Abstract
Hepatocyte nuclear factors-3 (Foxa-1–3) are winged forkhead transcription factors that regulate gene expression in the liver and pancreatic islets and are required for normal metabolism. Here we show that Foxa-2 is expressed in preadipocytes and induced de novo in adipocytes of genetic and diet-induced rodent models of obesity. In preadipocytes Foxa-2 inhibits adipocyte differentiation by activating transcription of the Pref-1 gene. Foxa-2 and Pref-1 expression can be enhanced in primary preadipocytes by growth hormone, suggesting that the antiadipogenic activity of growth hormone is mediated by Foxa-2. In differentiated adipocytes Foxa-2 expression leads to induction of gene expression involved in glucose and fat metabolism, including glucose transporter-4, hexokinase-2, muscle-pyruvate kinase, hormone-sensitive lipase, and uncoupling proteins-2 and -3. Diet-induced obese mice with haploinsufficiency in Foxa-2 (Foxa-2+/–) develop increased adiposity compared with wild-type littermates as a result of decreased energy expenditure. Furthermore, adipocytes of these Foxa-2+/– mice exhibit defects in glucose uptake and metabolism. These data suggest that Foxa-2 plays an important role as a physiological regulator of adipocyte differentiation and metabolism.
Introduction
Obesity results from a massive expansion of white adipose tissue and recruitment of adipocyte precursor cells and is a common cause of insulin resistance and diabetes (1, 2). Obesity may arise from increased size of individual adipose cells due to lipid accumulation or increased number of adipocytes arising from differentiation of adipose precursor cells to mature adipocytes (2). Studies in rodents indicate a lifelong ability to make new fat cells in response to high-fat diet and reveal that preadipocytes continue to undergo differentiation under the appropriate nutritional and hormonal stimuli (3). In addition to serving as an energy store, adipocytes also secrete hormones (e.g., leptin, adiponectin, resistin) that regulate energy balance, metabolism, and the neuroendocrine response to altered nutrition (4).
The process of adipocyte differentiation is the focus of extensive research and has identified a cascade of transcription factors that are responsible for this conversion (1). Factors that counterregulate adipogenesis include the extracellular-signaling molecules IL-1, TNF-α, and the cell surface protein preadipocyte factor–1 (Pref-1) (5–7). Recent studies have suggested that forkhead transcription factors may also play a regulatory role in adipocyte differentiation and metabolism. Forkhead transcription factors are characterized by their 100–amino acid winged-helix motif that is responsible for monomeric recognition of specific DNA target sites (8). The expression of forkhead transcription factor Foxo-1 is induced in late stages of adipocyte differentiation, and a constitutive active form of Foxo-1 can inhibit adipogenesis in a preadipocyte cell line, suggesting that Foxo-1 functions as a negative regulator of adipocyte differentiation (9). The related factor Foxc-2 is also expressed in preadipocytes and adipocytes, and transgenic mice overexpressing Foxc-2 in adipocytes exhibit altered insulin action, differentiation, metabolism, and sensitivity to adrenergic stimuli that protect them against symptoms related to obesity, including hypertriglyceridemia and diet-induced insulin resistance (10).
The hepatocyte nuclear factor-3 (HNF-3)/forkhead family of transcription factors in mammals include three genes, designated as FOXA1 (HNF-3α), FOXA2 (HNF-3β), and FOXA3 (HNF-3γ) (11). FOXA proteins were first identified as a binding activity to an enhancer region of the transthyretin gene in nuclear liver extracts (12). They are also expressed in the stomach, intestine, pancreas, and lung (13). Mouse genetic studies have revealed important roles of Foxa genes in development and metabolism. Mice with a targeted disruption of the Foxa-1 gene develop a complex catabolic phenotype that is characterized by persistent hypoglycemia, wasting, and neonatal mortality due to insufficient synthesis and secretion of glucagon (13, 14). Mice homologous for a null mutation in Foxa-2 develop an embryonic-lethal phenotype, lack a notochord, and exhibit defects in foregut and neural tube development (15, 16), while heterozygous Foxa-2 mice have normal glucose and energy homeostasis (17). Foxa-3–deficient embryos develop normally with no impairment of glucose homeostasis, but they have decreased expression levels of the rate-limiting enzyme of gluconeogenesis, phosphoenolpyruvate carboxykinase (18).
In this study we investigated the role of Foxa transcription factors in adipocyte differentiation and metabolism. Our data indicate that Foxa-2 is induced de novo in adipose tissue of obese rodents and acts as a crucial inhibitor of adipocyte differentiation and activator of insulin-sensitizing genes in adipocytes, thereby counter-regulating adipogenesis.
Methods
Plasmids.
The hPREF-1 promoter (681 bp) was cloned from a human lambda Fix II library (Stratagene, La Jolla, California, USA). The murine muscle isoform of pyruvate kinase (M2Pk; 989 bp), hexokinase-2 (Hk-2; 771 bp), lipoprotein lipase (Lpl; 1,011 bp), hormone-sensitive lipase (Hsl; 599 bp), and uncoupling protein-2 (Ucp-2; 877 bp) promoters were amplified using site-specific primers (sequence available on request) and genomic DNA as templates and cloned into the pGL2-enhancer vector (Promega Corp., Madison, Wisconsin, USA). The sequences of all the clones were confirmed by dideoxynucleotide sequencing.
Animals and metabolic cages.
All animal models were maintained in a C57Bl/6J background and maintained on a 12-hour light/12-hour dark cycle in a pathogen-free animal facility. Oxygen consumption, CO2, heat production, and food and water intake were simultaneously determined for four mice per experiment in an Oxymax metabolic chamber system (Columbus Instruments, Columbus, Ohio, USA). Individual mice (15 weeks old) were placed in a chamber with an airflow of 0.6 l/min, and one reading per mouse was taken at 4-minute intervals over 24 hours. Resting metabolic parameters were determined by integrating values at periods of no activity.
Electrophoretic mobility-shift assay.
Nuclear extracts were prepared as described with minor modifications (19). Visceral fat from wild-type and ob/ob animals was washed in prechilled PBS supplemented with protease inhibitor cocktail (Roche, Indianapolis, Indiana, USA) and homogenized in sucrose buffer (20 mM HEPES, pH 7.9, 25 mM KCl, 2 M sucrose, 20% vol/vol glycerol, 1 mM EDTA, protease inhibitor cocktail) using a Dounce homogenizer. The homogenized tissue was centrifuged over a sucrose buffer cushion (100,000 g, 40 minutes), and the nuclei were resuspended in lysis buffer (20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, protease inhibitor cocktail, 25% vol/vol glycerol). After 30 minutes, incubation nuclear extracts were centrifuged (45,000 g, 30 minutes) and the supernatant was snap-frozen in liquid nitrogen. Protein content was measured by bichinonic acid test.
Nuclear extracts from ob/ob fat (20 μg) were incubated with 32P-labeled double-stranded oligonucleotide probes with either the wild-type or a mutated putative Foxa-binding sites from the PREF-1, Lpl, and Ucp-2 promoter. The reaction was performed in a mixture containing HEPES buffer (20 mM, pH 7.9), KCl (40 mM), MgCl2 (1 mM), EGTA (0.1 mM), DTT (0.5 mM), 4% Ficoll and poly(dIdC) at room temperature for 10 minutes. Competition analysis was performed by incubating the cellular extracts and the probe with the unlabeled oligonucleotide. Supershift analysis was carried out by incubating the nuclear extracts with either anti–Foxa-1 or anti–Foxa-2 Ab (20). The reaction mixture was loaded on a 6% nondenaturing polyacrylamide gel in TBE buffer (0.023 M Tris-borate, 0.5 mM EDTA) and run at 4°C. Bands were visualized by autoradiography. The sequence of the binding sites are: PREF-1, 5′-GTGTGTAATTATGTGCTTAG-3′; Lpl, 5′-CTTATTTGCATATTTCCAGT-3′; Ucp-2, 5′-CAGGTTGCCTGTTTGTTTTC-3′.
Cell culture.
