Abstract
Hepatitis delta virus (HDV) can dramatically worsen liver disease in patients coinfected with hepatitis B virus (HBV). No effective medical therapy exists for HDV. The HDV envelope requires HBV surface antigen proteins provided by HBV. Once inside a cell, however, HDV can replicate its genome in the absence of any HBV gene products. In vitro, HDV virion assembly is critically dependent on prenyl lipid modification, or prenylation, of its nucleocapsid-like protein large delta antigen. To overcome limitations of current animal models and to test the hypothesis that pharmacologic prenylation inhibition can prevent the production of HDV virions in vivo, we established a convenient mouse-based model of HDV infection capable of yielding viremia. Such mice were then treated with the prenylation inhibitors FTI-277 and FTI-2153. Both agents were highly effective at clearing HDV viremia. As expected, HDV inhibition exhibited duration-of-treatment dependence. These results provide the first preclinical data supporting the in vivo efficacy of prenylation inhibition as a novel antiviral therapy with potential application to HDV and a wide variety of other viruses.
Introduction
Hepatitis delta virus (HDV) is an important cause of acute and chronic liver disease (1–5) for which there is no effective medical therapy. Here we sought to test the hypothesis that specific insights gained from the study of HDV molecular virology can be translated into a novel type of in vivo antiviral therapy.
The HDV virion is composed of three general elements: an RNA genome, delta antigens — the only proteins known to be encoded by the genome — and an envelope that surrounds the other two elements. The lipid envelope is embedded with hepatitis B virus (HBV) surface antigen (HBsAg) proteins that are provided by a coinfecting HBV. They provide a means of exit and, presumably, entry for HDV, and this explains why delta infections are always found in the presence of a coexisting HBV infection (3, 6). Once inside a cell, however, HDV can replicate its genome in the absence of any HBV gene products (7–9). The HDV genome is a 1.7-kb single-stranded circular molecule (10). There are two major isoforms of delta antigen, termed small and large (4). They are identical in sequence, except that the large delta antigen has an extra 19 amino acids at its C terminus.
The presence of these extra C-terminal amino acids dramatically changes the function of delta antigen. For example, while the small delta antigen promotes HDV genome replication, the large delta antigen is a potent transdominant inhibitor (11–13). The two isoforms also have differences in their ability to transactivate heterologous genes (14, 15). Perhaps the most striking functional difference between these isoforms has emerged from studies of HDV assembly.
A complex of newly replicated HDV genome and delta antigens must acquire an envelope to complete the assembly process. While both delta antigen isoforms are found in mature virions, only the large delta antigen is capable of promoting particle formation with the HBsAg envelope proteins; the small delta antigen alone cannot (16–18). The molecular basis for this selective role in assembly lies within the 19 amino acids unique to large delta antigen. In particular, the last four amino acids constitute a “CXXX box,” where C = cysteine and X = one of the last three amino acids at the carboxyl terminus of a protein (19–21). This sequence motif is the substrate for a family of enzymes, termed prenyltransferases, which catalyze the covalent addition of a 15-carbon (farnesyl) or 20-carbon (geranylgeranyl) prenyl lipid onto the CXXX box cysteine. These prenyl lipids, the products of synthetic pathways originating with mevalonic acid, have been found to modify the CXXX boxes of a growing collection of proteins (19–21). Prenylation of proteins such as Ras renders the modified protein more lipophilic and promotes its association with membranes. Molecular genetic mutation of large delta antigen’s CXXX box cysteine→serine not only prevents prenylation of large delta antigen, but also abolishes large delta antigen’s ability to form virus-like particles (VLPs) with HBsAg in vitro (22).
The essential role of prenylation in HDV assembly suggests that disruption of this modification might form the basis for a novel anti-HDV strategy. Because the type of prenyl lipid found on delta antigen is farnesyl (23), farnesyltransferase inhibitors (FTIs), which target the transfer of fully formed farnesyl to substrates such as large delta antigen, represent attractive candidate drugs for this strategy. Precisely such compounds already have been developed to inhibit the farnesylation of Ras (24, 25) and in doing so prevent H-RasV12-mediated transformation of cultured cells (26) or Ras-dependent tumor growth in nude mice (27, 28). The low–side effect profile of FTIs in phase I/II oncology trials (29, 30) suggests that these compounds originally developed as anticancer agents might have an entirely new application as antivirals for use against HDV and other viruses similarly dependent on prenylation (31). Although in vitro studies with simple assembly models of HDV VLPs (32) or transfected cells (33) have been encouraging, the potential efficacy of FTIs as in vivo antivirals has been questioned. Moreover, practical animal models in which to evaluate such a strategy have been lacking. Besides being either endangered species or somewhat difficult to handle, the acquisition and maintenance costs of the chimpanzee or woodchuck models (34) limit the number of animals that can be used, affecting the statistical significance of any potential results. The relatively large size and genetic variability of these animals impose additional limitations for experiments that require reproducible host factors (35) or that are designed to study experimental compounds that are often expensive and synthesized in small quantities.
