Abstract
Transcription is controlled in part by the dynamic acetylation and deacetylation of histone proteins. The latter process is mediated by histone deacetylases (HDACs). Previous analysis of the regulation of HDAC activity in transcription has focused primarily on the recruitment of HDAC proteins to specific promoters or chromosomal domains by association with DNA-binding proteins. To characterize the cellular function of the recently identified HDAC4 and HDAC5 proteins, complexes were isolated by immunoprecipitation. Both HDACs were found to interact with14-3-3 proteins at three phosphorylation sites. The association of 14-3-3 with HDAC4 and HDAC5 results in the sequestration of these proteins in the cytoplasm. Loss of this interaction allows HDAC4 and HDAC5 to translocate to the nucleus, interact with HDAC3, and repress gene expression. Regulation of the cellular localization of HDAC4 and HDAC5 by 14-3-3 represents a mechanism for controlling the transcriptional activity of these class II HDAC proteins.
In eukaryotic cells, DNA is packaged into nucleosomes, which consist of octamers of histone proteins. These nucleosomal arrays are organized into higher-order structures to form chromatin. The incorporation of DNA into chromatin creates a barrier to fundamental cellular processes such as replication and transcription. At least two mechanisms have evolved to regulate this repressive environment by altering chromatin structure: nucleosome remodeling and post-translational modification of histones, including the dynamic acetylation and deacetylation of the amino termini. The latter process is controlled by histone acetyltransferase and histone deacetylase (HDAC) enzymes, respectively (reviewed in ref. 1).
Two distinct histone deacetylase complexes in yeast containing separate HDAC enzymes, Rpd3 and Hda1, have been purified (2). Seven human HDACs have been identified thus far and have been divided into two classes based on their primary structure (3–11). The first class of HDACs (HDAC1, -2, and -3) is more closely related to yeast Rpd3p whereas the second class (HDAC4, -5, -6, and -7) is composed of significantly larger proteins with greater similarity to yeast Hda1p. All contain conserved catalytic domains and possess the ability to deacetylate histones in vitro (3, 4, 6, 9, 11). However, as in the case of the yeast enzymes, there is mounting evidence that the two classes perform distinct functions in cells.
Of the class I HDACs, HDAC1 and HDAC2 are the best characterized. Along with RbAp46 and RbAp48, these form the core of the mSin3A and NRD/NuRD complexes (9, 12–16). These complexes are recruited by a variety of transcription repressors, including nuclear hormone receptors (reviewed in ref. 17) and methylated DNA-binding proteins (18–20). Thus, HDAC1 and HDAC2 are involved in silencing expression of both specific genes and entire chromosomal domains.
Of the class II HDACs, HDAC4 has been shown to associate with transcription factors of the myocyte enhancer factor 2 (MEF2) family (7, 10). MEF2 proteins are involved in several processes, including muscle cell differentiation and cellular proliferation (reviewed in ref. 21). MEF2 interacts with the amino acid 118–188 region in HDAC4 (10), which is related in sequence (64% similarity) to the amino acid 123–206 region in HDAC5. Because HDAC4 and HDAC5 have been shown to interact with HDAC3 (6), it is possible that all three proteins are involved in mediating transcriptional repression by MEF2. HDAC4 and HDAC5 are expressed highly in muscle cells (6, 10) and thus may function in regulating muscle cell differentiation via MEF2. Furthermore, as with HDAC1 and HDAC2, HDACs 4, 5, and 7 interact with the corepressors NCoR and SMRT and hence may also repress transcription in association with nuclear hormone receptors (22, 23).
Aside from recruitment by DNA binding proteins, very little is known regarding the regulation of mammalian HDAC activity. In maize, there is evidence that HDAC phosphorylation alters substrate specificity and activity of the enzymes (24) whereas in the case of chicken HDACs, substrate specificity is modified by association with factors in the nuclear matrix (25).
To understand the cellular function of HDAC4 and HDAC5, complexes of these proteins were isolated by immunoprecipitation. Characterization of the associated proteins revealed the presence of two isoforms of the 14-3-3 protein. Further analysis revealed that 14-3-3 associates with HDAC4 and HDAC5 at three phosphorylation sites and that this interaction sequesters these proteins in the cytoplasm. Loss of 14-3-3 binding allows HDAC4 and HDAC5 to shuttle into the nucleus, associate with HDAC3, and repress gene transcription. Thus, the activity of both HDAC4 and HDAC5 are regulated by cellular localization, as mediated by 14-3-3.
Materials and Methods
DNA Constructs.
FLAG-epitope-tagged HDAC4 and HDAC5 constructs in the pBJ5 mammalian expression vector have been described previously (6). The HDAC4–enhanced green fluorescent protein (EGFP) clone in pBJ5 was made by ligation of a NotI-XbaI HDAC4-FLAG fragment to a XbaI-SalI EGFP fragment, which was generated by PCR from a plasmid containing EGFP (CLONTECH). The HDAC5-EGFP/pBJ5 construct was made similarly by using NotI-XbaI HDAC5-FLAG fragment. The C-terminally myc-epitope-tagged 14-3-3 β and ɛ were produced by PCR from a Jurkat cDNA library (Stratagene) and were subcloned into the NotI and SpeI sites of pBJ5. To generate mutant 14-3-3 proteins, R58 on 14-3-3 β and R57 in 14-3-3 ɛ were mutated to alanine. Note that these residues correlate with R56 of 14-3-3 η, which has been shown to be critical for phosphoserine binding (26). The mutations were generated by PCR overlap extension.
