The splicing activity of Group I and Group II introns can be harnessed as a molecular tool that may potentially revise any gene of interest. Although hurdles remain, progress has been made in the development of these ribozymes for therapeutic application.
While gene therapy is showing promise in the treatment of some diseases, many genes are regulated in ways that are not easily recapitulated by adding back a functional copy of the gene. Therefore, as an alternative to the traditional approach, it may be possible to correct the genetic information directly in the genome or in the messages transcribed from mutant genes. In this way, genetic information is left intact so that the cell may properly regulate the expression and processing of genetic information.
Several classes of catalytic RNAs, or ribozymes, have the capacity to revise genetic information. Group I and Group II ribozymes are derived from naturally occurring Group I and Group II introns, respectively (1). These introns are found in the genes of a variety of lower eukaryotes and prokaryotes. They differ fundamentally from spliceosomal introns since Group I and Group II introns self-splice from the precursor RNA independent of the spliceosome. The intron adopts the catalytic structure that is capable of cleaving RNA splice sites and ligating the flanking exons together. In addition to self-splicing from RNA precursors, some Group II introns are able to reverse-splice into DNA. The splicing activity of these introns can be harnessed as a molecular tool that may potentially revise any gene of interest.
The therapeutic revision of RNA by ribozymes is mechanistically different from that of DNA. To revise RNA sequences, a Group I intron from Tetrahymena thermophila has been adapted for trans-splicing (2). Trans-splicing ribozymes splice therapeutic RNA sequences onto a target transcript in a process called ribozyme-mediated RNA repair. Considerable progress has been made in the development of trans-splicing ribozymes for therapeutic applications such as treating sickle cell disease and cancer. The spliceosome has also been shown to trans-splice RNA transcripts (3). Spliceosomal-mediated RNA trans-splicing (SMaRT) is discussed by Mariano Garcia-Blanco (4) in this Perspective series. To modify genomic DNA, a Group II ribozyme has recently been developed that reverse-splices into target genes of interest (5, 6). If applicable to mammalian cell genomes, this exciting technology may allow the therapeutic insertion of genetic information into a specific target locus. In this Perspective series article, we report on the progress made in development of Group I and Group II ribozymes as tools to revise RNA and DNA. We will also discuss the hurdles that must be overcome to make clinical use of these molecular therapeutics.
Self-splicing and trans-splicing ribozymes
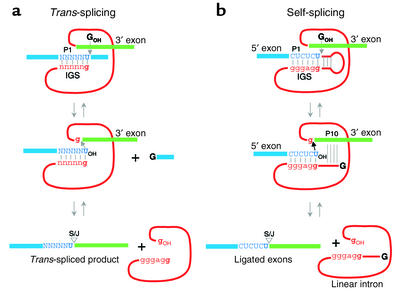
Group I introns are ribozymes that carry out two transesterification reactions in order to excise themselves from a precursor transcript (1). The self-splicing reaction pathway of the Tetrahymena intron is illustrated in Figure 1. The 5′ splice site is located in helix P1, which is formed by base-pairing between the exon sequence and the internal guide sequence (IGS) of the intron. The 5′ splice site, adjacent to a G-U pair in P1, is cleaved using exogenous guanosine in the first step of splicing. In the second step of splicing, the final nucleotide of the intron, also a guanosine, is positioned so that the 3′ exon may be ligated to the 5′ exon through the reverse of the first chemical step. Pairing between the 5′ end of the intron and the 3′ exon form P10, which has been implicated in facilitating 3′ splice site selection during the second step of splicing.
Figure 1.
Comparison of self-splicing versus trans-splicing. The trans-splicing reaction (a) is similar to the natural self-splicing reaction (b) except that the target RNA is not covalently attached to the ribozyme. Exon sequences (blue and green) are shown as boxes and capital letters. Intron sequences (red) are shown as solid lines with lowercase letters. IGS, P1, and P10 helices are shown. The targeted uridine residue is shown as a bold letter and the splicing junction (S/J) is indicated. The exogenous guanosine nucleophile and terminal guanosine nucleotide of the ribozyme are shown as G (black) and g (red) respectively. A trans-splicing ribozyme binds a substrate RNA by forming bp’s (N-n) with the targeted transcript.
