formation: one essential for the proper patterning of these primordia and the second required for their identity. The latter is at least partially involved in full activation of AP3 and PI expression, because coexpression of these genes from a heterologous promoter rescues some of the identity defects of a strong ufo mutant (8). Furthermore, ectopic expression of UFO causes B class gene activation in cells containing LFY (6, 7). Both pathways presumably depend on the UFO expression detected in middle whorls between stages 2–4, which coincides with the time AP3/PI expression patterns are established (11, 12). UFO mutations that interfere with these functions result in the strong phenotype.
After patterning and identity have been properly established, UFO is next required for initiating w2 primordia, correlating with the restriction of UFO expression to the petal primordia region beginning during stage 4–5, at or just before organ initiation (14). The weak ufo mutations identified here specifically affect this UFO activity, either by inhibiting expression of the gene or by interfering with some aspect of UFO protein function dedicated to this role. These mutants, therefore, specifically lack petals but are otherwise wild type, because the earlier UFO functions have already been executed. Note that this phenotype is virtually identical to the type IV flowers observed when UFO expression was prematurely terminated during development (38).
A working model of how w2 initiation/proliferation is regulated can be proposed (Fig. 7). Epistasis of ag-1 over weak ufo alleles suggests that petal initiation and/or proliferation is initially restrained through an AG-dependent inhibitory pathway. Consistent with this model, high-level ectopic AG expression blocks w2 development (35, 36), whereas petal cells accumulate more quickly in ag-1 than wild type (39). This model may also explain why w2 organs display a lag phase during their early development (14, 33).
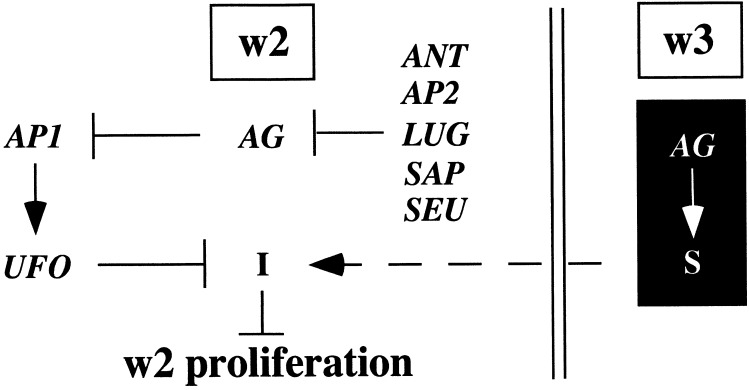
Fig. 7.
Model for mechanisms governing w2 proliferation. An AG-dependent inhibitory pathway (black box) regulates an unknown molecule (S) that can elicit a block in w2 proliferation. In wild type, S is produced in w3 (or w4) and then is transmitted to w2 (dashed line), where it activates an inhibitor (I). Note that the signaling molecule could be the inhibitor. Inhibition is overcome by two w2 promoting mechanisms. First, AP2, LUG, SAP, SEU, and ANT genes repress AG transcription in w2. Ectopic expression of AG antagonizes AP1 transcription (36, 44). Second, an AP1- and UFO-dependent pathway functions downstream of AG to overcome the action of I.
The w2 inhibitory pathway is opposed in wild type by two mechanisms. First, AG transcription is repressed through the activity of multiple factors, including AP2 (28), LEUNIG (40), STERILE APETALA (41), AINTEGUMENTA (42), and SEUSS (43). Mutations in these genes lead to varying w2 phenotypes, most of which appear to depend on AG. For example, strong ap2 mutations lead to loss of petals and display similar genetic characteristics as the weak ufo alleles with respect to w2 development. Like the ufo mutants, w2 organs are restored in ap2-2 ag-1 double mutants, although they are petaloid stamens (1). The w2 requirement for AP2 is also organ type independent, because ap2-2 ap3-1 or ap2-2 pi-1 flowers lack the w2 sepals normally found in either B class mutant (1).