The 3T3-L1 cells were maintained in DMEM with 4.5 g/l glucose, 10% FCS, 2 mM glutamine, 1 mM pyruvate, and penicillin/streptomycin (Life Technologies Inc., Carlsbad, California, USA) in a humidified incubator at 5% CO2. Cells were subcultured at a split ratio of 1:4. Adipocyte differentiation was induced as described by treating with 1 μM dexamethasone (Sigma-Aldrich, St. Louis, Missouri, USA) and 0.5 mM MIX (Sigma-Aldrich) for 8 days in the presence or absence of insulin (5 nM) (7). Incorporation of lipids was visualized by staining with oil red O (Sigma-Aldrich).
Ab’s.
Anti–Pref-1 Ab’s were generated by synthesis of two peptides: Pref-1-N, CVDLEKGQYECSCTPG, and Pref-1-C, SSFSTSAKPGCPTCAT. Peptides were conjugated to KLH and used for immunization of rabbits (Bethyl Laboratories, Montgomery, Texas, USA). Ab’s were affinity purified and shown to recognize Pref-1 protein in Western blot analysis.
Recombinant adenoviruses.
The rat full-length Foxa-2 cDNA was cloned into vector Ad5CMV K-NpA (ViraQuest Inc., North Liberty, Iowa, USA) and used for the generation of recombinant adenovirus. AdEmpty (ViraQuest Inc.) does not contain a transgene and was used as control. All adenoviruses express the GFP. The lowest titer that was found to transduce more than 95% of cells was determined and used for all experiments.
Transfection and transactivation assays.
The 3T3-L1 cells were grown to 60–70% confluence and subsequently transfected with the reporter genes (0.5 μg), pCMV-β-gal as internal reference (0.5 μg), and the expression vectors for Foxa-1 and Foxa-2 (0.5 μg) or pcDNA3 alone as control, using the transfection reagent Fugene6 according to the manufacturer’s protocol (Roche). Cells were grown for an additional 48 hours after transfection. Luciferase activity was measured using the Luciferase Detection System following the manufacturer’s protocol (Promega Corp.). Luciferase was normalized for transfection efficiency by the corresponding β-gal activity.
Generation of the stable cell lines.
The 3T3-L1 cells were plated at a density of 20,000 cells/cm2 and transfected with the expression vector pcDNA3–Foxa-1, –Foxa-2, and –Pref-1, using Fugene6 (Roche) as transfection reagent. The transfected cells were selected in 350 μg/ml of G418 (Life Technologies Inc.) and approximately 200 G418-resistant clones were pooled and expanded in selection medium. Expression of the stably transfected gene was confirmed by RT-PCR.
RT-PCR.
Total RNA was extracted from cells and embryoid bodies (EBs) with TRIzol, following the manufacturer’s instructions (Life Technologies Inc.). Contaminating genomic DNA was removed from the extract by treating it with 5 U of RNase-free DNase-I (Roche) per 10 μg of RNA. The cDNA was synthesized using Moloney leukemia virus reverse transcriptase with dNTPs and random hexamer primers (Stratagene). The cDNAs provided templates for PCRs using specific primers at annealing temperatures ranging between 60°C and 65°C in the presence of dNTPs, [α-32P]dCTP, and Taq DNA polymerase. PCR synthesis for each primer pair was quantified at 15, 20, 25, and 30 cycles in a test reaction to ensure that the quantitative PCR amplification was in the linear range. The primer sequences used for PCR are available upon request.
Northern blot analysis.
Specific DNA probes were generated using the Highprime DNA-labeling kit following the manufacturer’s instructions (Roche). Total RNA fat tissue was prepared using TRIzol as described by the manufacturer (Life Technologies Inc.) and separated (30 μg per lane) on a 1% agarose gel containing 5% formaldehyde. After blotting onto a positively charged nylon membrane (Schleicher & Schuell Inc., Keene, New Hampshire, USA), the blot was hybridized at 42°C, with the respective probe using Hybrisol hybridization buffer (Intergen Co., Purchase, New York, USA).
Western blot analysis.
Cytosolic protein extracts were separated by SDS-PAGE (13.5%) and transferred onto a nitrocellulose membrane (Schleicher & Schuell Inc.) by electroblotting. Adipocyte fatty acid binding protein was detected with anti-human adipocyte fatty acid–binding protein antiserum (aP2-antiserum; 1:500) (gift of F. Spener, Muenster, Germany) and goat anti-rabbit IgGs conjugated to HRP (1:10,000) in TBS supplemented with 5% nonfat dry milk. Foxa-2 was detected with anti–Foxa-2 antiserum (20) (1:1,000) and goat anti-rabbit IgG conjugated to HRP (1:10,000). All Ab’s were dissolved in 5% milk in TBS with 0.5% Tween-20. The blots were washed three times for 15 minutes between incubations. Membranes were incubated with primary Ab’s overnight at 4°C. Incubations containing the secondary Ab were performed at room temperature for 1 hour. For visualization, the Renaissance Chemiluminescence Substrate (NEN Life Science Products Inc., Boston, Massachusetts, USA) was used.
Preadipocyte and adipocyte isolation and metabolic studies.
Primary adipocytes and preadipocytes were isolated and differentiated as described previously (21, 22). For the assessment of lipogenesis, lipolysis, and glucose metabolism, a 10% isolated mature adipocyte cell suspension at 5 mM glucose was used. Glucose transport of isolated adipocytes was measured by incubation for 30 minutes with 3 μM U-[14C]glucose with or without insulin stimulation (22). The reaction was terminated by spinning through dinonyl phtalathe oil and the radioactivity quantified by scintillation counting. Glucose incorporated into triglycerides, lactate, and CO2 was measured after 2-hour incubation with 3 μM U-[14C]glucose in the absence or presence of 100 nM insulin as described (22, 23). Fatty acid de novo synthesis was analyzed by saponification of total lipids (24) and quantification of radioactive label into fatty acids. Incorporation into lipid glycerol could be calculated by subtracting the fatty acid radioactive label from the total lipid radioactive label. To quantify lipolysis, isolated adipocytes (200 μl of a 10% isolated fat cell suspension) were incubated in the presence of adenosine deaminase and 10 μM N6-[R-(-)-1-methyl-2-phenyl]adenosine, with or without 100 μM isoproterenol to produce maximal increase of lipolysis for 30 minutes in the presence or absence of insulin. Glycerol content of the incubation medium was determined using a radiometric assay as previously described (25).
Statistical analysis.
Results are given as mean plus or minus SD. Statistical analyses were performed by using a Student t test, and the null hypothesis was rejected at the 0.05 level. Linear regression was calculated using Origin (Microcal Software Inc., Northampton, Massachusetts, USA).
Results
De novo expression of Foxa-2 in adipocytes of obese mice.
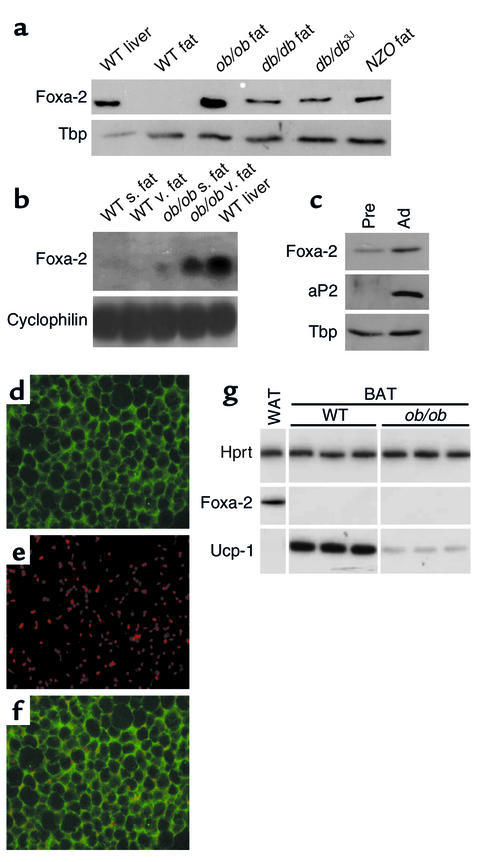
Gene expression analysis of genes encoding transcription factors revealed that the forkhead transcription factor Foxa-2 (HNF-3β), undetectable in fat tissue of wild-type mice, was expressed in adipose tissue of obese mice. Of the three Foxa genes, only Foxa-2 transcripts could be detected in adipose tissue of obese (ob/ob) mice (Figure 1a, data not shown). Expression of Foxa-2 was also found in other monogenic (db/db) and polygenic (NZO) animal models of obesity (Figure 1, a and b). Moreover, Foxa-2 was expressed in fat tissue of wild-type mice in which obesity was induced with a high-fat diet (see Figure 7b and Figure 8g). Foxa-2 was expressed in visceral and subcutaneous fat of ob/ob mice, but was enriched in visceral fat depots (Figure 1b). De novo expression of Foxa-2 was specific for adipocytes and was not observed in other insulin-sensitive tissues such as muscle (data not shown). Both the adipocyte fraction and the stromal fraction of adipocyte tissue containing preadipocytes expressed the Foxa-2 protein (Figure 1c). Foxa-2 protein was detected in the nucleus and cytoplasm of adipocytes of obese animals by immunohistochemistry analysis (Figure 1, d–f). Foxa-2 expression was not found in brown adipose tissue of lean or obese mice (Figure 1g).