Here we established a new and convenient mouse-based model of HDV that results in HDV viremia. This model allowed us to directly test the hypothesis that prenylation inhibitors represent a novel class of potent in vivo antiviral agents.
Methods
Hydrodynamic transfections.
HBV-transgenic FVB mice (36) or control FVB mice were injected with 25 μg total DNA in saline into the tail vein according to the method of Liu et al. (37). HDV-encoding plasmids pCMV·HDVI(+) or pCMV·HDVIII(+), encoding 1.2-unit-length antigenomes of HDV genotypes I and III, respectively (38), were injected along with pGEM4ayw.2×, which bears a head-to-tail dimer of the full-length HBV genome (33), or plain vector pcDNA3 (Invitrogen Corp., Carlsbad, California, USA), which was used for control. With experience, adequate levels of intrahepatic delivery considered acceptable for analysis (106 genome equivalents of HDV RNA per microgram of liver RNA) could be routinely achieved (i.e., over 95% of the time). Alternatively, 2 μg of plasmid pBS.ApoE.HCR.hAATp.hFIX+IntA.bpA encoding human α1-antitrypsin (hAAT) (39) was coinjected. Serum hAAT levels determined by ELISA assay (40) on day 2 after transfection were then used to identify successfully transfected mice, because day-2 hAAT level was found to correlate with the amount of day-7 HDV RNA in the liver.
Northern blot analysis.
Northern blot analysis was performed as described in Bordier et al. (33). Briefly, glyoxalated RNA samples were migrated on 1.5% agarose gels, transferred, and UV cross-linked with a Stratalinker (Stratagene, San Diego, California, USA) to a charged Zeta-Probe (Bio-Rad Laboratories, Richmond, California, USA) nylon membrane. The latter was hybridized with an [α-32P]UTP-labeled (3,000 Ci/mmol; Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) riboprobe specific for HDV genomic sense RNA. The membrane was washed, dried, and exposed to BioMax-MR films (Eastman Kodak, Rochester, New York, USA) or to PhosphorImager (Molecular Dynamics, Sunnyvale, California, USA) plates.
Western blot analysis.
Liver tissue samples were homogenized in lysis buffer (1× TBS with protease inhibitor cocktail; Boehringer Mannheim GmbH, Mannheim, Germany). After a 3-minute preclearing at maximum speed in an Eppendorf microfuge, supernatants were passed ten times through a 27-gauge needle. Following addition of an equal volume of 2× Laemmli sample buffer and incubation at 95°C for 5 minutes, aliquots were subjected to Western blot analysis essentially as described in Glenn et al. (22). Briefly, samples were subjected to PAGE on 12% gels followed by transfer to nitrocellulose. Blots were probed with a primary human Ab to delta antigen (22) followed by an HRP-conjugated anti-human secondary Ab (Promega Corp., Madison, Wisconsin, USA) and ECL detection reagent (Amersham Pharmacia Biotech).
Immunohistochemistry.
Immunohistochemistry was performed as described in Ohashi et al. (40). Briefly, formalin-fixed, paraffin-embedded liver sections were stained with a polyclonal human Ab against HDV delta antigen (22) as primary Ab at a dilution of 1:10,000. This Ab was detected by avidin-biotin complex immunoperoxidase technique and 3,3-diaminobenzine tetrahydrochloride development using an ABC Elite kit (Vector Laboratories, Burlingame, California, USA).
Sera processing.
Blood samples were allowed to clot at room temperature for 2–3 hours, spun in an Eppendorf microfuge, and the sera transferred to new tubes for storage at –80°C until use. HDV virions in serum aliquots were either first pelleted through a 20% sucrose cushion as described (33) or directly extracted with 1 ml of TRIzol reagent (Invitrogen Corp.) in the presence of 1 μl of a Pellet Paint (Novagen, Madison, Wisconsin, USA) coprecipitant following the manufacturer’s instructions. The resulting RNA pellets were resuspended in diethylpyrocarbonate-treated (DEPC-treated) sterile doubled-distilled water (ddH2O), treated for 1 hour at 37°C with RQ1 DNase (Promega Corp.), and extracted again with 1 ml TRIzol reagent. The final pellets were resuspended in DEPC-treated sterile ddH2O to the original volume of serum extracted.
RT-PCR.
Limiting dilution reactions were performed with the one-step RT-PCR kit (GIBCO-BRL; Life Technologies Inc., Grand Island, New York, USA) using primers (synthesized by Operon, Alameda, California, USA) designed to yield the desired products only from closed circular genomic HDV RNA as found in virions as follows: HDV genotype I, primers 5′-GGAACTCGACTTATCGTCCCATTAG-3′ and 5′-ACATCAGGGGAAACCAGGGATTTCA-3′, which yield a 556-bp product; HDV genotype III, primers 5′-AGCAGTTCCCATAGTATGGGTTTACC-3′ and 5′-GGGACGCCTCGGCCCTTCCTTAGCA-3′, which yield a 540-bp product. RT-PCR reactions followed the program of 30 minutes at 55°C, 2 minutes at 94°C, and then 35 cycles of 30 seconds at 94°C, 1 minute at 60°C, 1 minute at 72°C, and a last extension step of 6 minutes at 72°C. A CsCl gradient–purified standardized inoculum produced in cultured Huh-7 cells (33) was used for positive controls and quantitation standards.