A derivative of the Tetrahymena self-splicing intron, known as L-21 (lacking the first 21 nucleotides of the intron), binds to and cleaves oligonucleotide substrates in the trans configuration (Figure 1). The IGS of the Tetrahymena ribozyme may be modified to base pair with essentially any sequence as long as a G-U wobble pair is maintained at the splice site. If sequences are present downstream of the ribozyme, the ribozyme ligates this portion of the RNA transcript to the cleavage site of the substrate RNA. These 3′ exon sequences may be changed to potentially any nucleotide sequence. With the ability to “cut and paste” RNA sequences together, trans-splicing ribozymes were quickly recognized to have therapeutic potential in the revision of mutant RNAs.
Ribozyme-mediated RNA repair
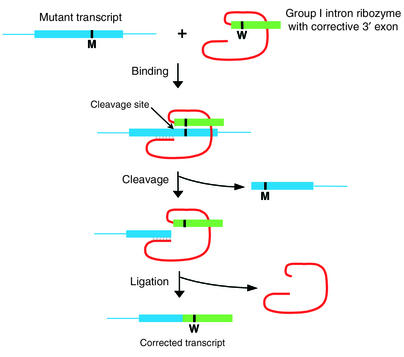
To use the activity of trans-splicing ribozymes to treat genetic disorders, a ribozyme IGS may be modified to base pair with a mutant messenger RNA upstream of the mutation in the target transcript. Therapeutic sequences are incorporated into the ribozyme transcript as a “3′ exon” that will be ligated to the cleaved target in the second step of splicing (Figure 2). Because the ribozyme modifies the RNA transcript posttranscriptionally, the natural transcriptional regulation of the gene is maintained.
Figure 2.
Schematic representation of RNA repair using a trans-splicing ribozyme. A Group I ribozyme delivers corrective sequences (3′ exon) to a mutant transcript. The ribozyme binds upstream of the mutation through base-pairing. Once bound, the ribozyme cleaves the target RNA, releases the downstream cleavage product, and splices the 3′ exon sequence onto the upstream cleavage product. M, mutant sequence; W, wild-type sequence corresponding to the mutated region.
The success of ribozyme-mediated RNA repair is contingent on many factors. First, the ribozyme transcripts must be delivered to or expressed in the appropriate cells. Like many gene therapy strategies, this may be accomplished using viral vectors to express the ribozyme transcript intracellularly. Also, trans-splicing ribozymes must be designed so that they specifically recognize the target transcript in a cell. Finally, trans-splicing must accurately revise its RNA target such that the repaired RNA may be functionally translated in the cell. We report on recent developments in evaluating the efficiency, specificity, and fidelity of trans-splicing ribozymes.
Recent advances in repair of mutant RNAs by trans-splicing ribozymes
The concept of ribozyme-mediated RNA repair was first demonstrated in Escherichia coli (2) and in cultured mammalian cells (7). The Group I intron from T. thermophila was engineered to repair truncated lacZ transcripts via targeted trans-splicing. Specifically, an L-21 ribozyme carrying a corrective lacZ “3′ exon” restored mutant lacZ target RNAs with high fidelity. In bacteria, complementation of β-galactosidase activity was achieved by translation of the product RNAs (2). Following cotransfection of ribozyme and substrate plasmids in mammalian cells, the ribozyme revised up to 49% of the truncated lacZ RNAs (8).