Figure 1.
Foxa-2 is expressed in adipose tissue of obese mice. (a) Liver cell extracts from wild-type mice and adipocyte extracts from wild-type, ob/ob, db/db, and NZO mice were separated by SDS-PAGE and analyzed by Western blotting for Foxa-2 expression. TATA-binding protein (Tbp) expression was measured as a control for loading. (b) Visceral (v.) and subcutaneous (s.) fat RNA from wild-type and ob/ob mice was analyzed for Foxa-2 expression by Northern blotting. Membrane was rehybridized with a probe for cyclophilin as control. (c) Preadipocyte (Pre) and adipocyte (Ad) protein extracts from ob/ob mice were separated by SDS-PAGE and analyzed by Western blotting for Foxa-2 and aP2 expression. (d–f) Confocal image immunostaining of visceral fat from ob/ob mouse using anti–Foxa-2 Ab’s (d) and TO-PRO-3 (molecular probes) for nuclear staining (e). Images were superimposed (f). Foxa-2 protein is present in cytoplasm and nuclei. (g) Semiquantitative RT-PCR of Foxa-2 and Ucp-1 in white (WAT) and brown (BAT) adipose tissue of three ob/ob and three wild-type littermate mice. Hprt, hypoxanthine phosphoribosyltransferase.
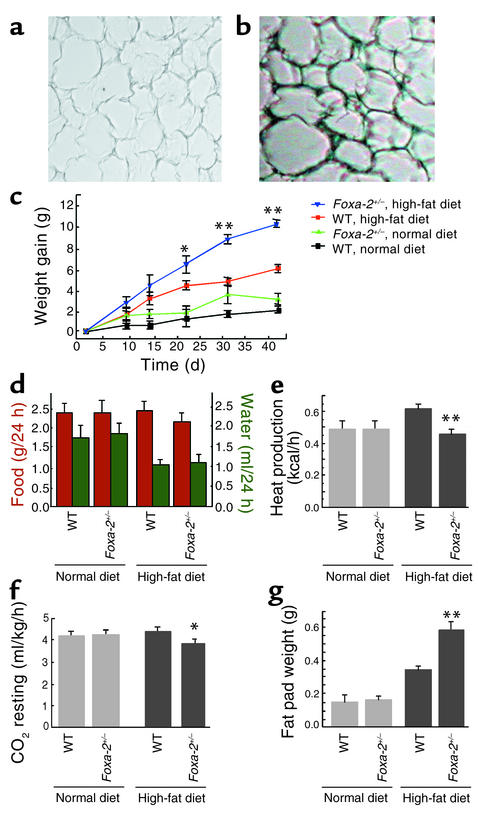
Figure 7.
Development of diet-induced obesity in Foxa-2+/– mice. (a and b) X-gal staining of adipose tissue of wild-type (a) and Foxa-2+/– (b) mice after 6 weeks of a high-fat diet. Absolute weight gain (c), food and water intake (d), heat production (e), resting CO2 production (f), and epidydimal fat pad weight (g) of Foxa-2+/– and wild-type littermates on chow and high-fat diets are shown. *P < 0.05; **P < 0.01.
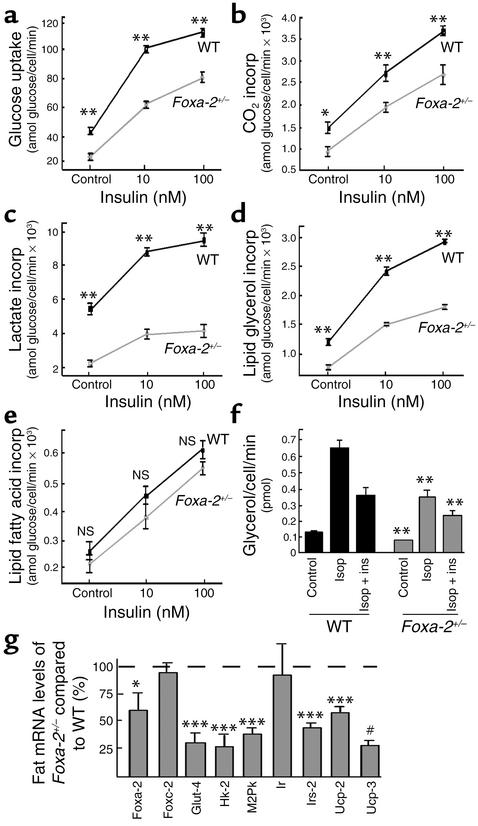
Figure 8.
Altered insulin sensitivity in primary adipocytes of Foxa-2+/– and wild-type littermates. Primary adipocytes were isolated from Foxa-2+/– and wild-type littermate animals that had received a high-fat diet for 42 days. Parameters of glucose metabolism, lipogenesis and lipolysis were measured. (a–e) Glucose metabolism into different pathways at 10 and 100 nM insulin in isolated adipocytes from Foxa-2+/– (gray) and wild-type (black) littermates. [U-14C]Glucose uptake in isolated adipocytes from epidydimal fat (a), [U-14C]glucose incorporation (incorp) into CO2 (b), lactate (c), lipid glycerol (d), and lipid fatty acids (e), measured after 2 hours of incubation in the absence (control) or presence of insulin. (f) Glycerol release from adipocytes in the presence or absence of insulin (ins) after stimulation of lipolysis with isoperenterol (Isop). (g) Measurements of relative gene expression levels of metabolic genes in adipocytes of Foxa-2+/– and wild-type littermates (100%) using semiquantitative RT-PCR. All mice were female, 15 weeks of age, n = 7, means ± SD. *P = 0.05; **P = 0.01; ***P = 0.001; #P = 10–5.
Foxa-2 inhibits of adipocyte differentiation by activation of Pref-1.