We propose a second mechanism for w2 initiation/proliferation that involves a UFO-dependent mechanism. The expression patterns of the ufo-11 and ufo-12 alleles indicate this pathway is activated during stage 4/5, when UFO transcription normally becomes restricted to the incipient w2 region. Evidence suggests that the A class transcription factor, AP1, is a key upstream activator of UFO in these cells. First, strong ap1 mutants also lead to loss of petals, and genetic analysis suggests that the two genes function in the same pathway. Like the weak ufo alleles, w2 petals are restored in ap1 double mutants with ag-1 (44). AP1 is also generally required for w2 development, because double mutants with either ap3 or pi produce flowers lacking the w2 sepals found in the B class single mutants (45). Thus, both UFO and AP1 are involved in promoting w2 development of any organ type and are essential only in the presence of AG. Second, UFO expression from the CaMV 35S or AP3 promoter restores organ formation in ap1-1 flowers, indicating that UFO is downstream of AP1 with respect to w2 development (I. Lee and D.W., unpublished results). Third, w2-specific UFO expression depends on AP1. In ap1-1 flowers, like ufo-11 and ufo-12, UFO expression up to stage 4 is nearly normal but is lost in the petal region thereafter (6). This AP1-dependent expression is potentially a direct effect based on the identification of five possible AP1-binding sites in the putative w2 enhancer that are disrupted by the ufo-11 and ufo-12 T-DNA insertions (Fig. 2). Therefore, a primary function of AP1 in w2 may be to activate UFO during stage 4–5, which in turn positively regulates w2 initiation/proliferation.
What might be the nature of the AG-dependent inhibitor, and how does UFO overcome its activity? On the basis of our results in ufo-12 flowers, AG RNA is apparently not ectopically expressed in w2 cells, indicating that UFO is not involved in repressing AG transcription. Further, ectopic expression of UFO throughout the flower does not lead to an ag phenotype (6), implying that UFO likely does not act directly on AG. The non-cell autonomy of the AG effect therefore suggests that a signal is generated in w3 (and possibly w4) and transmitted to w2 cells where it inhibits w2 initiation/proliferation. Note that AG has been shown to act noncell autonomously in w2 as well as between cell layers in w3 and w4 (46, 47). Because AG is expressed from stage 2/3 onward, the inhibitor is potentially active in w2 up until stage 5 when UFO expression becomes restricted to the w2 region allowing primordia development to occur. If UFO acts through an SCFUFO complex, it presumably recognizes a protein(s) necessary for inhibition and targets it for ubiquitin-mediated degradation, as has been suggested for its activation of B class gene expression (23). Initial studies with ufo-13 and -14 suggest that recognition of this putative target requires amino acids 298–442 of the UFO C-terminal domain.
Second whorl initiation also depends on the PETAL LOSS (PTL) gene, mutants of which have characteristics in common with the weak ufo alleles; they lead specifically to a loss of w2 organs and affect w2 development in an organ-independent manner (48). However, in contrast to ufo, ag ptl double mutants have an additive phenotype, suggesting that their pathways do not intersect. Although this result implies that UFO and PTL function independently, a dominant modifier of ptl, pmd-1d, maps very close to UFO on chromosome 1 (48).
w2 initiation/proliferation provides a system to dissect how key organ identity genes are linked during development. A major role of the A class genes, AP1 and AP2, likely is to block the action of the antagonistic AG-dependent pathway and not to provide organ identity functions per se. AP1 likely accomplishes this role through activation of UFO and repression of AG. The w2 role of AP2 can largely be attributed to its repression of AG transcription (28). However, the w2 petaloid stamens that form in ap2-2 ag-1 flowers (1) suggest that AP2 may also contribute to w2 identity. Regardless, a key w2 function of the A and C class genes is apparently to provide a mechanism coordinating the timing of petal development. This leaves the B class genes, in concert with one or more of the SEPALLATA genes (49), as the major factors specifying petal identity.
