Abstract
To reveal the structural principles determining substrate specificity of 4-coumarate:CoA ligase (4CL), the crystal structure of the phenylalanine activation domain of gramicidin S synthetase was used as a template for homology modeling. According to our model, 12 amino acid residues lining the Arabidopsis 4CL isoform 2 (At4CL2) substrate binding pocket (SBP) function as a signature motif generally determining 4CL substrate specificity. We used this substrate specificity code to create At4CL2 gain-of-function mutants. By increasing the space within the SBP we generated ferulic- and sinapic acid-activating At4CL2 variants. Increasing the hydrophobicity of the SBP resulted in At4CL2 variants with strongly enhanced conversion of cinnamic acid. These enzyme variants are suitable tools for investigating and influencing metabolic channeling mediated by 4CL. Knowledge of the 4CL specificity code will facilitate the prediction of substrate preference of numerous, still uncharacterized 4CL-like proteins.
The 4-coumarate:CoA ligase (4CL, EC 6.2.1.12) represents the branch point enzyme of the general phenylpropanoid pathway, which channels carbon flow from primary metabolism to different branch pathways of plant secondary metabolism (1). Typically, 4CLs catalyze the conversion of the three cinnamic acid derivatives 4-coumaric acid (4-hydroxycinnamic acid), caffeic acid (3,4-dihydroxycinnamic acid), and ferulic acid (3-methoxy-4-hydroxycinnamic acid) to their corresponding CoA esters in a two-step reaction. This catalytic process is characterized by the formation of a coumaroyl-adenylate intermediate, which is subsequently converted to the corresponding CoA ester. The activated phenolic acids serve as precursors for the biosynthesis of numerous plant secondary products such as flavonoids, isoflavonoids, coumarins, lignin, suberin, and wall-bound phenolics (1, 2). These compounds are important for plant growth and development by providing mechanical support and rigidity to cell walls, attracting insects for pollination, or protecting against biotic and abiotic stresses. In addition, several of these secondary plant products have been suggested to have a health-promoting function in human nutrition (3). Recently, the in vitro synthesis of various monoadenosine and diadenosine polyphosphates in the absence of CoA has been observed as an additional 4CL-catalyzed reaction (4). However, the biological significance of this second 4CL-associated reaction is still unknown, although the accumulation of (di)adenosine polyphosphates has been correlated with stress responses in bacteria and fungi and cell differentiation and apoptosis in cultured mammalian cells (5, 6).
The presence of a highly conserved putative AMP-binding domain signature has been used as the most important criterion to group enzymes such as 4CLs, firefly luciferases, acetyl-CoA synthetases, fatty acyl-CoA synthetases, and nonribosomal polypeptide synthetases in one superfamily of adenylate-forming enzymes (7). In good agreement with this theoretical classification, mutational analysis of the Arabidopsis 4CL isoform 2 (At4CL2) corroborated a close functional relationship between 4CL and the adenylation and substrate recognition domains of nonribosomal peptide synthetases (8). For three members of the above-described superfamily of adenylate-forming enzymes structural information derived from x-ray analysis of protein crystals is available. Firefly luciferase crystals were obtained in the absence of its substrate luciferin, whereas two bacterial proteins, the phenylalanine-activating domain (PheA) of gramicidin S synthetase and the 2,3-dihydroxybenzoic acid-activating enzyme DhbE were cocrystallized with ATP and their specific substrates phenylalanine and 2,3-dihydroxybenzoate, respectively (9–11). Despite significant differences in their primary sequences, all three enzymes exhibit similar overall folding patterns with a large N-terminal and a small C-terminal domain. For PheA, 10 amino acid residues lining the phenylalanine binding pocket have been identified. With the exception of Lys-517, which functions as the catalytic residue in adenylate formation and is localized in the small C-terminal domain, all of these amino acids are restricted to a 100-aa-residue-comprising region of the large N-terminal protein domain. This region is flanked by the conserved core motifs A3 and A6 of peptide synthetases, which correspond to the conserved box I and box II motifs of 4CLs (12).
4CL isoforms with distinct substrate conversion profiles have been reported for several plants, including soybean, petunia, pea, and Arabidopsis (13–17). We recently characterized three 4CL isoforms from Arabidopsis thaliana and demonstrated that At4CL1 and At4CL3 exhibit typical substrate utilization profiles with high conversion rates of coumaric acid, caffeic acid, and ferulic acid. In contrast, At4CL2 is unusual in that its preferred substrate is caffeic acid rather than coumaric acid, whereas the structurally related ferulic acid is not converted. Cinnamic acid is a poor substrate for all three At4CLs and sinapic acid (3,5-methoxy-4-hydroxycinnamic acid) is not activated at all (17). Because of At4CL2's unique biochemical properties and exceptional enzymatic stability, we and others have chosen it to investigate the structural principles generally determining 4CL substrate specificity (12, 18). In particular, the lack of appreciable ferulic acid and sinapic acid conversion and the very low activity with cinnamic acid obviously facilitate gain-of-function experiments, thereby making At4CL2 a well-suited experimental system. In soybean it was recently demonstrated that deletion of a single amino acid residue resulted in the generation of sinapic acid-converting 4CLs (19). However, a conclusive model explaining 4CL substrate specificity determination is not available.
Based on structural data obtained by homology modeling we postulate the existence of a 4CL signature motif consisting of 12 aa lining the substrate binding pocket (SBP), which may generally determine 4CL substrate specificity. Here we provide experimental evidence to substantiate this concept. By using the proposed specificity code as the basis for rational enzyme design we were able to create, in addition to recently described ferulic acid-activating At4CL2 variants, enzymes capable of converting sinapic acid and cinnamic acid.