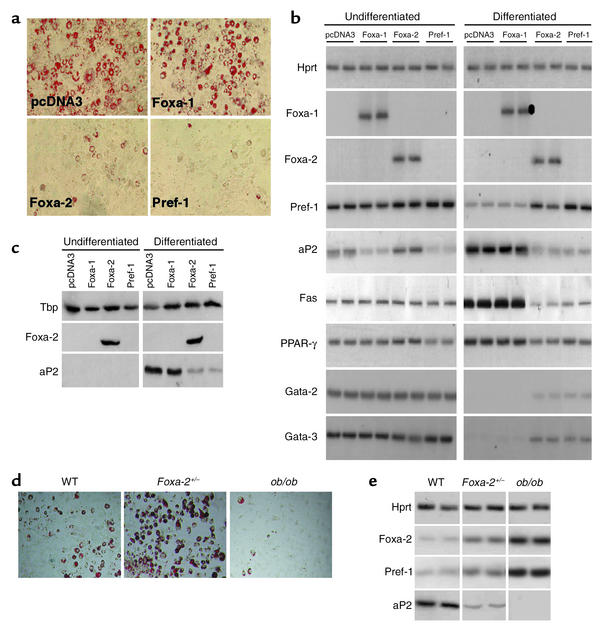
To study the physiological role of Foxa-2 we generated preadipocyte (3T3–L1) cell lines that express Foxa-1, Foxa-2, or Pref-1. All genes were expressed under the control of a constitutive promoter. After selection with neomycin, cells were induced to differentiate (Figure 2a). Immunoblot analysis and RT-PCR analysis revealed that Foxa-1 and Foxa-2 were expressed in transfected 3T3-L1 cells, but absent in untransfected cells (Figure 2, b and c). Expression of Foxa-2 or Pref-1 inhibited adipocyte differentiation in the presence of a prodifferentiation medium (Figure 2a). In contrast, cells expressing Foxa-1 or the empty expression vector (pcDNA3) were able to accumulate lipid droplets (Figure 2a). Consistent with the morphological differentiation, ectopic expression of Foxa-2 prevented the downregulation of Pref-1, Gata-2, and Gata-3, all of which have been shown to inhibit adipocyte differentiation (Figure 2b) (2, 26). Foxa-2 expression inhibited the induction of late markers of adipocyte differentiation such as PPARγ, aP2, and fatty acid synthase (Figure 3, b and c). We also isolated primary preadipocytes of Foxa-2+/–, wild-type littermates, and ob/ob mice and studied their ability to differentiate in vitro. Differentiated cells of the stromal vascular fraction of Foxa-2+/– animals showed significantly increased accumulation of lipid droplets compared with wild-type and ob/ob mice (Figure 2d). The differentiation status correlated well with expression levels of Foxa-2 and Pref-1, which were highest in ob/ob adipocytes, intermediate in wild-type cells, and lowest in Foxa-2+/– cells. Furthermore, we determined that expression levels of Foxa-2 and Pref-1 in preadipocytes are higher in ob/ob compared with wild-type animals (data not shown). Expression levels of aP2, a late marker of adipocyte differentiation, showed an inverse correlation to Foxa-2 and Pref-1 levels (Figure 2e). Taken together, these results indicate that Foxa-2 is a negative regulator of adipocyte differentiation.
Figure 2.
Foxa-2 is an inhibitor of adipocyte differentiation. (a) Inhibition of adipocyte differentiation in 3T3-L1 cells. Cells were transfected with vector pcDNA3 (control) or expression vectors containing cDNAs of Foxa-1, Foxa-2, and Pref-1. Pools of stable transfectants were induced with differentiation medium (not containing insulin). At day 8 after induction, cells were either stained for lipid accumulation using oil red O or mRNA, and total protein extracts were prepared. (b) Measurements of gene expression profiles using RT-PCR. Hprt expression was used as a loading control indicating that each sample contained similar amounts of mRNA. No products were amplified in the absence of reverse transcriptase. (c) Western blot analysis of cell extracts from undifferentiated and differentiated 3T3-L1 cell lines. Total protein was separated by SDS-PAGE and analyzed by immunoblotting for Foxa-2 and aP2 expression. TATA-binding protein (Tbp) expression was measured as a loading control. (d) Differentiation of primary preadipocytes of wild-type, mutant Foxa-2 (Foxa-2+/–), and ob/ob cells. (e) Gene expression of Foxa-2, Pref-1, and aP2 in differentiated primary preadipocytes of Foxa-2+/–, wild-type, and ob/ob mice.
Figure 3.
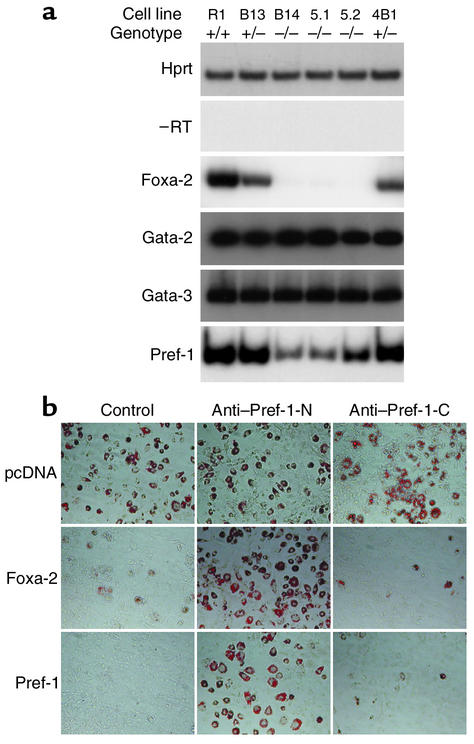
Foxa-2 regulates the expression of Pref-1 and inhibits adipocyte differentiation. (a) Pref-1 expression is reduced in Foxa-2–deficient EBs. Wild-type (R1; +/+), heterozygous (B13, 4B1; +/–), and homozygous (B14, 5.1, 5.2; –/–) Foxa-2 ES cells were differentiated to EBs as described previously (27) and assayed for Hprt, Foxa-2, Gata-2, Gata-3, and Pref-1 mRNA expression by RT-PCR. –RT, reaction in absence of RT. (b) Anti–Pref-1 Ab’s render Foxa-2–overexpressing 3T3-L1 preadipocytes sensitive to differentiation. The 3T3-L1 cells that were transfected with empty vectors (pcDNA3), pCMV-Foxa-2 (Foxa-2), or pCMV-Pref-1 (Pref-1) were differentiated in the absence (Control) or presence of anti–Pref-1 Ab’s that were directed against the extracellular (Anti–Pref-1-N) or intracellular (Anti–Pref-1-C) domains.
To investigate if Foxa-2 is a direct activator of the Pref-1, Gata-2, or Gata-3 genes, we compared the expression of these genes in differentiated wild-type (Foxa-2+/+), heterozygous (Foxa-2+/–), and null (Foxa-2–/–) ES cells (27). The wild-type and mutant Foxa-2 ES cells were differentiated to EBs, and gene expression was measured by semiquantitative RT-PCR. No differences in Gata-2 and Gata-3 gene expression were found in EBs of different Foxa-2 genotypes (Figure 3a). In contrast, Pref-1 expression was markedly reduced in Foxa-2–/– EBs compared with wild-type and Foxa-2+/– cells. To explore if the Pref-1 gene is a direct target of Foxa-2 we analyzed its promoter. The Pref-1 transcription start site was mapped by 5′RACE, and a 680-bp fragment of 5′ regulatory sequence was cloned into a reporter vector containing the luciferase gene (pPref-Luc). Sequence analysis of the promoter sequences identified two Foxa-binding sites at position –621 and –316 that were highly conserved between mouse and human (5′-GTGTAATTATGT-3′ and 5′-ACATGTGTGCGT-3′, respectively). Expression of Foxa-2 in 3T3-L1 cells in the presence of reporter construct pPref1-Luc, which contained a 680-bp promoter sequence upstream of the luciferase gene, revealed a sixfold activation compared with cells that do not express Foxa-2 (see Figure 6a). Deletion of the upstream Foxa element reduced the transcriptional activity 50% (data not shown), suggesting that this element is important for Pref-1 gene expression. In electrophoretic mobility shift assay (EMSA) analysis, 32P-labeled oligonucleotides that contain the upstream Foxa-binding site were able to bind to Foxa-2 protein from nuclear extracts of adipocytes from obese mice. Taken together, these data suggest that Foxa-2 is a direct transcriptional activator of the Pref-1 gene.
Figure 6.