L-21 trans-splicing ribozymes have also been used to repair faulty transcripts associated with common genetic diseases. Phylactou et al. showed that a trans-splicing ribozyme could be used to shorten the trinucleotide repeat expansion in the 3′ untranslated region of the human myotonic dystrophy protein kinase transcript found in individuals with myotonic dystrophy (9). Toward a treatment for sickle cell anemia, Lan et al. used a trans-splicing ribozyme to convert sickle β-globin transcripts into mRNAs encoding an anti-sickling form of globin. Remarkably, this repair was demonstrated in erythrocyte precursors isolated from patients with sickle cell disease (10).
Ribozyme designs in addition to L-21 ribozymes have been explored and show great promise for therapeutic application (11–13). These versions extend the IGS from six to nine nucleotides. Also, analogous to P10 of the self-splicing intron, four to six nucleotides of complementarity between the intron and the 3′ exon are maintained. Finally, to increase complementarity of the ribozyme and the target transcript, an antisense region was appended to the 5′ end of the ribozyme. These modifications result in an “extended guide” design that was shown to be more effective than L-21 trans-splicing ribozymes in bacteria (11). Using a trans-splicing ribozyme with an extended guide, cytotoxicity could be induced in yeast by specific trans-splicing of diphtheria toxin coding sequences onto a pathogenic viral target RNA (12). Ribozymes with an extended guide have also been tested in mammalian cells with encouraging success. A trans-splicing ribozyme was recently used to repair mutant p53 RNA in human cancerous cells (13). Of particular significance, the repaired transcripts were translated to produce functional wild-type p53 protein, as evidenced by activation of p53-responsive transcription units in a luciferase assay. This result was the first demonstration of protein activity produced by the translation of repaired RNA in human cells. Rogers et al. (14) repaired a mutant canine skeletal muscle chloride channel (cClC-1) with a trans-splicing ribozyme containing a large (4 kb) 3′ exon. Repair efficiency was low (1.2%) when ascertained by quantitative RT-PCR in a population of cells; however, patch-clamp analysis of individual cells yielded a wide range of repair efficiency, with 18% of cells showing some electrophysiological restoration and several cells showing complete restoration of wild-type function. These data suggest that efficiency varies from cell to cell and trans-splicing can restore protein function.
A new genetic approach using Group I trans-splicing ribozymes to destroy viral RNA to slow viral propagation while selectively destroying virally infected cells was developed to combat hepatitis C virus (HCV) infection (15). A trans-splicing ribozyme was targeted to a site in the HCV internal ribosome entry site (IRES) present in all viral RNAs and used for cap-independent translation of viral genes in mammalian cells. The 3′ exon contained the IRES sequence after the splice site fused in-frame to the diphtheria toxin A chain. After trans-splicing and IRES-mediated translation of the diphtheria toxin A chain, virally infected cells were destroyed by apoptosis. These data suggest that trans-splicing ribozymes can be used specifically and selectively to deliver new gene activities to virally infected cells. Finally, designing an extended guide in ribozymes that revised sickle β-globin mRNAs resulted in a greater fraction of substrate RNA repair. Up to 10% of sickle β-globin messages were converted to γ-globin transcripts when the reaction efficiency was measured in 293 cells (J. Byun et al., unpublished observations). These significant advances in using trans-splicing ribozymes in cell culture systems are promising for future animal studies.
Efficiency and specificity of ribozyme-mediated RNA repair
Characterization of repaired RNA demonstrates that the fidelity of trans-splicing is excellent in cell lines. In all examined cases, sequence analyses of the amended RNAs demonstrated that the trans-splicing ribozyme formed the proper splice junctions between the targeted transcript and the restorative sequence. However, success in animal studies and future clinical application will likely require improvements in the efficiency and specificity of the reaction in vivo.