Supplementary Material
Acknowledgments
We thank Steven Ruzin and the Center for Biological Imaging for excellent technical assistance and advice. We also thank the Meyerowitz laboratory (California Institute of Technology), Vivian Irish (Yale University, New Haven, CT), and John Bowman (University of California, Davis) for gifts of various reagents. This work was supported by Department of Energy Grant 88ER13882 and U.S. Department of Agriculture Grant 35304-09986 (to P.C.Z.) and by National Science Foundation Grant IBN-9723818 (to D.W.). T.D. was supported by National Institutes of Health Research Service Award Postdoctoral Fellowship GM16915.
Abbreviations: wn, whorl n; UFO, UNUSUAL FLORAL ORGANS; SAM, short apical meristem.
References
- 1.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112 1-20. [DOI] [PubMed] [Google Scholar]
- 2.Weigel, D. & Meyerowitz, E. M. (1994) Cell 78 203-209. [DOI] [PubMed] [Google Scholar]
- 3.Soltis, D. E., Soltis, P. S., Albert, V. A., Oppenheimer, D. G., dePamphilis, C. W., Ma, H., Frohlich, M. W. & Theissen, G. (2002) Trends Plant Sci. 7 22-31; discussion 31-34. [DOI] [PubMed] [Google Scholar]
- 4.Levin, J. Z. & Meyerowitz, E. M. (1995) Plant Cell 7 529-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson, M. D. & Haughn, G. W. (1995) Plant Cell 7 1485-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, I., Wolfe, D. S., Nilsson, O. & Weigel, D. (1997) Curr. Biol. 7 95-104. [DOI] [PubMed] [Google Scholar]
- 7.Parcy, F., Nilsson, O., Busch, M. A., Lee, I. & Weigel, D. (1998) Nature 395 561-566. [DOI] [PubMed] [Google Scholar]
- 8.Krizek, B. A. & Meyerowitz, E. M. (1996) Development (Cambridge, U.K.) 122 11-22. [DOI] [PubMed] [Google Scholar]
- 9.Long, J. A. & Barton, M. K. (1998) Development (Cambridge, U.K.) 125 3027-3035. [DOI] [PubMed] [Google Scholar]
- 10.Ingram, G. C., Goodrich, J., Wilkinson, M. D., Simon, R., Haughn, G. W. & Coen, E. S. (1995) Plant Cell 7 1501-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goto, K. & Meyerowitz, E. M. (1994) Genes Dev. 8 1548-1560. [DOI] [PubMed] [Google Scholar]
- 12.Jack, T., Brockman, L. L. & Meyerowitz, E. M. (1992) Cell 68 683-697. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J. P. & Lord, E. M. (1989) Can. J. Bot. 67 2922-2936. [Google Scholar]
- 14.Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. (1990) Plant Cell 2 755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86 263-274. [DOI] [PubMed] [Google Scholar]
- 16.Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K. & Matsui, M. (2002) Plant Cell Physiol. 43 1073-1085. [DOI] [PubMed] [Google Scholar]
- 17.Pouteau, S., Nicholls, D., Tooke, F., Coen, E. & Battey, N. (1998) Plant J. 14 235-246. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, S., Sandal, N., Polowick, P. L., Stiller, J., Stougaard, J. & Fobert, P. R. (2003) Plant J. 33 607-619. [DOI] [PubMed] [Google Scholar]
- 19.Patton, E. E., Willems, A. R. & Tyers, M. (1998) Trends Genet. 14 236-243. [DOI] [PubMed] [Google Scholar]
- 20.Ingram, G. C., Doyle, S., Carpenter, R., Schultz, E. A., Simon, R. & Coen, E. S. (1997) EMBO J. 16 6521-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samach, A., Klenz, J. E., Kohalmi, S. E., Risseeuw, E., Haughn, G. W. & Crosby, W. L. (1999) Plant J. 20 433-445. [DOI] [PubMed] [Google Scholar]