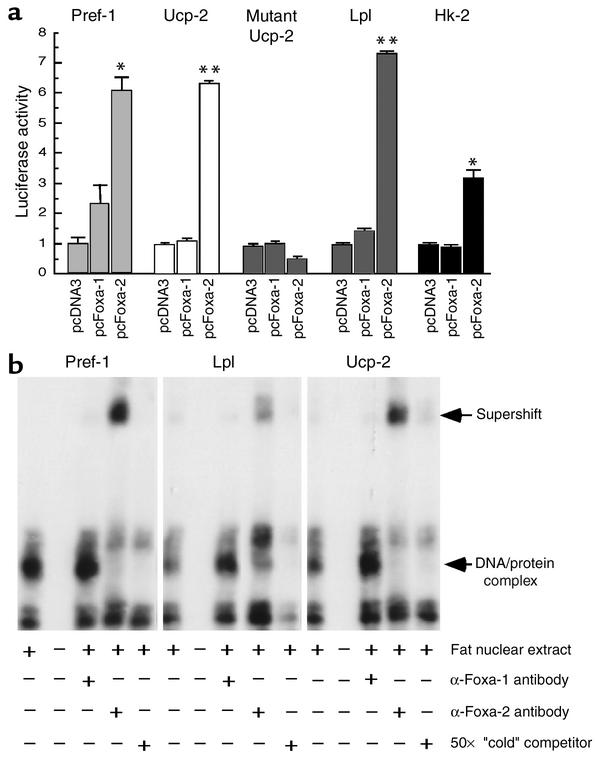
Foxa-2 is a transcriptional activator of Pref-1, Ucp-2, Lpl, and Hk-2 gene promoters in vitro. (a) Foxa-2 transactivates the promoters of Ucp-2, Lpl, Pref-1, and Hk-2 in 3T3-L1 cells. The 3T3-L1 cells were transfected with the indicated expression vectors, pCMV–β-gal, and the luciferase reporter genes. Luciferase activity was normalized to β-gal activity. Each value represents mean of nine independent experiments ± SD. *P < 0.01; **P < 0.001. (b) Foxa-2 from ob/ob fat nuclear extract binds to sites in the Pref-1, Lpl, and Ucp-2 promoters. 32P-labeled probes corresponding to putative Foxa-2–binding sites in the promoters of Pref-1, Lpl, and Ucp-2 were incubated with nuclear extracts from ob/ob fat in the presence of either unlabeled probe, anti–Foxa-1, or anti–Foxa-2 Ab (21). Protein/DNA complexes were separated on a 4% acrylamide gel and visualized by autoradiography. The radioactive probes (bottom of the gel) are not shown.
Constitutive expression of Pref-1 is known to inhibit 3T3-L1 adipocyte differentiation (7), and downregulation of Pref-1 expression promotes adipogenesis (28). Inhibition of adipocyte differentiation by Pref-1 is mediated by a proteolytically processed, approximately 50-kDa soluble form of the extracellular domain of the native Pref-1 protein (29). Ab’s against this ectodomain can block the inhibitory effects of Pref-1 on adipocyte differentiation (30). To investigate if Foxa-2 is able to inhibit adipogenesis in the absence of functional Pref-1 we differentiated 3T3-L1 cells that were transfected with pcDNA3 (control) Foxa-2 or Pref-1 expression vectors in the absence or presence of anti–Pref-1 Ab’s that recognize epitopes in the extracellular (anti–Pref-1-N) or intracellular (anti–Pref-1-C) domains of Pref-1. Anti–Pref-1 Ab’s directed against the extracellular domain were able to revert the inhibition of adipocyte differentiation in Foxa-2 and Pref-1 overexpressing 3T3-L1 cells. Furthermore, these cells could not be differentiated when grown in the absence of anti–Pref-1 Ab’s (control) or in the presence of an anti–Pref-1 Ab directed against an intracellular epitope of the Pref-1 protein (Figure 3b). This suggests that Foxa-2 inhibits adipocyte differentiation through activation of Pref-1 gene expression.
Growth hormone increases Foxa-2 expression in primary preadipocytes.
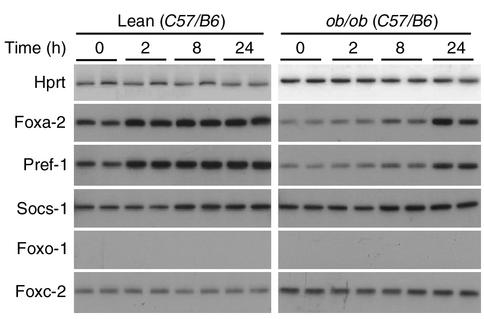
Growth hormone (GH) has been shown to be a potent inhibitor of differentiation in primary preadipocytes. This antiadipogenic activity is mediated by a GH-dependent activation of Pref-1, a known inhibitor of adipocyte differentiation (7, 21). To test the hypothesis that GH-dependent induction of Pref-1 expression is mediated by Foxa-2 we isolated primary preadipocytes from lean C57/B6 and ob/ob mice and cultured these cells in the presence or absence of growth hormone (500 ng/ml). Gene expression profiles of Pref-1, Foxa-2, Foxc-2, Foxo-1, and Socs-1, a known target gene of GH, were measured in stromal-vascular cells after 2, 8, and 24 hours. We found that Foxa-2, Pref-1, and Socs-1 mRNA levels in preadipocytes of lean mice were induced twofold in stromal vascular cells that were treated with GH for 2 hours (Figure 4). Expression levels of these genes were also induced in preadipocytes of ob/ob mice, although induction occurred somewhat later and was strongest after 24 hours. In contrast, we did not detect significant expression levels of Foxo-1 in stromal vascular cells, and mRNA levels of Foxc-2 did not change with GH treatment (Figure 4). Taken together, these data indicate that Foxa-2 is a target gene of GH in primary preadipocytes and that the GH-dependent inhibitory activity on adipocyte differentiation may be mediated by induction of Foxa-2 and Pref-1 expression.
Figure 4.
GH stimulates the expression of Foxa-2 in primary preadipocytes of lean and obese mice. Analysis of gene expression in the stromal vascular fraction (containing preadipocytes) of wild-type (lean, C57/B6) and ob/ob mice that were cultured in the absence (0) or presence of 500 ng/ml growth hormone for 2, 8, and 24 hours. Foxa-2 and Pref-1 expression levels were increased (2.5 and 3.5, respectively; P ≤ 0.05) in 2-, 8-, and 24-hour time points compared with untreated cells. Socs-1 expression was increased at 8 and 24 hours (1.9 ± 0.2; P < 0.05). Expression levels in ob/ob preadipocytes were significantly increased after 8- and 24-hour stimulation compared with unstimulated cells (Foxa-2: 2.6 ± 0.2 and 4.8 ± 0.4, P ≤ 0.05 and P ≤ 0.01, respectively; and Pref-1: 2.4 ± 0.3 and 2.3 ± 0.1, respectively, P ≤ 0.05). Socs-1 was increased at 8 and 24 hours (1.9 ± 0.1 and 2.1 ± 0.3, respectively, P < 0.05). No significant changes in expression levels of Foxc-2 were detected.
Foxa-2 activates expression of insulin-sensitizing genes in adipocytes.
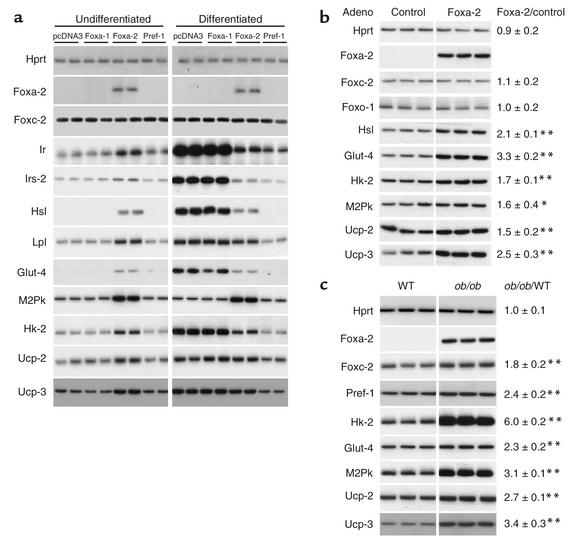
Foxa proteins regulate the expression of many metabolic genes through its interaction with specific binding sites in promoters/enhancers that lead to chromatin remodeling and transcriptional activation (31, 32). To test if Foxa-2 is a transcriptional regulator in adipocytes, expression analysis of genes that have putative Foxa-binding sites in their 5′-regulatory sequences were performed in Foxa-2–expressing preadipocytes and in differentiated adipocytes (Figure 5a). We found that mRNA levels of the insulin receptor (Ir), insulin receptor substrate-2 (Irs-2), Hsl, Lpl, glucose transporter-4 (Glut-4), M2Pk, Hk-2, and Ucp-2 and Ucp-3 were increased in 3T3-L1 cells that expressed Foxa-2 (Figure 5a). The expression of Ucp-1, mitochondrial ATP-citrate lysase, GAPDH, acyl-CoA carboxylase, Srebp-1c, mitochondrial pyruvate decarboxylase, mitochondrial carnitine transporter, and Irs-1 were unaffected by Foxa-2 (data not shown). The observed expression changes were Foxa-2 specific since they are not present in preadipocytes expressing Foxa-1 (Figure 5a). Differentiation of cells transfected with pcDNA3 (control) or expressing Foxa-1 led to a differentiation-dependent increase in the expression of Ir, Irs-2, Hsl, Lpl, Glut-4, Hk-2, Ucp-2, and Ucp-3. Foxa-2 expression blocked adipocyte differentiation and thereby prevented the induction of these genes. These observations underline the important effect of the status of adipocyte differentiation state on gene expression. Interestingly, we found that steady-state mRNA levels of M2Pk were not affected by the differentiation state of 3T3-L1 cells, and expression levels remained markedly increased in Foxa-2–expressing cells. This suggests that Foxa-2 is also an important activator of metabolic genes whose expression is not affected by adipocyte differentiation per se.