One factor that contributes to the overall efficiency of ribozyme-mediated RNA repair is the ribozyme’s accessibility to its target RNA. One way to enhance repair efficiency may be to colocalize the ribozyme with the substrate (16). Once the ribozyme is in proximity to its target, the ribozyme must also be able to access the targeted uridine residue. Some uridines may not be accessible due to the nature of the RNA secondary structure and/or bound proteins. Strategies to determine which uridines are most accessible in vivo have been reported (10, 13, 17). The results from these studies indicated that multiple conformations of substrate RNAs exist in vivo and that only some of these conformations allow access to a trans-splicing ribozyme. In support of this observation, the use of two ribozymes targeting different sites on the same transcript was shown to enhance overall repair efficiency (17).
Another factor affecting RNA repair efficiency is the efficient expression of the ribozyme. One of the challenges for constructing ribozyme expression cassettes is to design transcription units that will not reduce the ribozyme’s efficacy. Pertinent to RNA polymerase II–mediated ribozyme expression, two recent studies have evaluated the ability of Group I introns to fold into a catalytically competent conformation (18, 19). It was found that the extent of self-splicing is greatly influenced by the sequences flanking the intron and presumably reflects differences in the intron’s ability to fold into an active conformation inside cells.
The specificity of ribozyme targeting is a challenge to therapeutic application, as it is in other trans-splicing strategies. Although trans-splicing should ideally occur only at the intended target RNA, such a high degree of specificity was not observed when using a ribozyme with a six-nucleotide IGS in the cellular milieu (7). Low specificity may result in unwanted effects on the cell and reduce the efficiency of repair. Nevertheless, recent studies show promising advances toward increasing the specificity of trans-splicing ribozymes. Based on in vitro characterization of reaction kinetics (20), rationally designed ribozyme mutants were shown to have increased specificity (21). In yeast, trans-splicing ribozymes with an extended guide delivered a cytotoxic peptide sequence to viral transcripts. In this model system, specificity appeared very high since toxicity was not observed in the absence of the intended target (12). In the case of therapeutic anti-sickling ribozymes, providing additional complementarity with five bp’s of antisense, in addition to the nine-bp extended P1, was sufficient to achieve a certain level of discrimination of sickle β-globin transcript from wild-type β-globin transcript (J. Byun et al., unpublished observations).
Future directions
Despite the challenges of optimizing specificity and efficiency in vivo, results with trans-splicing ribozymes look promising. Trans-splicing ribozymes have now been successfully used to repair pathogenic transcripts in clinically relevant settings such as primary erythrocyte precursors from sickle cell patients and human tumor cell lines (10, 13). Several technical hurdles must be overcome before ribozyme-mediated repair of mutant RNAs becomes useful in the clinic. Many of these obstacles are common to other gene therapy strategies, including development of appropriate gene delivery vehicles and efficient expression of the therapeutic ribozyme transcripts. RNA repair efficiency would be increased by optimizing RNA polymerase II–mediated ribozyme expression, which includes determining the fraction of the ribozyme that is folded into active conformation, as well as expression levels and localization of transcripts. Use of other promoters might be useful because the type of promoter and its context can determine the intracellular compartmentalization of the ribozyme. Also, more systematic experiments are needed to determine the proper level of ribozyme expression — one that will ensure reasonable efficiency while maintaining a high specificity. These hurdles are certainly surmountable given the progress in developing experimental systems to characterize and optimize ribozyme-mediated repair in cells.
Revision of DNA by Group II introns
A subclass of Group II introns consists of mobile genetic elements that insert into an intronless allele through a process called retrohoming (22). Redirecting retrohoming introns so they insert into new target sites in DNA may allow Group II introns to be engineered as therapeutics (23). One strategy would be to retarget introns to disrupt pathogenic genes. Alternatively, if exogenous sequences are embedded within the intron, new genetic information may be introduced into the genomic locus of interest. Although redirecting Group II introns is a technology still in its infancy, the potential to apply Group II retrohoming to mammalian genomes is an exciting prospect.