Methods
Mutagenesis of At4CL2. The pQE30-based At4CL2 expression plasmid that was used for mutagenesis has been described (12). Point mutations were introduced into At4CL2 by PCR-based amplification of the entire At4CL2 expression plasmid by using two mutated oligonucleotide primers, each complementary to opposite strands of the vector. Conditions for PCR-based mutagenesis were as follows: 1 cycle at 94°C (1 min), 16 cycles at 94°C (1 min), 59°C (1 min), 68°C (14 min), and one final polymerization step at 68°C (14 min). All components necessary for this mutagenesis procedure were included in the commercial QuikChange kit (Stratagene). Double and triple mutations were introduced in the same way by using already existing mutants as template.
Domain Exchange Experiments. To serve as template for domain exchange experiments, the At4CL2 reading frame was modified by introduction of a StuI and BglII recognition site into the DNA segment encoding box I and box II, respectively. Point mutations were introduced as described above. One primer sequence of each pair of complementary mutagenesis primers is listed here (5′-CCTTTCTCATCCGGCACGACAGGCCTCCCCAAAGGAGTGATGC-3′; 5′-GCATCACTCCTTTGGGGAGGCCTGTCGTGCCGGATGAGAAAG-3′). For substitution of the At4CL2 SBP by the SBP of gramicidin synthetase S (PheA), genomic DNA of Bacillus brevis was isolated according to the method described for Brevibacterium ammoniagenes (20). Subsequently, the PheA SBP was PCR-amplified by using the primer pair PheAStuIB and PheABglIIB (5′-GTACAACAGGCCTTCCAAAAGGTACAATGCTG-3′;5′-ACCAATACAGATCTCACCAGCTTCACCAACCG-3′). During this PCR amplification the StuI and BglII sites required for domain exchange were introduced. For substitution of the At4CL2 SBP by the SBP of the Arabidopsis gene At3g21230, total RNA of Arabidopsis Col0 was isolated by using the Qiagen RNA DNA maxi-kit (Qiagen, Hilden, Germany). Synthesis of cDNA and subsequent PCR amplification of the full-length At3g21230 reading frame was performed with the Titan one tube RT-PCR kit (Roche Diagnostics) according to the manufacturer's instructions with the primer pair c112 and c118 (5′-CACACGCATGCTGCTCCAACAACAAACGCA-3′; 5′-GAAACTCTCTTGTGTCTATTTAGAGC-3′). The resulting full-length cDNA was used as template for PCR amplification of the SBP encoding region using the primer pair 4CL2-likeA and 4CL2-likeB (5′-GGAACAACAGGCCTTCCAAAGGGAGTG-3′; 5′-CCTCGGACGCAGATCTCGCCAG-3′). During this second round of PCR amplification the StuI and BglII sites required for the domain exchange were introduced.
Purification of At4CL Proteins and Enzyme Assays. Expression and purification of At4CL2 proteins was performed as described (8). 4CL activity was determined with the spectrophotometric assay as described, using cinnamic acid, coumaric acid, caffeic acid, ferulic acid, and sinapic acid as phenolic substrates (17, 21).
Homology Modeling of At4CL2. The 3D model of At4CL2 comprising all nonhydrogen atoms was generated by using the MODELLER 4 program package. This program is based on a distance restraint algorithm with spatial restrains extracted from the alignment of the target sequence with the template structure and from the CHARMM-22 force field (22, 23). The thereby obtained model was subjected to a short simulated annealing refinement protocol available in the MODELLER program package. The stereochemical quality of the model was evaluated by using PROCHECK (24). Superposition of the model before the restrained optimization with default parameters (implemented in the MODELLER program package) and the final model after molecular dynamics and energy minimization revealed no significant change in the overall protein structure (rms deviation of 0.51 Å for all Cα atoms), indicating that the model is near the energetic minimum.
Phylogenetic Analysis of At4CL-Like Proteins. The Munich Information Center for Protein Sequences A. thaliana database (MatDB) was searched with the FASTA algorithm for 4CL-like proteins by using the three bona fide At4CLs (17). A multiple sequence alignment was generated with the PILEUP program of the GCG program package, version 10.2 (25), which was restricted to proteins sharing at least 25% sequence identity. Based on this alignment, a maximum parsimony analysis was performed (26) by using the PAUP 3.1.1 program (Smithsonian Institution, Washington, DC). The most parsimonious tree was found by using the heuristic search option with the tree bisection reconnection branch-swapping algorithm (27). For statistical analysis, 500 bootstrap replications (28) were analyzed.
Results
Design of a 3D Model of the At4CL2 SBP. As template for homology modeling of At4CL2 we chose the PheA of gramicidin S synthetase (10). The crystal structure of PheA has been determined in complex with its substrates, ATP and l-phenylalanine at a high resolution of 1.9 Å (10). We assumed that PheA represents a well-suited template for modeling the structure of 4CL, because of the established functional relatedness of the enzymes (8) and the obvious structural similarity of their respective substrates, phenylalanine and coumaric acid. Based on a recently published sequence alignment (12), the 3D structure of At4CL2 was calculated by using the structural prediction program MODELLER 4 with the standard settings.
The predicted fold of At4CL2 consists of a large N-terminal domain and a small C-terminal domain. The SBP of At4CL2 is formed mainly by amino acid residues of the large N-terminal protein domain. Only the catalytic Lys-540, which is known to stabilize the transition state during adenylate formation, is part of the C-terminal domain. From previous work it is known that the At4CL2 WT protein and the At4CL2 double mutant M293P+K320L differ drastically in their substrate preferences (12). Whereas the At4CL2 WT enzyme readily converts caffeic acid and is almost inactive toward ferulic acid, the latter substrate is efficiently activated by the double mutant (Table 1). It has been suggested that a size exclusion mechanism is responsible for the inactivity of At4CL2 WT toward ferulic acid, i.e., steric hindrance between the bulky residues Met-293 and Lys-320, and the 3-methoxy group of ferulic acid prevents correct binding of this substrate in the SBP (12).