Figure 5.
Foxa-2 regulates genes involved in glucose uptake, glycolysis, lipolysis, and energy dissipation. (a) Undifferentiated and differentiated 3T3-L1 cells containing the indicated expression vector were assayed for Hprt, Foxa-2, Foxc-2, Ir, Irs-2, Hsl, Lpl, Glut-4, M2Pk, Hk-2, Ucp-2, and Ucp-3 mRNA expression by RT-PCR. Fold regulation in undifferentiated, control 3T3-L1 cells (pcDNA3) versus Foxa-2–expressing cells: Ir: 3.3 ± 0.5*; Irs-2: 1.7 ± 0.3*; Lpl: 2.6 ± 0.4**; M2Pk: 3.3 ± 0.2***; Hk-2: 3.2 ± 0.3***; Ucp-2: 2.1 ± 0.7*; Ucp-3: 2.3 ± 0.4*. Expression levels of M2Pk were increased (3.4 ± 0.2**) in differentiated 3T3-L1–expressing Foxa-2 compared with control (pcDNA3). (b) Increased expression of metabolic genes in differentiated 3T3-L1 cells with enforced expression of Foxa-2 by adenoviral (Adeno) transduction. The 3T3-L1 cells were differentiated for 7 days and infected with control (no transgene) and recombinant adenovirus expressing Foxa-2. Cells were harvested after 48 hours, and gene expression was measured by RT-PCR. Experiments were carried out in triplicate. (c) Increased expression of metabolic enzymes in adipose tissue of ob/ob mice. Gene expression in fat tissue in ob/ob animals and wild-type littermates was measured by RT-PCR. Each lane indicates a different animal. Quantitative measurements of gene expression were obtained by densitometry, and ob/ob/WT indicates the ratio of adipocyte mRNA expression levels of the means of WT and ob/ob mice. The levels of significance of the comparison of WT versus ob/ob are shown on the right. *P = 0.05; **P = 0.01; ***P = 0.005.
Since Foxa-2 has strong effects on adipocyte differentiation, which is known to influence gene expression, we studied putative target genes of Foxa-2 independent of adipocyte differentiation. The 3T3-L1 cells were differentiated and transduced with either a recombinant adenovirus expressing Foxa-2 or a control adenovirus. Gene expression was analyzed after 48 hours by RT-PCR. We found a significant increase in the expression of Hsl, Glut-4, Hk-2, M2Pk, Ucp-2, and Ucp-3 in adipocytes expressing Foxa-2 compared with cells that were infected with the control virus (Figure 5b). These results were consistent with gene expression profiles in stable transfected 3T3-L1 cells, suggesting that Foxa-2 increases the expression of these genes in preadipocytes and adipocytes. We also determined the expression levels of these genes in adipocytes of ob/ob mice and lean littermate controls. A striking correlation was observed between Foxa-2–expressing adipocytes of ob/ob animals and increased expression of putative Foxa-2 target genes, including Pref-1, Hk-2, M2Pk, Glut-4, and Ucp-2 and Ucp-3 (Figure 5c).
To further explore if Foxa-2 can directly activate the above-mentioned genes we characterized their promoters. The promoters of Ucp-2, Lpl, Hk-2, and Pref-1 were cloned upstream of a luciferase reporter gene, and transcriptional activation was assayed in the absence (pcDNA3) and presence of Foxa-1 and Foxa-2. We detected a dose-dependent activation of all promoters when cotransfected with a plasmid expressing Foxa-2 (Figure 6a). Transactivation of the reporter gene was completely lost when the Foxa-binding site in the Ucp-2 promoter was selectively mutated, suggesting that the Foxa-binding site in the Ucp-2 promoter is functionally important (Figure 6a). EMSAs were performed to investigate whether Foxa-2 can bind to the putative binding sites in Ucp-2, Lpl, and Pref-1 promoters (Figure 6b). Nuclear extracts of adipose tissue from wild-type and ob/ob mice were prepared and tested for DNA-binding activity with 32P-labeled oligonucleotides that contained the Foxa-binding sites of the respective promoters. A major DNA/protein complex was detected with nuclear extracts from ob/ob but not wild-type mice (Figure 6b, data not shown). This binding activity could be competed by an unlabeled excess of “cold” Foxa binding oligonucleotides. Furthermore, supershifts of the complexes were detected after preincubation of the complexes with a monospecific Foxa-2 Ab but not with an anti–Foxa-1 antiserum. The supershifts of the DNA/protein complexes were almost complete, suggesting that Foxa-2 is a major forkhead transcription factor that binds to these sites in adipose tissue of ob/ob animals.
Increased obesity and decreased glucose metabolism in adipocytes of diet-induced Foxa-2+/– mice.
Comparisons of gene expression profiles in 3T3-L1 cells that either express or lack Foxa-2 indicate that Foxa-2 is a potent transcriptional activator of genes involved in adipocyte differentiation (Pref-1), glucose uptake (Glut-4), glucose metabolism (Hk-2, M2Pk), insulin signaling (Ir, Irs-2), lipid metabolism (Hsl), and, possibly, energy dissipation (Ucp-2, Ucp-3) and can be predicted to influence adipogenesis (33–36). To test this hypothesis in vivo we studied mutant Foxa-2 mice that have one inactivated Foxa-2 allele (Foxa-2+/–) by targeted insertion of the LacZ gene (15). We chose to study haploinsufficient Foxa-2 mice because Foxa-2–null mice have an early embryonic lethal phenotype (at approximately embryonic day 7.5), and heterozygous mice exhibit normal glucose and lipid metabolism (15–17). Foxa-2+/– mice and wild-type littermates were fed a high-fat (55% fat) diet and studied metabolically. RT-PCR and X-gal staining of fat from Foxa-2+/– animals confirmed that these mice lacked Foxa-2 in adipocytes at the beginning of the study but induced expression during 7 weeks of high-fat diet (Figure 7, a and b; data not shown). Fasting blood glucose, insulin, TNF-α, free fatty acid, and triglyceride levels were similar between Foxa-2+/– and control animals (data not shown). Mice on a high-fat diet increased their fasting plasma insulin levels approximately fourfold compared with animals on a chow diet. Foxa-2+/– mice exhibited a markedly increased weight gain compared with control mice when kept on a high-fat diet (10 g versus 6 g after 6 weeks, respectively) despite similar food and water intake and physical activity (Figure 7, c and d). Resting heat and CO2 production was diminished, however, in Foxa-2+/– mice, indicating that they are hypometabolic (Figure 7, e and f). Foxa-2+/– animals developed noticeably increased pericardial, intraperitoneal, and subcutaneous fat deposits compared with littermate animals. The adipose mass of the epidydimal fat pad was approximately double in Foxa-2+/– mice compared with littermate controls after a 7-week high-fat diet (Figure 7g). The increase in adipocyte mass in Foxa-2+/– mice was due to an increase in fat cell number; the size distribution of adipocytes was similar between mutant and wild-type animals (data not shown). To test if adipocytes of Foxa-2+/– mice have altered glucose metabolism we studied [U-14C]glucose uptake and metabolism into lactate, CO2, lipid glycerol, and fatty acids. Glucose uptake and glucose incorporation into CO2, lactate, and triglyceride glycerol was strikingly reduced in adipocytes in Foxa-2+/– mice compared with wild-type littermates (Figure 8, a–d). In contrast, no differences in adipocyte metabolism were observed in mutant and wild-type adipocytes of lean mice (that do not express Foxa-2) on a normal chow diet (data not shown). Adipocytes of Foxa-2+/– mice did not exhibit a significant reduction of glucose incorporation into fatty acids, a finding that is consistent with similar expression of genes of the fatty acid synthesis in Foxa-2+/– and control adipocytes (Figure 8e; data not shown). We measured reduced glycerol release from adipocytes of Foxa-2+/– mice, however, after maximal stimulation with isoproterenol and following inhibition with insulin, suggesting that lipolysis is decreased in adipocytes of Foxa-2+/– compared with control littermates (Figure 8f). The defect in metabolism in Foxa-2+/– adipocytes correlated with reduced expression of Foxa-2 target genes. Steady-state mRNA levels of Foxa-2, Glut-4, Hk-2, M2Pk, Irs-2, Ucp-2, and Ucp-3 were reduced approximately 50% and more in fat cells of diet-induced obese Foxa-2+/– compared with diet-induced obese wild-type mice (Figure 8g). Taken together, these data demonstrate that Foxa-2 is an important metabolic regulator of glucose metabolism and energy dissipation in adipocytes of diet-induced obese mice.