Southern blot analysis.
Aliquots of RT-PCR reactions were migrated on 2% agarose gels, denatured 45 minutes in 1.5 M NaCl and 0.5 N NaOH, neutralized 30 minutes in 1.5 M NaCl, 0.5 M Tris-HCl, pH 7.2, 1 mM EDTA, and transferred and UV cross-linked to charged nylon membranes (Hybond-N+; Amersham Pharmacia Biotech). The latter were hybridized 18 hours at 70°C in 5 ml of 5× SSPE (0.75 M NaCl, 0.05 M NaH2PO4, 5 mM EDTA, pH 7.0), 5× Denhardt’s solution, 0.5% (wt/vol) SDS, 20 μg/ml salmon sperm DNA (Life Technologies Inc.), and a PCR-generated probe made with the same primers as the RT-PCR reaction, [α-32P]dCTP (3,000 Ci/mmol, Amersham Pharmacia Biotech), and an appropriate HDV-encoding DNA template. Membranes were dried at 70°C and exposed as for Northern blots.
CsCl gradients.
A pool of transfected mice sera (250 μl) was centrifuged on a CsCl gradient (1.14–1.40 g/cm3, 22 hours, 150,000 g, 4°C) with a Beckman SW41Ti rotor. Gradient fractions collected by bottom puncture were analyzed by refractometry and quantitative RT-PCR for HDV genomic RNA.
Drug treatments.
FTI-277 and FTI-2153 were synthesized as described previously (41). Aliquots were diluted into sterile saline to final concentrations of 6.25 mg/ml 1× carrier (5% DMSO, 0.5 mM DTT for FTI-277, and 5% DMSO for FTI-2153) immediately prior to the single daily intraperitoneal injections dosed at 50 mg/kg/d. Control mice received the same volume of 1× carrier in sterile saline.
Alanine transaminase assays.
Alanine transaminase (ALT) assays were performed by automated chemistry on an Express Plus machine (Chiron Corp., Emeryville, California, USA) in the Stanford University Department of Comparative Medicine’s Diagnostic Lab.
Results
Establishment of HDV intrahepatic replication in HBV-transgenic mice.
To evaluate the potential in vivo efficacy of prenylation inhibitors against HDV, we first sought to establish an immunocompetent mouse model of HDV infection. Similar to the work of Chang et al. (42), we delivered plasmids capable of initiating HDV replication (33) by hydrodynamic transfection (37) to mice. HDV nucleic acid delivered in this manner can initiate apparently authentic replication cycles in mouse hepatocytes. In the absence of coexisting HBV, however, production of progeny HDV virus cannot occur, and no viremia is observed (42). We reasoned that supplying a source of HBV envelope protein — such as by using an HBV transgenic mouse as recipient — could allow the completion of the HDV viral life cycle and lead to viremia.
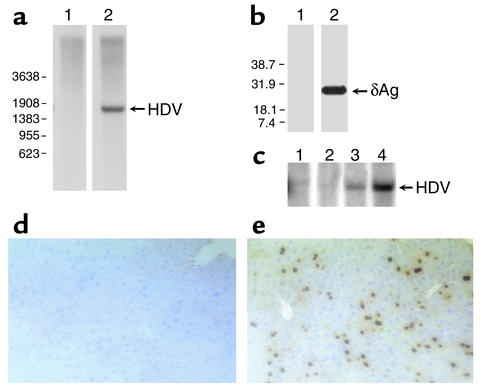
As seen in Figure 1a, Northern blot analysis of liver RNA shows that replicated HDV genomic RNA is readily detectable in the livers of HDV hydrodynamically transfected mice, but not in controls. Similarly, Western blot analysis revealed that delta antigen was abundantly expressed in the HDV-transfected mice, but not in controls (see Figure 1b). This indicates the delivered HDV-encoding plasmid was able to successfully initiate HDV genome replication. The time course for appearance of replicating RNA (Figure 1c) was similar to that observed in transfected cells in culture (12) and seen previously in mice transfected with HDV-encoding plasmids or RNA (42).
Figure 1.