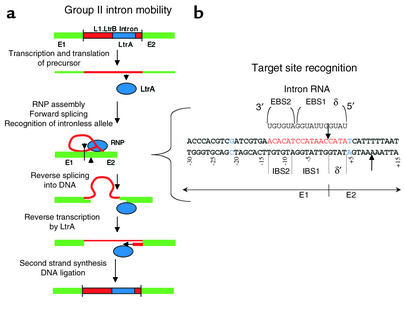
The Group II intron L1.LtrB, located in the ltrB gene of Lactococcus lactis, has been identified as a promising candidate for therapeutic development (23). The mechanism of Group II intron mobility for L1.LtrB is outlined in Figure 3. Following transcription of the LtrB precursor RNA, an intron-encoded protein called LtrA is translated. This multifunctional protein initially facilitates self-splicing of the catalytic intron (24–26). The excised intron and LtrA then form a ribonucleoprotein (RNP) complex capable of mobilizing the intron into DNA. Both components of the RNP participate in cleaving double-stranded DNA at the site of insertion (24). The intron RNA recognizes the target site via base-pairing between the exon-binding site (EBS) in the intron and the intron-binding site (IBS) in an intronless allele (Figure 3). The LtrA protein also interacts with the target site, at nucleotides flanking the EBS-IBS pairing (6). Following target recognition, the intron reverse-splices into the DNA. Then LtrA cleaves the DNA strand opposite the insertion site, nine nucleotides downstream of the intron. From this priming site, the intron is reverse-transcribed by the LtrA protein. Presumably, cellular machinery completes second-strand synthesis and ligation of the DNA to complete the retrohoming of the L1.LtrB into double-stranded DNA target sites (23).
Figure 3.
Group II intron mobility and revision of DNA. Mechanism of Group II L1.LtrB intron mobility. (a) A Group II intron (red) is found in the ltrB gene (green) of Lactococcus lactis between exons E1 and E2. The intron encodes the LtrA gene product (blue). Following transcription of the ltrB gene, the LtrA protein is translated from the intron-containing precursor. The LtrA protein facilitates self-splicing of the L1.LtrB intron. The excised intron is in the form of a lariat that is bound to the LtrA protein in an RNP particle. The assembled RNP targets an intronless allele for insertion into the DNA. The RNA component reverse-splices into the sense strand at the junction between E1 and E2. The LtrA protein then cleaves the antisense strand downstream of the intron insertion site, providing a template for reverse transcription by the LtrA protein. Second-strand synthesis and DNA repair complete the intron’s mobilization into the target DNA. (b) Target-site recognition by the retrohoming RNP is shown in greater detail. The EBSs of the intron (EBS1, EBS2, and δ) pair with IBSs IBS1, IBS2, and δ′ in the target (shown in red). Two bp’s, at –21 and +5 (shown in blue), strongly influence the activity of the LtrA protein during retrohoming (6). Cleavage sites are shown by the arrows. The top (sense) strand is cleaved by the RNA. LtrA cleaves the bottom strand following reverse splicing of the intron.
Retargeting the L1.LtrB intron to insert into a gene of interest
Because the target site is specified mainly by base-pairing between the intron and DNA, Group II introns may be retargeted to new target sites in DNA by altering the sequence of the EBS. However, a few constraints must be met for a new target sequence to be recognized by the LtrA protein. Based on the biochemical characterization of LtrA interactions with the DNA, the intron was retargeted to the E. coli thyA gene (6). Although the efficiency of the reaction was low, this demonstrated that the L1.LtrB intron may be retargeted to potentially any gene of interest by rational design.