Table 1. Kinetic properties of At4CL2 variants designed to activate sinapic acid.
|
Coumaric acid
|
Caffeic acid
|
Ferulic acid
|
Sinapic acid
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At4CL2 variant | Km, μM | Vmax, nkat/mg | SA, nkat/mg | Km, μM | Vmax, nkat/mg | SA, nkat/mg | Km, μM | Vmax, nkat/mg | SA, nkat/mg | Km, μM | Vmax, nkat/mg | SA nkat/mg |
| WT | 233 ± 15 | 475 ± 94 | 249 ± 26 | 22 ± 5 | 236 ± 30 | 199 ± 32 | n.d. | n.d. | 1.8 ± 0.2 | n.c. | n.c. | n.c. |
| PL (M293P + K320L) | 22 ± 8 | 321 ± 46 | 276 ± 44 | 41 ± 8 | 267 ± 91 | 218 ± 64 | 30 ± 7 | 247 ± 37 | 210 ± 35 | n.c. | n.c. | n.c. |
| PL + ΔV355 | 47 ± 6 | 187 ± 42 | 146 ± 28 | 75 ± 20 | 111 ± 3 | 68 ± 21 | 77 ± 7.9 | 104 ± 13 | 79 ± 10 | 382 ± 72 | 45 ± 9 | 22 ± 7 |
| PL + ΔL356 | 33 ± 3 | 142 ± 33 | 119 ± 26 | 109 ± 39 | 95 ± 15 | 60 ± 5 | 55 ± 1.7 | 78 ± 8 | 64 ± 3 | 168 ± 16 | 31 ± 2 | 17 ± 2 |
| PL + ΔV355 + ΔL356 | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. | n.c. |
| WT + ΔV355 | 111 ± 24 | 41 ± 6 | 28 ± 2 | 75 ± 8 | 80 ± 19 | 58 ± 14 | n.d. | n.d. | 5 ± 1 | n.c. | n.c. | n.c. |
SA, specific activity with 200 μM phenolic substrate; n.c., no conversion; n.d., not determinable, enzyme activity too low
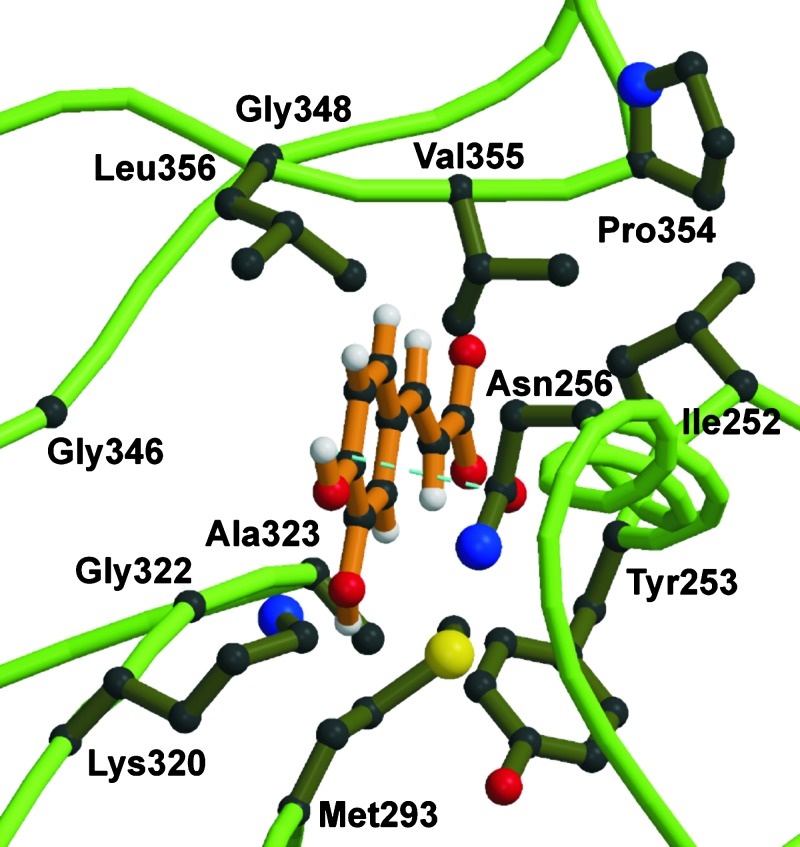
These biochemical data were essential for refinement of our model. Although the PheA structure has been determined in complex with its substrate phenylalanine, it was a critical step to predict the precise orientation of caffeic acid within the At4CL2 SBP as cinnamic acid derivatives are planar and more rigid than phenylalanine. We therefore chose two parameters for positioning the substrate within the 4CL SBP. On the one hand, the carboxyl group of the respective cinnamic acid derivative should be located in a distance of ≈3 Å to the ε-amino group of the catalytic residue Lys-540 to facilitate adenylate formation. For PheA a similar distance (3.04 Å) has been determined between the carboxyl group of phenylalanine bound in the SBP and the corresponding lysine residue (Lys-517) of the active center (10). On the other hand, insertion of ferulic acid into the SBP of the At4CL2 WT enzyme should provoke a steric conflict between the 3-methoxy group of this substrate and the amino acid residues Met-293 and Lys-320. The model depicted in Figs. 1 and 2 fulfills both criteria and is therefore in accordance with the available experimental data.
Fig. 1.
3D model of the At4CL2 SBP. The 3D structure of At4CL2 was calculated with the program package modeller 4 and visualized by using molscript. The presentation is restricted to the SBP harboring the substrate molecule caffeic acid. The 12 amino acid residues located at a distance of not more than 6 Å from the center of caffeic acid are shown. The catalytic lysine residue 540, which interacts with the carboxyl group of the substrate molecule, is not included in this presentation. The dotted line represents a hydrogen bond.