Intrahepatic replication of HDV following hydrodynamic transfection. Mice transgenic for HBV were hydrodynamically transfected with vector DNA (pcDNA3) or a vector bearing HDV replication-inducing sequences [pCMV·HDVI(+)]. Seven days later, mice were sacrificed and liver samples were analyzed for HDV RNA and protein. (a) Samples of total liver RNA from a mouse transfected with pcDNA3 (lane 1) or with pCMV·HDVI(+) (lane 2) were analyzed for HDV genomic RNA using Northern blots. Positions of molecular-weight markers are on the left; arrow indicates position of the replicated 1.7-kb RNA genome. (b) Samples of total liver RNA from a mouse transfected with pcDNA3 (lane 1) or with pCMV·HDVI(+) (lane 2) were analyzed for delta antigen using Western blots. Positions of molecular-weight markers are on the left; arrow indicates position of delta antigen (δAg). (c) Representative time course for HDV replication in hydrodynamically transfected mice. Mice transgenic for HBV were hydrodynamically transfected with pcDNA3 (lane 1) or pCMV·HDVI(+) (lanes 2–4) and were sacrificed at day 2 (lane 2), day 4 (lane 3), or day 7 (lanes 1 and 4) after transfection. Liver samples were analyzed for HDV RNA using Northern blots. To account for potential differences in transfection efficiency, a plasmid encoding hAAT was included in the transfection, and HDV replication was detected in mice who had similar serum levels of hAAT on day 2 after transfection. (d and e) Liver sections from the same mice as in a were fixed in formalin and stained by immunohistochemistry for detection of HDV delta antigen. (d) Mouse transfected with pcDNA3. (e) Mouse transfected with pCMV·HDVI(+). Brown-red spots indicate characteristic nuclear staining pattern of delta antigen.
To assess the percentage of cells supporting HDV replication, immunohistochemical analysis of liver sections for delta antigen was performed. As shown in Figure 1e, about 20–30% of hepatocytes displayed the characteristic nuclear staining pattern of HDV-infected cells. This percentage of HDV-positive nuclei is similar to that typically observed in liver biopsies of HDV-infected patients (43) and indicates that a relatively robust HDV burden is established in the livers of HBV-transgenic mice.
Interestingly, to date we have not observed a dramatic difference in ALT level between mice hydrodynamically transfected with HDV-encoding versus control plasmids. Also, our histopathologic evaluations reveal a less obvious cellular immune response than has been recently reported in mice hydrodynamically transfected with HBV (44). Perhaps this reflects that the role of specific immunity may be less prominent in response to HDV than HBV (45). Indeed, we did not detect specific anti-HDV Ab’s in the serum of our hydrodynamically transfected mice (our unpublished observations).
Production of HDV viremia.
We next wished to determine whether the replicating HDV RNA in the hepatocytes could be packaged with HBsAg envelope proteins into virions and released into the serum. For this we performed RT-PCR analysis with primers specific for virion circular genomic RNA on serum aliquots. The latter were taken from mice 2 days after transfections (when no viremia was yet expected) and upon sacrifice (when replicated HDV RNA was readily detected in the liver, as in Figure 1a). As seen in Figure 2a (lanes 4, 6, and 8), when replicated RNA and delta antigen are complemented with a source of their natural envelope protein, HBsAg, assembly and release of HDV virions into the mouse serum were readily detected. That no signal was yet detected on day 2 after transfection provides additional confirmation that the signal measured at later time points indeed reflects authentic viremia, as opposed to artifactual detection of residual input plasmid DNA. No HDV RNA was detected in mice injected with control vector or in non–HBV-transgenic recipients of HDV-encoding plasmids. The RT-PCR–amplified DNA can be examined using Southern blots with HDV-specific probes to allow quantitation by PhosphorImager analysis for increased sensitivity. Typically, titers of 107 genome equivalents per milliliter are observed, and the assay is linear to our detection limit of 50 genome equivalents per reaction. Recently, with a newer vector (to be described in detail by the authors in a future publication), titers are an additional log higher (our unpublished observations).
Figure 2.
HDV viremia following hydrodynamic transfection of HBV transgenic mice. (a) After hydrodynamic transfection, mice sera were tested for HDV genomic RNA by RT-PCR. Ethidium bromide gel of sera from HBV-transgenic mice transfected with vector alone assayed at day 2 (lane 1) and day 7 (lane 2); sera from three transgenic mice transfected with pCMV·HDVIII(+) and pGEM4ayw.2× assayed at day 2 (lanes 3, 5, and 7) and at day 7 (lanes 4, 6, and 8); sera from a nontransgenic mouse transfected with pCMV·HDVIII(+) and assayed at day 2 (lane 9) and day 7 (lane 10). (b) Aliquots of day-7 sera from mice as analyzed in lanes 4, 6, and 8 of a were banded in a CsCl gradient, and fractions were assayed for HDV RNA by quantitative RT-PCR (filled circles) and CsCl density by refractometry (gray line).
We observe a good correlation between the level of intrahepatic replication and HDV virion RNA in the serum. To date, in the majority of hydrodynamically transfected mice most of the HDV RNA in the livers and serum is gone by day 21. At present, we do not know what factors are responsible for clearance of the virus or why residual HDV is present at later time points in some mice. For the period of time in the experiments presented here, however, the course of HDV replication is very reproducible and robust with easily detectable viral markers. Further evidence that the HDV RNA detected in serum is contained in true virions was provided by sedimentation analysis of HDV-positive mouse sera. Specifically, isopycnic banding of the mouse-produced virions in CsCl gradients (Figure 2b) yielded a sedimentation profile characteristic of HDV virions derived from infected human patients (46).
To our knowledge, this is the first demonstration of HDV viremia in an immunocompetent mouse, and the percentage of mouse hepatocytes expressing delta antigen is tenfold to 50-fold greater than reported for earlier, nonhydrodynamic, transfection-based mouse experiments (45). This model thus represents an ideal system in which to test the hypothesis that HDV assembly in vivo is sensitive to prenylation inhibitors.