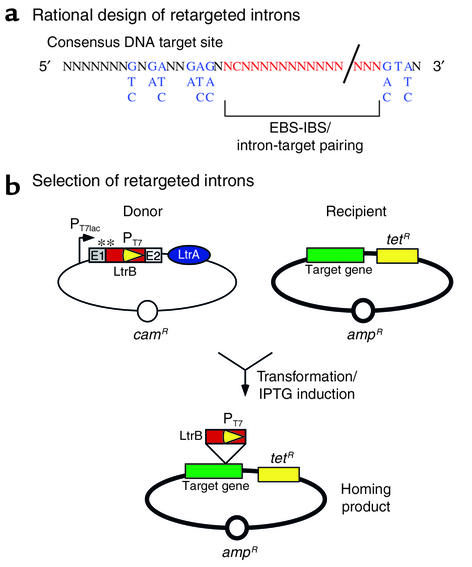
An elegant study has recently refined the rules for retargeting the L1.LtrB intron (5). A plasmid-based mobility assay in E. coli was used to probe partially randomized target sites. A consensus target site was derived from the successful mobilization events (Figure 4a). Any gene of interest may be analyzed for a potential target sequence and the corresponding EBS sequence can be engineered into the intron. It is estimated that the consensus sequence will appear in the human genome every several hundred nucleotides. Thus, a powerful new approach is available for the rational design of retargeted L1.LtrB introns.
Figure 4.
Rational design and selection of retargeted introns using a mobility assay in E. coli. (a) Rational design. Putative target sites may be searched in a gene of interest using the consensus sequence shown (N is any nucleotide). At positions where a subset of nucleotides is preferred by the LtrA protein, each preferred nucleotide is listed (in blue). The EBS of the intron is designed to base pair with the predicted IBS of the target (in red). Mobility is assessed in E. coli using donor and recipient plasmids as shown in b. (b) Selection of target sites. Retrohoming into the recipient plasmid activates expression of the promoterless tetracycline resistance gene (tetR, yellow box). The donor plasmid expresses the intron (L1.LtrB, in red) and associated protein (LtrA, in blue) following transformation into E. coli and isopropyl-β-D-thiogalactopyranoside (IPTG) induction of the T7lac promoter (PT7lac). The intron is flanked by exon sequences (E1 and E2) required for self-splicing. **A library of introns is generated by randomizing EBS sequences in the intron. IBS sequences in E1 are also randomized to facilitate forward splicing of the library of precursors. A T7 promoter (yellow arrowhead) is located in domain IV of the intron. Selectable markers on donor and recipient plasmids are shown (ampR, camR). Transcription termination signals prevent TetR expression in the absence of retrohoming. Retargeted introns are recovered from ampR, tetR colonies.
In addition to rational design, a selection strategy has been developed in E. coli to discover suitable target sites for the L1.LtrB intron in a gene of interest (Figure 4b) (5). A library of Group II introns was generated by randomizing EBS sequences in the intron expression cassette. Corresponding IBS sequences in the cassette were also randomized to facilitate self-splicing of the transcribed precursors. As described in Figure 4, successfully retargeted introns drive the expression of the tetracycline resistance gene in a recipient plasmid containing the target gene. Retargeted introns can thus be recovered from tetracycline-resistant bacteria. Both approaches to retarget the L1.LtrB intron — rational design and selection — were successful in revealing suitable target sites in therapeutically relevant genes (5). The ability to use assays in bacteria to find intron sequences that insert efficiently into a mammalian gene of interest is a significant advance in the development of Group II introns as therapeutics.
Insertion into therapeutically relevant genes
Two genes were chosen to demonstrate the capacity of retrohoming Group II introns to insert into therapeutically relevant targets (5). HIV-1 pol and human CCR5 genes are required for HIV replication or infection of macrophages, respectively. Disruption of the pol gene in infected cells could protect them from destruction by the virus, while disruption of the CCR5 gene could render cells resistant to infection. These target genes were scanned for consensus recognition sites by computer analysis. In addition, introns that successfully integrated into the targets were identified using the selection scheme and a library of introns having randomized EBS sequences (Figure 4). Retargeted introns obtained by both strategies were shown to mobilize into DNA containing HIV pol and CCR5 genes in E. coli (5). Several retargeted introns achieved mobilization frequencies (50–70%) that approached the mobility efficiency of the wild-type intron into its cognate target (>90%). These introns were chosen for further studies in mammalian cells.