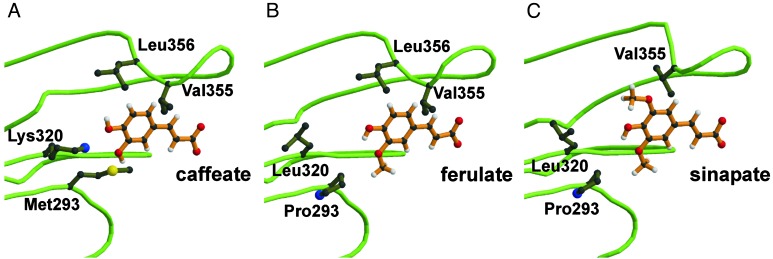
Fig. 2.
3D models of SBPs of different At4CL2 variants. For each enzyme variant the bulkiest substrate that can be fitted into the respective SBP is depicted. In comparison to Fig. 1, the viewpoint has been changed for a better visualization of the respective substrate molecules. In addition, the complexity of the presentation has been reduced by focusing on four amino acid residues of special consideration. (A) SBP of WT enzyme with caffeic acid. (B) SBP of double mutant M293P+K320L with ferulic acid. (C) SBP of triple mutant M293P+K320L+ΔL356 with sinapic acid.
According to the derived model, 12 amino acid residues of the large N-terminal protein domain are located at a distance of not >6 Å from the center of caffeic acid fitted into the At4CL2 SBP. The 6-Å distance was chosen for the identification of SBP constituents as this distance includes all putatively formed hydrogen bonds, all van der Waals interactions, and most ionic interactions between SBP lining amino acid residues and caffeic acid. We therefore propose that the 12 amino acid residues mentioned above form the substrate specificity code of 4CL. In At4CL2 this code comprises the residues Ile-252, Tyr-253, Asn-256, Met-293, Lys-320, Gly-322, Ala-323, Gly-346, Gly-348, Pro-354, Val-355, and Leu-356 (Fig. 3). According to our model, the oxygen atom of the amide group of Asn-256 is located at a distance of 3.1 Å from the hydrogen atom of the 4-hydroxyl group of caffeic acid. This finding indicates that a hydrogen bond stabilizes the orientation of the substrate within the SBP (Fig. 1). The residues Met-293 and Lys-320 form a kind of clamp around the 3-hydroxyl group of caffeate. On the opposite side of the SBP, a comparable structure is formed by the residues Val-355 and Leu-356. In fact, the latter residues are in such close proximity to the phenyl ring that they would spatially interfere with potential substituents at ring position 5. In Fig. 2 A and B the modeled SBPs of the At4CL2 WT enzyme and the At4CL2 mutant M293P+K320L are represented together with their substrates, caffeic acid and ferulic acid, respectively. In this presentation of reduced complexity only the protein backbone and four amino acid residues of special consideration are shown.
Fig. 3.
Amino acid sequence comparison of different 4CLs and a 4CL-like protein. The presentation is reduced to the region flanked by the conserved peptide motifs box I and box II (highlighted in yellow). The 12 amino acid residues proposed to function as the 4CL substrate specificity code are marked in red. Residues that were deleted from At4CL2 to create sinapic acid-converting enzymes are marked by an *. Gm4CL1, 4CL isoform 1 from G. max; At3g21230, predicted amino acid sequence of the corresponding Arabidopsis gene.
Model-Based Design of Sinapic Acid-Converting At4CL2 Variants. To evaluate the reliability of our model of the 4CL SBP, we set out to create a sinapic acid-converting At4CL2 variant. When we initiated this approach, sequence information of sinapic acid-converting 4CL isoforms was not available, although corresponding enzyme activities have been reported for several plants (13–15, 29). Because sinapic acid carries two methoxyl groups, in the three and five positions, we supposed that again a size exclusion mechanism is responsible for the incapacity of At4CL2 to activate this substrate. According to our model, cinnamic acid derivatives are positioned in the At4CL2 SBP in a strictly orientated manner. This finding led us to three conclusions: (i) successful design of a sinapic acid-converting At4CL2 variant should be initiated with the ferulic acid-activating mutant M293P+K320L as template for mutagenesis; (ii) the region for modification is defined by the geometry of both SBP and substrate and should include amino acid residues Val-355 and Leu-356; and (iii) substitution of Val-355 or Leu-356 by smaller residues appeared as a promising approach to create space for the large substrate sinapic acid (Fig. 2B). However, substitution of Val-355 and/or Leu-356 by alanine residues in the At4CL2 double mutant background did not result in a sinapic acid-converting enzyme. In fact, the L356A substitution did not significantly influence the conversion rates of coumaric acid, caffeic acid, or ferulic acid, whereas the V355A substitution and the double substitution L356A+V355A both resulted in enzymes with generally reduced activity (data not shown).