Discussion
During states of positive energy balance, hormonal and other factors are believed to provide stimuli for the conversion of preadipocytes into mature fat cells and to increased lipid accumulation in adipocytes (1). The proadipogenic actions of several hormones and transcription factors have been well studied and may contribute to the pathogenesis of obesity by enhancing adipocyte differentiation and modulation of insulin sensitivity in adipocytes (3, 4). Counter-regulatory mechanisms that enhance the effects of insulin signaling in adipocytes and protect mammals from extreme fat accumulation may also exist but have not been well studied. In this report we show that Foxa-2 is induced de novo in adipocytes of monogenic, polygenic, and diet-induced rodent models of obesity. This transcriptional induction of Foxa-2 is adipocyte specific and may be an important counterregulatory mechanism to prevent excessive adipogenesis. This notion is supported by the potent inhibitory effect of Foxa-2 on adipocyte differentiation in vitro, transcriptional activation of genes involved in adipocyte metabolism, and a genetic haploinsufficiency in Foxa-2 that leads to increased adipogenesis in diet-induced obesity and defects in adipocyte glucose metabolism. Our data therefore identify Foxa-2 as an obesity-induced gene that mediates a negative-feedback mechanism on adipocyte differentiation and a feed-forward mechanism on gene expression of glucose and lipid-metabolizing genes.
It has been suggested recently that Foxc-2, a winged-helix transcription factor, can counteract obesity and diet-induced insulin resistance. Transgenic mice overexpressing Foxc-2 in adipose tissue exhibit a lean and insulin-sensitive phenotype and a pleiotropic effect on gene expression that includes an induction of the brown adipocyte marker Ucp-1 (10). A constitutive active form of forkhead transcription factor Foxo-1 can also prevent the differentiation of preadipocyte cell lines (9). Several differences exist in the physiological properties of these transcription factors. Foxc-2 is expressed in primary preadipocytes and adipocytes of lean and obese mice and is not markedly induced in obesity. We could not detect Foxo-1 expression in stromal vascular cells, and expression of Foxo-1 seems to be restricted to differentiated adipocytes. Foxa-2, however, is expressed in preadipocytes of lean and obese mice, and expression in adipocytes is restricted to obese animals. Furthermore, GH stimulation leads to the activation the Foxa-2 and Pref-1 in preadipocytes, whereas expression levels of Foxc-2 and Foxo-1 are unresponsive to GH stimulation.
In contrast to Foxa-2+/– animals, Foxo-1+/– mice do not develop increased obesity on a high-fat diet compared with wild-type mice and are protected against the development of diet-induced insulin resistance (9). Opposite roles of Foxa-2 and Foxo-1 have been found in pancreatic β cells (where Foxa-2 acts as a transcriptional activator of the Pdx-1 gene), whereas Foxo-1 seems to inhibit Pdx-1 activity (37–39). It is possible that the resistance of haploinsufficient Foxo-1 mice have in developing diabetes results primarily from reduced expression of Foxo-1 in the liver and pancreatic islets, thereby repressing gluconeogenesis and maintaining pancreatic islet mass and insulin secretion (40). Foxo-1 and Foxa-2 are believed to bind to similar binding sites in promoters of target genes and regulate their expression. Differences in the biological functions of these forkhead transcription factors are incompletely understood but may be attributed to differential activation of Foxa-2 and Foxo-1 by coactivators/repressors, low expression levels of Foxo-1 relative to Foxa-2 in adipocytes of obese animals, and differences in the regulation of transcriptional activity by posttranscriptional modifications (e.g., phosphorylation) through extracellular stimuli.
Our results suggest that Foxa-2 is a potent inhibitor of adipocyte differentiation in vitro and that this effect is mediated by transcriptional activation of the Pref-1 gene by Foxa-2. We found increased expression levels of Foxa-2 and Pref-1 in primary preadipocytes of ob/ob animals compared with wild-type littermates, which may explain the reduction in differentiation potential of preadipocytes from ob/ob mice (ref. 41; Figure 2, d and e). The increased ability of Foxa-2+/– preadipocytes to differentiate compared with wild-type cells also provides further evidence for a role of Foxa-2 in adipocyte differentiation in vivo. Foxa-2 can activate Pref-1 gene expression in vitro, and transcriptional activation is likely mediated by two conserved Foxa-binding sites in the Pref-1 promoter. Pref-1 is required for the inhibitory actions of Foxa-2 on adipocyte differentiation since Ab’s against the bioactive, soluble form of Pref-1 can prevent the block of adipocyte differentiation in 3T3-L1 cells with constitutive expression of Foxa-2 (30). Interestingly, GH has been shown to exhibit antiadipogenic activity in primary preadipocytes by increasing the expression levels of Pref-1. In primary preadipocytes and 3T3-L1 cells expression of Pref-1 is high and decreases to undetectable levels after differentiation. Forced expression of Pref-1 in 3T3-L1 cells by stable transfection renders these cells resistant to differentiation, indicating that downregulation of Pref-1 in preadipocytes is required for differentiation to occur (7). The reduced expression of Pref-1 expression is probably an early event in the differentiation process, since several cytokines (e.g., IL-11, TNF-α, FGF) that inhibit adipocyte differentiation reduce the expression of PPAR-γ, but do not increase Pref-1 expression (6). In this study we tested if Foxa-2 may mediate the actions of GH since it was shown previously that GH increases the expression of Pref-1 in preadipocytes and pancreatic islets (21, 42). We show that GH stimulates the expression of Foxa-2 in preadipocytes of lean and obese mice and that the induction of Pref-1 expression coincides with Foxa-2 induction. Furthermore, the stimulation of Pref-1 expression seems to be the result of a direct transcriptional activation of the Pref-1 gene through a highly conserved Foxa-binding site in the promoter. Taken together, these data indicate that the inhibitory action of GH on adipocyte differentiation is mediated by Foxa-2.
The observed inhibitory effects of Foxa-2 on adipocyte differentiation in vitro are reflected by ex vivo primary preadipocytes from haploinsufficient Foxa-2 mice that show increased differentiation. In vivo, we found a profound increase in fat mass in mutant Foxa-2 mice compared with wild-type littermates that were fed a high-fat diet. It is likely that this increased adipocyte differentiation contributes to the observed phenotype. In addition, we found that mutant Foxa-2 mice had comparable food and water intake but exhibited significant reductions in resting CO2 and heat production. This suggests that mutant Foxa-2 animals increase fat deposits through impaired metabolism. This notion is supported by profound changes in insulin-mediated glucose uptake and metabolism. Furthermore, these physiological changes correlate with reductions in gene expression involved in glucose uptake and metabolism.