To demonstrate that retargeted RNPs retain their retrohoming activity in mammalian cells, RNPs have been prepared in vitro and transfected directly into cells along with plasmids containing the target genes. Gene disruption of the HIV pol and human CCR5 genes was observed using PCR amplification and sequence analysis (5). Although many questions about using Group II introns remain, these results are encouraging for the development of Group II introns to revise DNA in mammalian cells.
Therapeutic potential of retrohoming Group II introns
Exciting progress has been made in the design and engineering of Group II introns that can be inserted into new target sites. However, seminal questions remain unanswered with respect to using retargeted Group II introns in mammalian cells. Foremost, Group II introns must be shown to integrate site-specifically into a genomic DNA target. To achieve this result, retargeted RNPs will need to be efficiently delivered to the nucleus of cells by either direct transfection or intracellular expression of the LtrA protein and the intron precursor RNA. Once localized to the nucleus, it remains uncertain whether RNPs will be able to access a target site that is packaged in eukaryotic genomic DNA. It is encouraging that bacterial protein endonucleases involved in cleaving both strands of DNA during homing of Group I introns are active in mammalian cells (27). However, it is not certain that mammalian cells will complete the repair of DNA after reverse splicing and reverse transcription of the intron in genomic DNA. Mobility assays in mammalian cells where retrohoming integration products can be readily isolated will greatly facilitate such studies.
If retrohoming Group II introns can insert into genomic DNA, both the efficiency of the reaction and its specificity will need to be addressed and optimized. Fortunately, because the revised gene should be stably replicated and passed on to daughter cells, it may be possible to isolate and expand the modified cells ex vivo. In contrast to RNA repair, transient expression of the RNP should be sufficient to permanently alter a cell population.
Finally, we envision Group II introns to be effective therapeutics, not only for their ability to disrupt gene function but also to act as carriers of new genetic information. The LtrA protein is normally encoded within domain IV of the L1.LtrB intron. Other sequences may be added to or replace the LtrA open reading frame in domain IV and still retain Group II splicing and reverse-splicing activity (5, 23). This raises the possibility that cDNAs or particular exons may be incorporated into the intron and delivered to a specific genomic locus. If realized, this technology could revolutionize genomic studies in mammalian cells as well as have a profound impact on the treatment of genetic disorders.
RNA-mediated alteration of gene expression is a promising addition to gene therapy protocols whereby the Group I intron can be used to alter cellular information at the RNA level and the Group II intron can be used to alter cellular information at the DNA level. However, several hurdles need to be overcome in the near future, including delivery, expression, and efficacy of the RNA therapeutic, for potent repair to occur in mammalian cells. In addition to their potential therapeutic value, these techniques can be used to investigate gene expression patterns in a cell or across a chromosome, or possibly even to address questions about genome organization.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: spliceosome-mediated RNA trans-splicing (SMaRT); internal guide sequence (IGS); hepatitis C virus (HCV); internal ribosome entry site (IRES); ribonucleoprotein (RNP); exon-binding site (EBS); intron-binding site (IBS).
References
- 1.Cech, T.R. 1993. Structure and mechanism of the large catalytic RNAs: Group I and group II introns and ribonuclease P. In The RNA World. R.F. Gesteland and J.F. Atkins, editors. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 239–269.