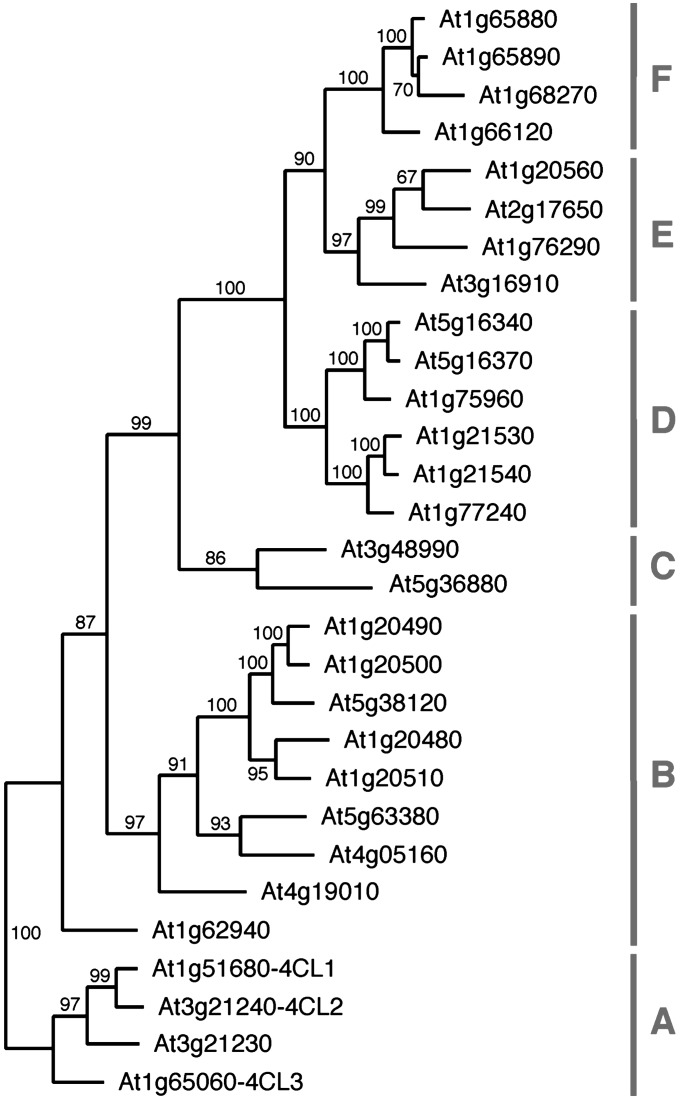
These results forced us to re-evaluate our structural model by analysis of enzymes that are capable of activating sinapic acid. To get access to that kind of protein we devised a domain substitution approach, which is based on the observation that all SBP-forming amino acid residues of 4CLs, with the exception of Lys-540, are encoded in a defined region flanked by the conserved sequence motifs box I and box II (Fig. 3). We modified both box I- and box II-encoding DNA segments of At4CL2 such that they harbor unique restriction sites, thereby allowing substitution of the intervening region by DNA fragments amplified from different sources using corresponding box I and box II primers. As sources for alternative SBPs we selected gramicidin S synthetase (PheA) and the Arabidopsis gene At3g21230 encoding a 4CL-like protein, which in a phylogenetic reconstruction grouped together with the three known Arabidopsis 4CL isoforms (Fig. 4). Although the At4CL2 variant carrying the SBP from PheA showed no conversion of any cinnamic acid derivative, the chimeric protein consisting of the At4CL2 backbone and the SBP derived from the gene At3g21230 was indeed capable of activating sinapic acid with a Km of 136 ± 55 μM and a Vmax of 12 ± 5 nkat/mg. Evaluation of the enzymatic properties of the complete protein encoded by At3g21230 uncovered that it represents a novel Arabidopsis 4CL isoform that efficiently converts sinapic acid (Km 23 ± 8 μM, Vmax 83 ± 13 nkat/mg).
Fig. 4.
Phylogenetic relationship of At4CLs and At4CL-like proteins. Based on the amino acid sequence alignment that was generated with the pileup program, the most parsimonious tree was found by using the heuristic search algorithm with the program paup. Bootstrap values (500 replicates) for each clade are shown at the branches. The tree has a consistency index of 0.61. Capital letters and bars define groups of related proteins.
An amino acid sequence alignment of the SBPs encoded by At3g21230, a recently described naturally occurring sinapic acid-converting 4CL isoform from Glycine max (16), and the three Arabidopsis 4CLs that are inactive toward sinapic acid, revealed that the region specifying sinapic acid conversion involves the amino acid residues Val-355 and Leu-356, as predicted by our model (Fig. 1). However, in contrast to our initial strategy to reduce the size of these residues by substitution, the deletion of either Val-355 or Leu-356 appeared as a promising approach for generating a sinapic acid-converting enzyme (Fig. 3). To experimentally verify this conclusion, we deleted Val-355, Leu-356, or both by using the At4CL2 double mutant M293P+K320L as template. Both single amino acid deletions, ΔV355 and ΔL356, generated enzymes that retained activity against coumaric, caffeic, and ferulic acid, and as a new activity acquired the capacity to activate sinapic acid (Table 1). In contrast, the deletion of both amino acids resulted in a completely inactive enzyme (Table 1).
As stated above, our structural model predicts a strictly defined orientation of cinnamic acid derivatives within the 4CL SBP. Correspondingly, deletion of Val-355 in the background of the At4CL2 WT enzyme did not significantly improve ferulic acid activation nor allow sinapic acid conversion (Table 1).