It should be noted that the stimuli that induce the de novo expression of Foxa-2 in mature adipocytes of obese mice are currently unknown. We have ruled out several candidate factors whose expression correlates with obesity, including TNF-α, adiponectin, dexamethasone, fatty acids, and IL-6. The identification of the elusive factor(s) may be a novel way to counteract adipogenesis by inhibiting adipocyte differentiation and by enhancing the expression of glucose-metabolizing genes and possibly improving insulin sensitivity in adipocytes. Furthermore, it will be of interest to investigate if genetic or environmental factors exist that affect the “physiological” threshold of Foxa-2 induction in the development of obesity since Foxa-2 expression levels in adipocytes could influence the development/progression of obesity. We propose that the Foxa-2 regulatory pathway of adipocyte differentiation and metabolism may serve as a new target for pharmaceutical interventions of obesity and diabetes.
Acknowledgments
We would like to thank J. Friedman for advice. This work was supported by NIH grant RO1 DK55033-04 (M. Stoffel) and Medical Scientists Training Program grant GM07739 (D.Q. Shih), the Deutsche Forschungsgemeinschaft (C. Wolfrum), and the Human Science Frontier Organization (M. Stoffel).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: protein preadipocyte factor–1 (Pref-1); hepatocyte nuclear factor 3 (HNF-3); hexokinase-2 (Hk-2); muscle isoform of pyruvate kinase (M2Pk); lipoprotein lipase (Lpl); hormone-sensitive lipase (Hsl); uncoupling protein-2 (Ucp-2); embryoid bodies (EBs); adipocyte fatty acid–binding protein (aP2); electrophoretic mobility shift assay (EMSA); growth hormone (GH); insulin receptor (Ir); insulin receptor substrate-2 (Irs-2); glucose transporter-4 (Glut-4).
References
- 1.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 2.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 3.Ahima RS, Flier JS. Adipose tissue as an endocrine organ. Trends Endocrinol. Metab. 2000;11:327–332. doi: 10.1016/s1043-2760(00)00301-5. [DOI] [PubMed] [Google Scholar]
- 4.Felber JP, Golay A. Pathways from obesity to diabetes. Int. J. Obes. Relat. Metab. Disord. 2002;26(Suppl. 2):S39–S45. doi: 10.1038/sj.ijo.0802126. [DOI] [PubMed] [Google Scholar]
- 5.Gregoire F, et al. Interferon-gamma and interleukin-1 beta inhibit adipoconversion in cultured rodent preadipocytes. J. Cell Physiol. 1992;151:300–309. doi: 10.1002/jcp.1041510211. [DOI] [PubMed] [Google Scholar]
- 6.Xing H, et al. TNF alpha-mediated inhibition and reversal of adipocyte differentiation is accompanied by suppressed expression of PPARgamma without effects on Pref-1 expression. Endocrinology. 1997;138:2776–2783. doi: 10.1210/endo.138.7.5242. [DOI] [PubMed] [Google Scholar]
- 7.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 8.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakae J, et al. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 10.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 11.Kaestner KH. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol. Metab. 2000;11:281–285. doi: 10.1016/s1043-2760(00)00271-x. [DOI] [PubMed] [Google Scholar]
- 12.Costa RH, Grayson DR, Darnell JE., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstein DC, et al. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 16.Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 17.Shih DQ, et al. Profound defects in pancreatic beta-cell function in mice with combined heterozygous mutations in Pdx-1, Hnf-1alpha, and Hnf-3beta. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3818–3823. doi: 10.1073/pnas.062605899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaestner KH, Hiemisch H, Schutz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3gamma results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 1998;18:4245–4251. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuempfle KJ, Koptides M, Karinch AM, Floros J. Preparation of transcriptionally active nuclear extracts from mammalian tissues. Biotechniques. 1996;21:48–50, 52. doi: 10.2144/96211bm09. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz i Altaba A, Prezioso VR, Darnell JE, Jessell TM. Sequential expression of HNF-3 beta and HNF-3 alpha by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech. Dev. 1993;44:91–108. doi: 10.1016/0925-4773(93)90060-b. [DOI] [PubMed] [Google Scholar]
- 21.Hansen LH, Madsen B, Teisner B, Nielsen JH, Billestrup N. Characterization of the inhibitory effect of growth hormone on primary preadipocyte differentiation. Mol. Endocrinol. 1998;12:1140–1149. doi: 10.1210/mend.12.8.0154. [DOI] [PubMed] [Google Scholar]
- 22.Tozzo E, Shepherd PR, Gnudi L, Kahn BB. Transgenic GLUT-4 overexpression in fat enhances glucose metabolism: preferential effect on fatty acid synthesis. Am. J. Physiol. 1995;268:E956–E964. doi: 10.1152/ajpendo.1995.268.5.E956. [DOI] [PubMed] [Google Scholar]
- 23.Rodbell M. Metabolism of isolated fat cells. J. Biol. Chem. 1964;239:375–380. [PubMed] [Google Scholar]
- 24.Shakir KM, Sundaram SG, Margolis S. Lipid synthesis in isolated intestinal cells. J. Lipid Res. 1978;19:433–442. [PubMed] [Google Scholar]
- 25.Susulic VS, et al. Targeted disruption of the beta 3-adrenergic receptor gene. J. Biol. Chem. 1995;270:29483–29492. doi: 10.1074/jbc.270.49.29483. [DOI] [PubMed] [Google Scholar]
- 26.Tong Q, et al. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 27.Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 28.Sul HS, Smas C, Mei B, Zhou L. Function of pref-1 as an inhibitor of adipocyte differentiation. Int. J. Obes. Relat. Metab. Disord. 2000;24(Suppl.):S15–S19. doi: 10.1038/sj.ijo.0801494. [DOI] [PubMed] [Google Scholar]
- 29.Mei B, Zhao L, Chen L, Sul HS. Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem. J. 2002;364:137–144. doi: 10.1042/bj3640137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smas CM, Chen L, Sul HS. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol. Cell. Biol. 1997;17:977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaya D, Hayamizu T, Bustin M, Zaret KS. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 2001;276:44385–44389. doi: 10.1074/jbc.M108214200. [DOI] [PubMed] [Google Scholar]
- 32.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiegelman BM, Choy L, Hotamisligil GS, Graves RA, Tontonoz P. Regulation of adipocyte gene expression in differentiation and syndromes of obesity/diabetes. J. Biol. Chem. 1993;268:6823–6826. [PubMed] [Google Scholar]
- 34.Boss O, Hagen T, Lowell BB. Uncoupling proteins 2 and 3: potential regulators of mitochondrial energy metabolism. Diabetes. 2000;49:143–156. doi: 10.2337/diabetes.49.2.143. [DOI] [PubMed] [Google Scholar]
- 35.Olefsky JM. The effects of spontaneous obesity on insulin binding, glucose transport, and glucose oxidation of isolated rat adipocytes. J. Clin. Invest. 1976;57:842–851. doi: 10.1172/JCI108360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tozzo E, Gnudi L, Kahn BB. Amelioration of insulin resistance in streptozotocin diabetic mice by transgenic overexpression of GLUT4 driven by an adipose-specific promoter. Endocrinology. 1997;138:1604–1611. doi: 10.1210/endo.138.4.5043. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura T, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J. Clin. Invest. 2002;110:1839–1847. doi:10.1172/JCI200216857. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerrish K, et al. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 39.Lee CS, et al. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- 40.Nakae J, et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 41.Black MA, Begin-Heick N. Growth and maturation of primary-cultured adipocytes from lean and ob/ob mice. J. Cell. Biochem. 1995;58:455–463. doi: 10.1002/jcb.240580408. [DOI] [PubMed] [Google Scholar]
- 42.Carlsson C, et al. Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology. 1997;138:3940–3948. doi: 10.1210/endo.138.9.5408. [DOI] [PubMed] [Google Scholar]