Prenylation inhibitors abolish HDV viremia.
To this end, HBV-transgenic mice hydrodynamically transfected to initiate HDV viremia were treated with a single daily dose of prenylation inhibitor or vehicle control for 1 week. Serum aliquots were then analyzed for HDV virion RNA. Because delta antigen is modified by the prenyl lipid farnesyl (23), we chose to test two farnesyltransferase inhibitors, FTI-277 (47) and FTI-2153 (41) (see Figure 3). FTI-277 is methylester prodrug of a Cys-Val-Ile-Met (the CXXX box of KB-Ras) peptidomimetic, wherein the dipeptide Val-Ile is replaced by 2-phenyl-4-aminobenzoic acid. FTI-2153 is a nonthiol containing peptidomimetic, wherein the tripeptide Cys-Val-Ile is replaced by an imidazole derivative of 2-(2-methylphenyl)-4-aminomethylbenzoic acid. Removing the thiol moiety obviates the need for a reducing agent to achieve activity. These compounds have been used successfully to inhibit the prenylation of Ras (41). We chose to administer 50 mg/kg/d per mouse because this dose has been shown previously to inhibit farnesyltransferase function in mice (27). (For a recent review of in vivo pharmacodynamics and efficacy of FTIs, see ref. 48).
Figure 3.
Design and structures of prenylation inhibitors used to treat HDV viremia. (a) Cys-Val-Ile-Met CXXX box tetrapeptide as found in K-Ras4B, a substrate of farnesyltransferase. Two peptidomimetics of this CXXX box were designed as, and shown to be, competitive inhibitors of farnesyltransferase, (b) FTI-277, wherein the dipeptide Val-Ile is replaced by 2-phenyl-4-aminobenzoic acid, and (c) FTI-2153, wherein the tripeptide Cys-Val-Ile is replaced by an imidazole derivative of 2-(2-methylphenyl)-4-aminomethylbenzoic acid.
As shown in Figure 4a, while HDV viremia was readily detectable in mice receiving vehicle controls, we were unable to detect HDV virions in the serum of parallel cohorts of FTI-treated mice. Both compounds were effective in inhibiting viremia. As hypothesized, inhibition appeared to be at the prenylation-dependent step of virus assembly, since HDV RNA levels were comparable in the livers of all mice (see Figure 4b). As a control for liver toxicity, serum ALT levels were also determined. As shown in Figure 4c, there were no significant differences in ALT levels between treated and control mice.
Figure 4.
In vivo treatment of HDV with prenylation inhibitors. (a) Mice hydrodynamically transfected to produce HDV viremia were treated with carrier alone (lanes 1 and 6), carrier plus FTI-277 (lanes 2–5), or carrier plus FTI-2153 (lanes 7–10) for 7 days. Serum aliquots were then assayed for HDV RNA by RT-PCR. (b) Corresponding liver samples were analyzed for HDV RNA using Northern blots. (c) Serum aliquots were also analyzed for ALT. (d) Mice hydrodynamically transfected to produce HDV viremia were treated with carrier alone (controls, filled circles) or carrier plus FTI-2153 (open circles) for the indicated number of days prior to sacrifice. Serum HDV RNA was quantitated by RT-PCR. HDV RNA per microgram of total liver RNA was quantitated by Northern blots and used to normalize for any differences in transfection efficiency. The mean serum HDV RNA genome equivalent (geq) for each group of mice is plotted (see Methods for additional details).
Before the initiation of treatment with FTIs, a pool of prenylated large delta antigen can accumulate. Because this pool is available to participate in virus particle assembly, we would predict that the efficiency of HDV clearance from the serum should be proportional to the length of treatment with prenylation inhibitor. As shown in Figure 4d, this is indeed the case. A 2-day course of therapy was associated with approximately 85% reduction in serum HDV titer as compared with mock-treated controls. By 4 days of treatment serum levels of HDV were reduced approximately 95%, and they were undetectable after 7 days of therapy.
Discussion
To enable an evaluation of the in vivo efficacy of prenylation inhibitors against HDV we established a novel mouse model of HDV that results in authentic HDV viremia. We then used this model to demonstrate the potent in vivo antiviral efficacy of two representative prenylation inhibitors. Indeed, despite a large intrahepatic burden of HDV, these compounds were able to completely clear HDV viremia to below the limit of detection. These results thus translate a previous in vitro observation (22) into what we believe to be a relatively dramatic and clear first in vivo confirmation of the potential of this novel class of antiviral agents.
Using the delivery technique of hydrodynamic transfection and HBV-transgenic mice as recipients, we sought to recapitulate several key aspects of HDV superinfection of human chronic carriers of HBV. A relatively high intrahepatic burden of HDV was achieved, as reflected both in the total amount of HDV RNA (Figure 1a) and the percentage of infected cells (Figure 1e) within the livers of recipient mice. By also providing the replicating HDV with a source of its natural envelope protein, we were able to observe the production and release of authentic HDV virions into the serum (Figure 2). Because HDV virion assembly in vitro is absolutely dependent on prenylation of the HDV large delta antigen protein, we hypothesized that administration of a specific inhibitor of the enzyme known to be responsible for delta antigen prenylation would result in abrogation of the HDV life cycle at the critical step of assembly and release into the serum. The presence of HDV viremia in our mouse model allowed us to test this hypothesis directly. When HDV-viremic mice were treated with either of two prenylation inhibitors at doses known to inhibit prenylation in vivo, rapid and efficient clearing of HDV from the serum was observed.