Design of At4CL2 Variants with Improved Cinnamic Acid Conversion Rates. Unsubstituted cinnamic acid is a generally poor substrate of 4CL, although its conversion by different 4CL isoforms varies considerably (16, 17). Because cinnamic acid is the smallest and most unpolar 4CL substrate, we assumed that not size restrictions but rather the overall hydrophobicity of the At4CL2 SBP is important for the efficiency of cinnamic acid conversion. Inspection of the proposed substrate specificity motif of At4CL2 revealed that it contains one positively charged (Lys-320), two polar (Tyr-253, Asn-256), and nine hydrophobic amino acid residues (Fig. 3). Obviously, the most promising approach for improving cinnamic acid conversion by At4CL2 seemed to be the replacement of Lys-320 by a hydrophobic amino acid residue. Indeed, the mutant enzyme carrying the K320L substitution showed a reduced Km for cinnamic acid (1,010 μM) in comparison to At4CL2 WT enzyme (6,642 μM), whereas the Vmax values were not significantly different (Table 2). Introduction of bulky, aromatic phenylalanine residues at positions 293 and 320, which are known to control the usage of 3-methoxylated 4-hydroxycinnamic acid derivatives, resulted in an enzyme variant with a capacity to activate cinnamic acid comparable to the mutant K320L (Table 2). In contrast to its enhanced activity with cinnamic acid, the capacity of the double mutant M293F+K320F for activating the 3-hydroxylated substrate caffeic acid was strongly reduced. This observation again indicates that hydrophobicity of the SBP is the most important criterion regulating cinnamic acid conversion, whereas size exclusion is the predominant mechanism controlling the conversion of substituted cinnamic acid derivatives. However, both modes of substrate selectivity cannot strictly be separated, as is obvious from the caffeic acid conversion rates of the mutant K320L (Table 2). The K320L substitution has a moderately negative influence on caffeic acid activation, arguing against substrate selectivity being solely based on size exclusion. The 4-fold improvement of cinnamic acid conversion observed with the double mutant M293P+K320L (Vmax/Km = 0.66), in comparison to K320L (Vmax/Km = 0.15), was rather unexpected and therefore demonstrates the limitation of theoretical predictions. Because our model of the At4CL2 SBP indicates that the asparagine residue at position 256 forms a hydrogen bond with the 4-hydroxy group of caffeic acid (Fig. 1), we suspected that this polar amino acid residue contributes to substrate specificity either by negative interference with the hydrophobic substrate cinnamic acid or by positive selection of 4-hydroxylated cinnamic acid derivatives. By introducing a N256A substitution into the M293P+K320L double mutant background, we were in fact able to design an enzyme with moderately enhanced cinnamic acid conversion rates but an 11-fold reduced activation of caffeic acid (Table 2). This observation clearly indicates that positive selection of 4-hydroxylated cinnamic acid derivatives by a polar residue at position 256 has a significant influence on substrate specificity determination. In conclusion, targeted substitution of three selected amino acid residues of the At4CL2 substrate specificity code resulted in a 30-fold improved conversion of cinnamic acid by the triple mutant N256A+M293P+K320L (Vmax/Km = 0.9) in comparison to the At4CL2 WT enzyme (Vmax/Km = 0.03).
Table 2. Kinetic properties of cinnamic acid-activating At4CL2 variants.
|
Cinnamic acid
|
Caffeic acid
|
|||
|---|---|---|---|---|
| At4CL2 variant | Km, μM | Vmax, nkat/mg | Km, μM | Vmax, nkat/mg |
| WT | 6,642±972 | 203±23 | 22±5 | 236±30 |
| K320L | 1,010±135 | 150±19 | 62±11 | 188±27 |
| M293F + K320F | 930±83 | 113±33 | 361±188 | 90±20 |
| PL (M293P + K320L) | 286±13 | 190±44 | 41±8 | 267±91 |
| PL + N256A | 163±37 | 145±21 | 166±12 | 93±17 |
Discussion
According to the 3D model of the At4CL2 SBP, 12 amino acid residues form a signature motif determining 4CL substrate specificity. In fact, targeted substitution or deletion of five selected residues of the substrate specificity code allows the design of ferulic acid-, sinapic acid-, and cinnamic acid-activating At4CL2 variants. As previously reported, ferulic acid activation by At4CL2 depends on the presence of a small amino acid residue at either position 293 or 320, whereas bulky residues at both positions prevent ferulic acid activation by steric interference with its 3-methoxy group (12). The structural model presented in this article additionally indicates that correct orientation of sinapic acid in the At4CL2 SBP is prevented by the amino acid residues Val-355 and Leu-356, which interfere with the 5-methoxy group of the substrate. Although the importance of both amino acids has been correctly predicted, it was not directly obvious that deletion rather than substitution of Val-355 or Leu-356 was a suited strategy to create gain-of-function mutants, but was inferred from recently identified sinapate-converting 4CL isoforms from G. max and Arabidopsis (ref. 16 and this article), which harbor deletions at the respective positions. Modeling the SBPs for both deletion variants shows that a conformational change of the loop, which significantly reduces the available space within the WT At4CL2 SBP, alleviates the steric hindrance for the 5-methoxy group of sinapic acid (Fig. 2C). Experimental evidence that one of these amino acids can influence substrate discrimination by 4CL was provided by Lindermayr et al. (19), who demonstrated that deletion of a single valine residue (corresponding to V355 in At4CL2) from two of the soybean 4CL isoforms, Gm4CL2 and Gm4CL3, generated enzymes with the capacity to activate sinapic acid.
We demonstrated that accessibility of the At4CL2 SBP for monomethoxylated and dimethoxylated cinnamic acid derivatives is regulated mainly by size exclusion, whereas cinnamic acid conversion is controlled by the overall hydrophobicity of the At4CL2 SBP. The capacity of naturally occurring 4CLs to activate cinnamic acid can also be correlated with the hydrophobicity of their SBPs. The signature motif of At4CL2 consists of one charged (Lys-320), two polar (Tyr-253, Asn-256), and nine hydrophobic amino acid residues, whereas the Gm4CL4 substrate specificity motif contains no charged, one polar, and 11 unpolar residues (16, 17). Correspondingly, both enzymes differ substantially in their capacity to activate cinnamic acid with Km values of 6,642 μM for At4CL2 and 260 μM for Gm4CL4 (Table 2 and ref. 16). Our model predicts that Asn-256 in At4CL2, which is highly conserved in all 4CLs, interacts with the 4-hydroxy group of coumaric acid and its derivatives by forming a hydrogen bond (Fig. 1). Data from the peptide synthetase system support this interpretation. Adenylation domains of nonribosomal peptide synthetases responsible for the activation of phenylalanine carry a hydrophobic residue at the position corresponding to Asn-256 in At4CL2, whereas adenylation domains that activate tyrosine carry an amino acid residue capable of forming a hydrogen bond (30). Based on these data, we postulate that a bona fide cinnamate:CoA ligase should carry a hydrophobic residue at the position corresponding to Asn-256 in At4CL2.