- 2.Sullenger BA, Cech TR. Ribozyme-mediated repair of defective mRNA by targeted trans-splicing. Nature. 1994;371:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- 3.Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotech. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Blanco, M. 2003. Messenger RNA reprogramming by spliceosome mediated RNA trans-splicing. J. Clin. Invest. In press. [DOI] [PMC free article] [PubMed]
- 5.Guo H, et al. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- 6.Mohr G, Smith D, Belfort M, Lambowitz AM. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev. 2000;14:559–573. [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JT, Lee S-W, Sullenger BA. Tagging ribozyme reaction sites to follow trans-splicing in mammalian cells. Nat. Med. 1996;2:643–648. doi: 10.1038/nm0696-643. [DOI] [PubMed] [Google Scholar]
- 8.Jones JT, Sullenger BA. Evaluating and enhancing ribozyme reaction efficiency in mammalian cells. Nat. Biotech. 1997;15:902–905. doi: 10.1038/nbt0997-902. [DOI] [PubMed] [Google Scholar]
- 9.Phylactou LA, Darrah C, Wood MJA. Ribozyme-mediated trans-splicing of a trinucleotide repeat. Nat. Genet. 1998;18:378–381. doi: 10.1038/ng0498-378. [DOI] [PubMed] [Google Scholar]
- 10.Lan N, Howrey RP, Lee S-W, Smith CA, Sullenger BA. Ribozyme-mediated repair of sickle β-globin mRNA’s in erythrocyte precursors. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 11.Kohler U, Ayre BG, Goodman HM, Haseloff J. Trans-splicing ribozymes for targeted gene delivery. J. Mol. Biol. 1999;285:1935–1950. doi: 10.1006/jmbi.1998.2447. [DOI] [PubMed] [Google Scholar]
- 12.Ayre BG, Kohler U, Goodman HM, Haseloff J. Design of highly specific cytotoxins by using trans-splicing ribozymes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3507–3512. doi: 10.1073/pnas.96.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Sullenger BA. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8490–8494. doi: 10.1073/pnas.150104097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers CS, Vanoye CG, Sullenger BA, George AL. Functional repair of a mutant chloride channel using a trans-splicing ribozyme. J. Clin. Invest. 2002;110:1783–1789. doi:10.1172/JCI200216481. doi: 10.1172/JCI200216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu K-J, Kim J-H, Lee S-W. Ribozyme-mediated selective induction of new gene activity in hepatitis C virus internal ribosome entry site-expressing cells by targeted trans-splicing. Mol. Ther. 2003;7:386–395. doi: 10.1016/s1525-0016(02)00063-1. [DOI] [PubMed] [Google Scholar]
- 16.Sullenger BA. Colocalizing ribozymes with substrate RNAs to increase their efficacy as gene inhibitors. Appl. Biochem. Biotechnol. 1995;54:57–61. doi: 10.1007/BF02787911. [DOI] [PubMed] [Google Scholar]
- 17.Lan N, et al. Enhancing RNA repair efficiency by combining trans-splicing ribozymes that recognize different accessible sites on a target RNA. Mol. Ther. 2000;2:245–255. doi: 10.1006/mthe.2000.0125. [DOI] [PubMed] [Google Scholar]
- 18.Long MB, Sullenger BA. Evaluating group I intron catalytic efficiency in mammalian cells. Mol. Cell. Biol. 1999;19:6479–6487. doi: 10.1128/mcb.19.10.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagen M, Cech TR. Self-splicing of the Tetrahymena intron from mRNA in mammalian cells. EMBO J. 1999;18:6491–6500. doi: 10.1093/emboj/18.22.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarrinkar PP, Sullenger BA. Probing the interplay between the two steps of group I intron splicing: competition of exogenous guanosine with ω G. Biochemistry. 1998;37:18056–18063. doi: 10.1021/bi982193x. [DOI] [PubMed] [Google Scholar]
- 21.Zarrinkar PP, Sullenger BA. Optimizing the substrate specificity of a group I intron ribozyme. Biochemistry. 1999;38:3426–3432. doi: 10.1021/bi982688m. [DOI] [PubMed] [Google Scholar]
- 22.Curcio MJ, Belfort M. Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell. 1996;84:9–12. doi: 10.1016/s0092-8674(00)80987-3. [DOI] [PubMed] [Google Scholar]
- 23.Cousineau B, et al. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 24.Matsuura M, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- 26.Saldanha R, et al. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 27.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]