This model has several attractive features compared with other HDV animal models (34). Because the mice are transgenic for HBV and therefore immunotolerant of these viral antigens, experimental HDV infection of these animals shares several important aspects with HDV superinfection of human chronic HBV carriers. As opposed to chimpanzees or woodchucks (34), the highly inbred nature of the mouse can help avoid problems such as the wide spectrum of liver disease observed among animals receiving a common inoculum (35). Our mouse model may prove particularly useful for studying host responses to viral replication in the setting of limited animal-to-animal genetic variability and, by crossing with other strains, specific genetic backgrounds. Because the mice are inoculated with cloned DNA, we have the ability to introduce HDV genomes with any desired mutations or genotype and thereby evaluate the effect of the latter on infection or pathogenesis. Finally, in contrast to xenotransplanted immunodeficient mice (40), the relative technical simplicity and low cost of this new model, coupled with the small size of the host animal, is ideal for experiments involving precious compounds available in only limited quantities.
As hypothesized, inhibition of HDV viremia appeared to be at the prenylation-dependent step of virus assembly and release, since intrahepatic HDV RNA levels were comparable in mice treated with drug or vehicle control. The observed clearance of viremia was not simply a nonspecific hepatotoxic effect of the FTIs because ALT levels were also comparable among treatment groups. Rather, as observed in HDV in vitro cell culture systems (32, 33), it is a critical stage of the viral life cycle — the ability of replicated HDV RNA to be packaged and released in the form of progeny virions — that is specifically and dramatically disrupted by the prenylation-inhibiting compounds.
Our results support our initial hypothesis and demonstrate that prenylation inhibitors can indeed effectively inhibit HDV viremia. These results have obvious clinical relevance and importance for human HDV infections against which effective medical therapy has been lacking to date. In our mouse model, newly produced virions cannot infect new cells because the mouse is not a natural host for HDV. Presumably, mouse hepatocytes lack the receptor for HDV. In human HDV infections, however, the ability to break a critical step in the HDV life cycle, such as virus assembly and release, would be expected to have a major impact on the subsequent spread and course of HDV infection and associated liver disease. Our results now provide the experimental foundation for testing this hypothesis directly in a larger animal model whose hepatocytes are naturally permissive for HDV infection, such as the woodchuck (49).
It is interesting to note that the antiviral strategy we have outlined represents a paradigm different from classical antiviral approaches, since we are not actually directly targeting a viral protein. Rather, the prenylation inhibitors are designed to deprive the virus access to a host function, namely farnesyltransferase activity. Thus, in contrast to traditional antiviral agents that bind a viral enzyme capable of mutating to develop resistance, it may prove to be particularly challenging for HDV to develop resistance to prenylation inhibition, especially because the relevant genetic loci are not under the virus’ control. Moreover, because the farnesyl moiety on the delta antigen may act as a specific ligand (50), its function might not be readily substituted by another prenyl lipid such as geranylgeranyl.
Considering that farnesyltransferase is a host cell enzyme, it is surprising that FTIs are so well tolerated even in in vitro studies (51). Perhaps this results from the existence of a family of prenyltransferase enzymes and the ability of different prenyltransferases to occasionally cross-prenylate substrates. It appears, however, that HDV is unable to benefit from either of these back-up mechanisms. It is exciting that orally available FTIs have been developed (29) and have been used in human phase I/II trials (52) with relative lack of toxicity.
Finally, our use of prenylation inhibitors as antiviral agents represents a prototype for an antiviral strategy that may be applicable to a wide variety of viruses that may also use prenylation in key aspects of their respective life cycles. These viruses include a collection of other medically important viruses (31), as well as potential agents of bioterrorism.
Acknowledgments
This work was supported by a Veterans Administration Merit Review Award (to J.S. Glenn) and NIH grant (CA67771 to S.M. Sebti and A.D. Hamilton). J.S. Glenn is also the recipient of an Amgen/American Association for the Study of Liver Diseases/American Liver Foundation Award and a Burroughs Wellcome Fund Career Award.
Footnotes
See the related Commentary beginning on page 319.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Hepatitis delta virus (HDV); HBV surface antigen (HBsAg); hepatitis B virus (HBV); carboxyl (C); virus-like particle (VLP); farnesyltransferase inhibitor (FTI); human α1-antitrypsin (hAAT); diethylpyrocarbonate (DEPC); doubled-distilled water (ddH2O); alanine transaminase (ALT).