In the Arabidopsis genome >25 genes can be identified that encode proteins of similar length and at least 25% sequence identity to the three bona fide 4CLs (Fig. 4). In addition, ≈200 genes code for proteins containing the so-called AMP-binding domain. The assumed function of all of these proteins is the formation of specific adenylate intermediates, which subsequently might be converted to their corresponding CoA esters or alternative yet unknown end products. For example, the Arabidopsis Jar1 gene, which originally has been suggested to encode a regulatory protein of jasmonic acid-dependent stress responses, was recently cloned (31). JAR1 contains an AMP-binding domain, structurally resembles firefly luciferase, and correspondingly was demonstrated to support the formation of an adenylate intermediate when incubated with jasmonic acid and ATP. However, the end product remains unknown (31). For one of the 4CL-like proteins, encoded by the gene At3g21230, we demonstrated that it represents a yet uncharacterized bona fide Arabidopsis 4CL with the capacity to activate sinapic acid. In contrast, the biochemical function of the remaining 4CL-like proteins is still unknown.
Putative substrates of 4CL-like proteins comprise a wide range of organic acids such as amino acids, cinnamic acids, benzoic acids, and all kinds of fatty acids. To identify specific protein functions it would be desirable to be able to predict at least the class of substrate converted by a particular enzyme. We expect that knowledge of the 4CL specificity code allows such kind of prediction. Comparative analysis of PheA, the 2,3-dihydroxybenzoic acid-activating enzyme DhbE, and At4CL2 indicates that the amino acid residue corresponding to Asp-235 in PheA, Ile-252 in At4CL2, and Asn-235 in DhbE participates in discrimination between different substrate classes. In peptide synthetases, the acidic residue stabilizes the orientation of the substrate in the SBP by ionic interaction with its α-amino group (10). In bacterial 2,3-dihydroxybenzoate:CoA-ligases and salicylate:CoA-ligases Asn-235 fulfills a comparable function by forming a hydrogen bond with either the 3-hydroxy or 4-hydroxy group of the respective benzoic acid derivative (11). None of the 4CL-like proteins carries an acidic residue at the position corresponding to Ile-252 in At4CL2 and therefore it is unlikely that an amino acid is a substrate of any of these enzymes. However, 11 of the 4CL-like proteins contain a polar residue at the corresponding position, indicating that a hydroxylated benzoic acid derivative could be a substrate. Using DhbE as structural template, we modeled the SBPs of three candidate proteins. According to these models, both 2,3-dihydroxybenzoic acid and salicylic acid can be fitted into the SBPs encoded by the group B genes At1g20500 and At5g38120 (Fig. 4). In contrast, limitation of space in the SBP encoded by the group C gene At3g48990 probably will prevent the accommodation of benzoic acid derivatives.
For fatty acyl-CoA synthetases of primary metabolism a sequence of 25 aa has been suggested to form a signature motif that regulates the preference for fatty acids of different chain length (32). Adjacent to this chain length specificity motif, nine amino acids have been identified, which might be involved in substrate binding (33). Surprisingly, linear alignments of conserved domains of peptide synthetases, 4CLs, and fatty acyl-CoA synthetases indicate that the 34-aa comprising region involved in fatty acid binding and chain length selectivity is not part of the SBP as defined for 4CLs and peptide synthetases. Instead, it is located directly C terminal of the box II motif of 4CL. Therefore, it is obviously not possible to correlate the properties of single SBP lining residues of 4CL-like proteins with their capacity to activate fatty acids. However, searching the Arabidopsis genome with the 34-aa comprising sequence motif identified all 4CL-like proteins of group F as putative fatty acyl-CoA synthetases (Fig. 4).
A challenging, but yet unresolved, question concerns the specific biological function of each of the 4CL isoforms present in Arabidopsis. Based on the structural relationships between 4CLs from different plants, the existence of two functionally divergent classes of enzyme has been proposed (17). The Arabidopsis class I isoforms, At4CL1 and At4CL2, are constitutively expressed in lignified bolting stems of adult plants and at the onset of lignin deposition in seedling roots and cotyledons, respectively (17, 34). Class I 4CL isoforms from aspen and tobacco are likewise highly expressed in lignifying tissues (35, 36). Contrasting expression patterns have been found for class II 4CL isoforms. For example, At4CL3 from Arabidopsis is expressed in all light-exposed organs such as leaves, flowers, and siliques, and Pt4CL1 from aspen is restricted to epidermal cells of leaves and stem, suggesting that both enzymes are associated with nonlignin-related phenylpropanoids that function as UV protectants (17, 35).
Although the above evidence supports the notion that At4CL1 and At4CL2 contribute mainly to lignin biosynthesis, whereas At4CL3 provides the precursor for flavonoid biosynthesis via chalcone synthase, it is still an open question whether distinct 4CL isoforms indeed have specialized functions in channeling substrates to different metabolic pathways. Expression of At4CL2 variants with new substrate specificity under the control of the native At4CL2 promoter in a recently available At4CL2 knockout line will avoid interference with the catalytic activity of the endogenous protein. Using this approach, comparative analyses of At4CL2 transgenic lines, with At4CL2 WT and knockout lines with respect to content and composition of phenolic products derived from ferulic acid, sinapic acid, and cinnamic acid precursors, should provide information on the metabolic network or pathway that is specifically controlled or affected by At4CL2.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Grant KO 1192/6 (to E.K. and H.-P.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: 4CL, 4-coumarate:CoA ligase; At4CL2, Arabidopsis 4CL isoform 2; PheA, phenylalanine-activating domain; SBP, substrate binding pocket.