- 22.Zhao, D., Yang, M., Solava, J. & Ma, H. (1999) Dev. Genet. 25 209-223. [DOI] [PubMed] [Google Scholar]
- 23.Zhao, D., Yu, Q., Chen, M. & Ma, H. (2001) Development (Cambridge, U.K.) 128 2735-2746. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann, K. (1991) Plant J. 1 71-82. [Google Scholar]
- 25.Sessions, R. A. (1997) Am. J. Bot. 84 1179-1191. [PubMed] [Google Scholar]
- 26.Drews, G. N. (1995) Arabidopsis Molecular Genetics Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 27.Irish, V. F. & Yamamoto, Y. T. (1995) Plant Cell 7 1635-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drews, G. N., Bowman, J. L. & Meyerowitz, E. M. (1991) Cell 65 991-1002. [DOI] [PubMed] [Google Scholar]
- 29.Durfee, T., Becherer, K., Chen, P. L., Yeh, S. H., Yang, Y., Kilburn, A. E., Lee, W. H. & Elledge, S. J. (1993) Genes Dev. 7 555-569. [DOI] [PubMed] [Google Scholar]
- 30.Schultz, E. A. & Haughn, G. W. (1991) Plant Cell 3 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huala, E. & Sussex, I. M. (1992) Plant Cell 4 901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F. & Meyerowitz, E. M. (1992) Cell 69 843-859. [DOI] [PubMed] [Google Scholar]
- 33.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1989) Plant Cell 1 37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roe, J. L., Nemhauser, J. L. & Zambryski, P. C. (1997) Plant Cell 9 335-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizukami, Y. & Ma, H. (1992) Cell 71 119-131. [DOI] [PubMed] [Google Scholar]
- 36.Jack, T., Sieburth, L. & Meyerowitz, E. (1997) Plant J. 11 825-839. [DOI] [PubMed] [Google Scholar]
- 37.Bossinger, G. & Smyth, D. R. (1996) Development (Cambridge, U.K.) 122 1093-1102. [DOI] [PubMed] [Google Scholar]
- 38.Laufs, P., Coen, E., Kronenberger, J., Traas, J. & Doonan, J. (2003) Development (Cambridge, U.K.) 130 785-796. [DOI] [PubMed] [Google Scholar]
- 39.Crone, W. & Lord, E. M. (1994) Can. J. Bot. 72 384-401. [Google Scholar]
- 40.Liu, Z. & Meyerowitz, E. M. (1995) Development (Cambridge, U.K.) 121 975-991. [DOI] [PubMed] [Google Scholar]
- 41.Byzova, M. V., Franken, J., Aarts, M. G., de Almeida-Engler, J., Engler, G., Mariani, C., Van Lookeren Campagne, M. M. & Angenent, G. C. (1999) Genes Dev. 13 1002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krizek, B. A., Prost, V. & Macias, A. (2000) Plant Cell 12 1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franks, R. G., Wang, C., Levin, J. Z. & Liu, Z. (2002) Development (Cambridge, U.K.) 129 253-263. [DOI] [PubMed] [Google Scholar]
- 44.Gustafson-Brown, C., Savidge, B. & Yanofsky, M. F. (1994) Cell 76 131-143. [DOI] [PubMed] [Google Scholar]
- 45.Irish, V. F. & Sussex, I. M. (1990) Plant Cell 2 741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenik, P. D. & Irish, V. F. (2000) Development (Cambridge, U.K.) 127 1267-1276. [DOI] [PubMed] [Google Scholar]
- 47.Sieburth, L. E., Drews, G. N. & Meyerowitz, E. M. (1998) Development (Cambridge, U.K.) 125 4303-4312. [DOI] [PubMed] [Google Scholar]
- 48.Griffith, M. E., da Silva Conceicao, A. & Smyth, D. R. (1999) Development (Cambridge, U.K.) 126 5635-5644. [DOI] [PubMed] [Google Scholar]
- 49.Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E. & Yanofsky, M. F. (2000) Nature 405 200-203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.