References
- 1.Rizzetto, M., Ponzetto, A., and Forzani, I. 1991. Epidemiology of hepatitis delta virus: overview. In The hepatitis delta virus. J.L. Gerin, R.H. Purcell, and M. Rizetto, editors. Wiley-Liss Inc. New York, New York, USA. 1–20. [PubMed]
- 2.Alter, M.J., and Hadler, S.C. 1993. Delta hepatitis and infection in North America. In Hepatitis delta virus: molecular biology, pathogenesis, and clinical aspects. S.J. Hadziyannis, J.M. Taylor, and F. Bonino, editors. Wiley-Liss Inc. New York, New York, USA. 243–250. [PubMed]
- 3.Rizetto, M., and Ponzetto, A. 1995. Hepatitis delta virus infection: medical aspects. In The unique hepatitis delta virus. G. Dinter-Gottlieb, editor. R.C. Landes Publishing Co. Austin, Texas, USA. 125–139.
- 4.Lai MMC. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 5.Taylor JM. Human hepatitis delta virus: an agent with similarities to certain satellite RNAs of plants. Curr. Top. Microbiol. Immunol. 1999;239:107–122. doi: 10.1007/978-3-662-09796-0_6. [DOI] [PubMed] [Google Scholar]
- 6.Gerin, J.L., Casey, J.L., and Purcell, R.H. 2001. Hepatitis delta virus. In Fields virology. D.M. Knipe and P.M. Howley, editors. Lippincot& Wilkins. Philadelphia, Pennsylvania, USA. 3037–3050.
- 7.Kuo MY-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glenn JS, Taylor JM, White JM. In vitro synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J. Virol. 1990;64:3104–3107. doi: 10.1128/jvi.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai F-P, Chen P-J, Chang F-L, Chen D-S. Hepatitis delta virus cDNA monomer can be used in transfection experiments to initiate viral RNA replication. Virology. 1993;197:137–142. doi: 10.1006/viro.1993.1574. [DOI] [PubMed] [Google Scholar]
- 10.Wang K-S, et al. Structure, sequence and expression of the hepatitis delta viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 11.Chao M, Hsieh S-Y, Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glenn JS, White JM. Trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 1991;65:2357–2361. doi: 10.1128/jvi.65.5.2357-2361.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazinski DW, Taylor JM. Recent developments in hepatitis delta virus research. Adv. Virus Res. 1994;43:187–231. doi: 10.1016/s0065-3527(08)60049-4. [DOI] [PubMed] [Google Scholar]
- 14.Wei Y, Ganem D. Activation of heterologous gene expression by the large isoform of hepatitis delta virus. J. Virol. 1998;72:2089–2096. doi: 10.1128/jvi.72.3.2089-2096.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto T, et al. Large isoform of hepatitis delta antigen activates serum response factor-associated transcription. J. Biol. Chem. 2000;275:37311–37316. doi: 10.1074/jbc.M002947200. [DOI] [PubMed] [Google Scholar]
- 16.Chang F-L, Chen P-J, Tu S-J, Wang C-J, Chen D-S. The large form of hepatitis δ antigen is crucial for assembly of hepatitis δ virus. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C-J, Chen PJ, Wu JC, Patel D, Chen D-S. Small-form hepatitis B surface antigen is sufficient to help in the assembly of hepatitis delta virus-like particles. J. Virol. 1991;65:6630–6636. doi: 10.1128/jvi.65.12.6630-6636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu W-S, Bayer M, Taylor J. Assembly of hepatitis delta virus particles. J. Virol. 1992;66:2310–2315. doi: 10.1128/jvi.66.4.2310-2315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maltese WA. Posttranslational modification of proteins by isoprenoids in mammalian cells. FASEB J. 1990;4:3319–3328. doi: 10.1096/fasebj.4.15.2123808. [DOI] [PubMed] [Google Scholar]
- 20.Schafer WR, Rine J. Protein prenylation: genes, enzymes, targets, and functions. Annu. Rev. Genet. 1992;30:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- 21.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 22.Glenn JS, Watson JA, Havel CM, White JM. Identification of a prenylation site in delta virus large antigen. Science. 1992;256:1331–1333. doi: 10.1126/science.1598578. [DOI] [PubMed] [Google Scholar]
- 23.Otto JC, Casey PJ. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J. Biol. Chem. 1996;271:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 24.Barinaga M. From bench top to bedside. Science. 1997;278:1036–1039. doi: 10.1126/science.278.5340.1036. [DOI] [PubMed] [Google Scholar]
- 25.Sebti S, Hamilton AD. Inhibitors of prenyl transferases. Curr. Opin. Oncol. 1997;9:557–561. doi: 10.1097/00001622-199711000-00011. [DOI] [PubMed] [Google Scholar]
- 26.James GL, et al. Benzodiazepine peptidomimetics: potent inhibitors of Ras farnesylation in animal cells. Science. 1993;260:1937–1942. doi: 10.1126/science.8316834. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Qian Y, Hamilton AD, Sebti SM. Ras CAAX peptidomimetic FTI 276 selectively blocks tumor growth in nude mice of a human lung carcinoma with K-Ras mutation and p53 deletion. Cancer Res. 1995;55:4243–4247. [PubMed] [Google Scholar]
- 28.Liu M, et al. Effects of SCH 59228, an orally bioavailable farnesyl protein transferase inhibitor, on the growth of oncogene-transformed fibroblasts and a human colon carcinoma xenograft in nude mice. Cancer Chemother. Pharmacol. 1999;43:50–58. doi: 10.1007/s002800050862. [DOI] [PubMed] [Google Scholar]
- 29.Rowinsky EK, Windle JJ, Von Hoff DD. Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. J. Clin. Oncol. 1999;17:3631–3652. doi: 10.1200/JCO.1999.17.11.3631. [DOI] [PubMed] [Google Scholar]
- 30.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 31.Glenn, J.S. 1995. Prenylation and virion morphogenesis. In The unique hepatitis delta virus. G. Dinter-Gottlieb, editor. R.C. Landes Publishing Co. Austin, Texas, USA. 83–93.