References
- 1.Douglas, C. J. (1996) Trends Plant Sci. 1 171-178. [Google Scholar]
- 2.Weisshaar, B. & Jenkins, G. I. (1998) Curr. Opin. Plant Biol. 1 251-257. [DOI] [PubMed] [Google Scholar]
- 3.Dixon, R. A. & Steele, C. L. (1999) Trends Plant Sci. 4 394-400. [DOI] [PubMed] [Google Scholar]
- 4.Pietrowska-Borek, M., Stuible, H.-P., Kombrink, E. & Guranowski, A. (2003) Plant Physiol. 131 1401-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kisselev, L. L., Justesen, J., Wolfson, A. D. & Frolova, L. Y. (1998) FEBS Lett. 427 157-163. [DOI] [PubMed] [Google Scholar]
- 6.McLennan, A. G. (2000) Pharmacol. Ther. 87 73-89. [DOI] [PubMed] [Google Scholar]
- 7.Fulda, M., Heinz, E. & Wolter, F. P. (1994) Mol. Gen. Genet. 242 241-249. [DOI] [PubMed] [Google Scholar]
- 8.Stuible, H.-P., Büttner, D., Ehlting, J., Hahlbrock, K. & Kombrink, E. (2000) FEBS Lett. 467 117-122. [DOI] [PubMed] [Google Scholar]
- 9.Conti, E., Franks, N. P. & Brick, P. (1996) Structure (London) 4 287-298. [DOI] [PubMed] [Google Scholar]
- 10.Conti, E., Stachelhaus, T., Marahiel, M. A. & Brick, P. (1997) EMBO J. 16 4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May, J. J., Kessler, N., Marahiel, M. A. & Stubbs, M. T. (2002) Proc. Natl. Acad. Sci. USA 99 12120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuible, H.-P. & Kombrink, E. (2001) J. Biol. Chem. 276 26893-26897. [DOI] [PubMed] [Google Scholar]
- 13.Knobloch, K.-H. & Hahlbrock, K. (1975) Eur. J. Biochem. 52 311-320. [DOI] [PubMed] [Google Scholar]
- 14.Ranjeva, R., Boudet, A. M. & Faggion, R. (1976) Biochimie 58 1255-1262. [DOI] [PubMed] [Google Scholar]
- 15.Wallis, P. J. & Rhodes, M. J. C. (1977) Phytochemistry 16 1891-1894. [Google Scholar]
- 16.Lindermayr, C., Möllers, B., Fliegmann, J., Uhlmann, A., Lottspeich, F., Meimberg, H. & Ebel, J. (2002) Eur. J. Biochem. 269 1304-1315. [DOI] [PubMed] [Google Scholar]
- 17.Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E. & Kombrink, E. (1999) Plant J. 19 9-20. [DOI] [PubMed] [Google Scholar]
- 18.Ehlting, J., Shin, J. J. K. & Douglas, C. J. (2001) Plant J. 28 1-12. [DOI] [PubMed] [Google Scholar]
- 19.Lindermayr, C., Fliegmann, J. & Ebel, J. (2003) J. Biol. Chem. 278 2781-2786. [DOI] [PubMed] [Google Scholar]
- 20.Stuible, H.-P., Wagner, C., Andreou, I., Huter, G., Haselmann, J. & Schweizer, E. (1996) J. Bacteriol. 178 4787-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knobloch, K.-H. & Hahlbrock, K. (1977) Arch. Biochem. Biophys. 184 237-248. [DOI] [PubMed] [Google Scholar]
- 22.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234 779-815. [DOI] [PubMed] [Google Scholar]
- 23.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4 187-217. [Google Scholar]
- 24.Laskowski, R. A., McArthur, W. M., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291. [Google Scholar]
- 25.Devereux, J., Haeberli, P. & Smithies, O. (1984) Nucleic Acids Res. 12 387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitch, W. M. (1977) Am. Nat. 111 223-257. [Google Scholar]
- 27.Swoffort, D. L., Olsen, G. J., Waddell, P. J. & Hillis, D. M. (1996) in Molecular Systematics, eds. Hillis, D. M., Moritz, C. & Mable, B. K. (Sinauer, Sunderland, MA), pp. 407-514.
- 28.Felsenstein, J. (1985) Evolution 39 783-791. [DOI] [PubMed] [Google Scholar]
- 29.Grand, C., Boudet, A. & Boudet, A. M. (1983) Planta 158 225-229. [DOI] [PubMed] [Google Scholar]
- 30.Challis, G. L., Ravel, J. & Townsend, C. A. (2000) Chem. Biol. 7 211-224. [DOI] [PubMed] [Google Scholar]
- 31.Staswick, P. E., Tiryaki, I. & Rowe, M. L. (2002) Plant Cell 14 1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black, P. N., Zhang, Q., Weimar, J. D. & DiRusso, C. C. (1997) J. Biol. Chem. 272 4896-4903. [DOI] [PubMed] [Google Scholar]
- 33.Black, P. N., DiRusso, C. C., Sherin, D., MacColl, R., Knudsen, J. & Weimar, J. D. (2000) J. Biol. Chem. 275 38547-38553. [DOI] [PubMed] [Google Scholar]
- 34.Lee, D., Ellard, M., Wanner, L. A., Davis, K. R. & Douglas, C. J. (1995) Plant Mol. Biol. 28 871-884. [DOI] [PubMed] [Google Scholar]
- 35.Hu, W.-J., Kawaoka, A., Tsai, C.-J., Lung, J., Osakabe, K., Ebinuma, H. & Chiang, V. L. (1998) Proc. Natl. Acad. Sci. USA 95 5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, D. & Douglas, C. J. (1996) Plant Physiol. 112 193-205. [DOI] [PMC free article] [PubMed] [Google Scholar]