- 32.Glenn JS, Marsters JC, Jr, Greenberg HB. Use of a prenylation inhibitor as a novel antiviral agent. J. Virol. 1998;72:9303–9306. doi: 10.1128/jvi.72.11.9303-9306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordier BB, et al. A prenylation inhibitor prevents production of infectious hepatitis delta virus particles. J. Virol. 2002;76:10465–10472. doi: 10.1128/JVI.76.20.10465-10472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negro F. Animal models of hepatitis delta virus infection. Viral Hepatitis Reviews. 1996;2:175–185. [Google Scholar]
- 35.Schlipkoeter U, et al. Different outcomes of chronic hepatitis delta virus infection in woodchucks. Liver. 1990;10:291–301. doi: 10.1111/j.1600-0676.1990.tb00472.x. [DOI] [PubMed] [Google Scholar]
- 36.Marion, P.L., et al. 2003. A transgenic mouse lineage useful for testing antivirals targeting hepatitis B virus. Elsevier Science. Amsterdam, The Netherlands. 197–209 pp.
- 37.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 38.Casey JL, Gerin JL. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 1998;72:2806–2814. doi: 10.1128/jvi.72.4.2806-2814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen ZY, et al. Linear DNAs concatemerize in vivo and result in sustained transgene expression in mouse liver. Mol. Ther. 2001;3:403–410. doi: 10.1006/mthe.2001.0278. [DOI] [PubMed] [Google Scholar]
- 40.Ohashi K, et al. Sustained survival of human hepatocytes in mice: a model for in vivo infection with human hepatitis B and hepatitis delta viruses. Nat. Med. 2000;6:327–332. doi: 10.1038/73187. [DOI] [PubMed] [Google Scholar]
- 41.Sun J, et al. Antitumor efficacy of a novel class of non-thiol-containing peptidomimetic inhibitors of farnesyltransferase and geranylgeranyltransferase I: combination therapy with the cytotoxic agents cisplatin, Taxol, and gemcitabine. Cancer Res. 1999;59:4919–4926. [PubMed] [Google Scholar]
- 42.Chang J, Sigal LJ, Lerro A, Taylor J. Replication of the human hepatitis delta virus genome is initiated in mouse hepatocytes following intravenous injection of naked DNA or RNA sequences. J. Virol. 2001;75:3469–3473. doi: 10.1128/JVI.75.7.3469-3473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verme G, et al. A histological study of hepatitis delta virus liver disease. Hepatology. 1986;6:1303–1307. doi: 10.1002/hep.1840060613. [DOI] [PubMed] [Google Scholar]
- 44.Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netter HJ, Kajino K, Taylor JM. Experimental transmission of human hepatitis delta virus to the laboratory mouse. J. Virol. 1993;67:3357–3362. doi: 10.1128/jvi.67.6.3357-3362.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizzetto M, et al. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 1980;77:6124–6128. doi: 10.1073/pnas.77.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerner EC, et al. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic ras signaling by inducing cytoplasmic accumulation of inactive Ras/Raf complexes. J. Biol. Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- 48.Ohkanda J, Knowles DB, Blaskovich MA, Sebti SM, Hamilton AD. Inhibitors of protein farnesyltransferase as novel anticancer agents. Curr. Top. Med. Chem. 2002;2:303–323. doi: 10.2174/1568026023394281. [DOI] [PubMed] [Google Scholar]
- 49.Ponzetto A, et al. Transmission of the hepatitis B virus-associated δ agent to the eastern woodchuck. Proc. Natl. Acad. Sci. U. S. A. 1984;81:2208–2212. doi: 10.1073/pnas.81.7.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- 51.Dalton MB, et al. The farnesyl protein transferase inhibitor BZA-5B blocks farnesylation of nuclear lamins and p21ras but does not affect their function or localization. Cancer Res. 1995;55:3295–3304. [PubMed] [Google Scholar]
- 52.Sharma S, et al. A phase II trial of farnesyl protein transferase inhibitor SCH 66336, given by twice-daily oral administration, in patients with metastatic colorectal cancer refractory to 5-fluorouracil and irinotecan. Ann. Oncol. 2002;13:1067–1071. doi: 10.1093/annonc/mdf173. [DOI] [PubMed] [Google Scholar]