The S246A, S467A, and S632A single, double, and triple mutations in HDAC4 were generated by PCR overlap extension. The fragments were cloned into the NotI and SacII sites of the HDAC4-F/pBJ5 construct and were sequenced to ensure proper incorporation of the mutations. The reporter used in the luciferase assays contains three copies of the desmin MEF2 site in pGL2-E1b-luciferase and was generously provided by Eric Olson (University of Texas Southwestern Medical Center). The myc-epitope-tagged MEF2D/pSCT mammalian expression plasmid was kindly provided by Jun Liu (Massachusetts Institute of Technology).
Antibodies, Immunoprecipitation, and Western Blotting.
Polyclonal rabbit antibodies were generated to the N-terminal 19 residues of HDAC4 (Biosynthesis, Lewisville, TX). Antibodies against HDAC3 have been described (27). Isoform-specific antibodies against 14-3-3 ɛ and β were obtained from Santa Cruz Biotechnology whereas antibodies to importin α (α-Rch1) were acquired from Transduction Laboratories (Lexington, KY). α-FLAG M2 antibodies and α-mouse IgG Texas-red conjugated secondary antibodies for immunofluorescence were obtained from Sigma whereas α-c-myc antibodies were purchased from Upstate Biotechnology (Lake Placid, NY).
Forty-eight hours after transfection, cells were lysed in JLB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/10% glycerol/0.5% TritonX-100) containing a complete protease inhibitor mixture (Boehringer Mannheim) and phosphatase inhibitors [20 mM NaH2(PO4), pH 7.2/25 mM NaF/2 mM EDTA]. Lysis proceeded for 10 min at 4°C, after which the cellular debris was pelleted by centrifugation at 14,000 × g for 5 min. Recombinant proteins were immunoprecipitated from the supernatant by incubation with α-FLAG M2 agarose affinity gel (Sigma) for 1 h at 4°C. For Western blot analysis and silver staining, the beads were washed three times for 5 min at room temperature with MSWB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1 mM EDTA/0.1% Nonidet P-40), and the proteins were separated by SDS/PAGE. For enzyme activity assays, the beads were washed three times with JLB at 4°C.
Peptide Microsequencing.
The sequence analysis was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Finnigan LCQ quadropole ion trap mass spectrometer. Phosphoserine residues were identified by using targeted ion MS/MS.
HDAC Assays.
3[H]acetate-incorporated histones were isolated from butyrate-treated HeLa cells by hydroxyapitite chromatography as described (12). Immunoprecipitates were incubated with 1.4 μg (10,000 dpm) of histones for 2 h at 37°C. HDAC activity was determined by scintillation counting of the ethyl-acetate soluble 3[H] acetic acid (8).
Cell Culture and Transfections.
TAg-Jurkat cells were transfected by electroporation with 5 μg of DNA for expression of recombinant proteins or were mock-transfected without DNA as a negative control. Cells were harvested 48 h post-transfection. When required, cells were treated with 200 nM staurosporine (Calbiochem) or 20 nM calyculin A (Calbiochem) for 1.5 h before harvesting.
Immunofluorescence.
COS-7 or U20S cells were transfected with 1–2 μg of DNA by using the Lipofectamine PLUS system (GIBCO/BRL). Forty-eight hours later, cells were fixed with paraformaldehyde and were stained with antibodies and Hoechst dye (Molecular Probes), or live cells were stained with Hoechst and the EGFP was visualized directly by using a fluorescence microscope (Spencer Scientific Corporation, Derry, NH).
Reporter Gene Assays.
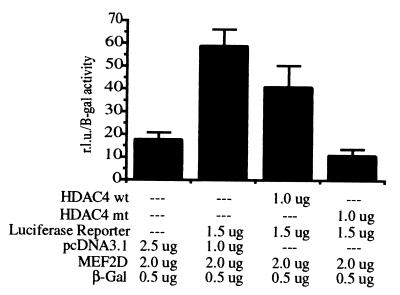
For each sample, 10 million TAg Jurkat cells were transfected with a total of 5 μg of DNA (see Fig. 7). A constitutive β-galactosidase expression vector was used as a control for protein expression levels in the luciferase assays. Thirty-eight hours after transfection, the samples were harvested and split into sets of three. Luciferase activity was determined according to the manufacturer's instructions (Promega), and β-galactosidase activity was determined by using a standard β-galactosidase assay. Luciferase values (relative light units) were normalized for transfection efficiency by dividing by β-gal activity. These assays were performed four times with similar results.
Figure 7.
Increased nuclear localization of HDAC4 enhances MEF2-dependent transcriptional repression. TAg-Jurkat cells were transfected with MEF2D, a MEF2-luciferase reporter construct, a constitutive β-gal expression construct, and either wild-type HDAC4 or HDAC4 with mutations in all three 14-3-3 binding sites (HDAC4 S246/466/632A). pcDNA3.1 (Invitrogen) was used to normalize the amount of DNA transfected. Thirty-eight hours after transfection, the samples were harvested and divided to perform the subsequent assays in triplicate. The amount of luciferase activity was measured (Promega) and divided by the amount of β-gal activity present to normalize for protein expression levels.
Results
Immunoprecipitation of HDAC4 and HDAC5 Complexes.
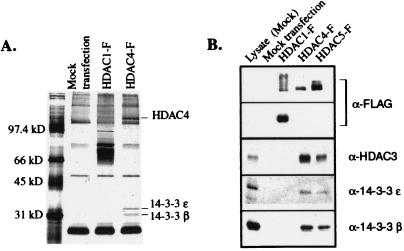
Recombinant, FLAG-epitope tagged HDAC1 and HDAC4 were transiently expressed in TAg Jurkat cells, and the α-FLAG immunoprecipitates were separated by SDS/PAGE and were visualized by silver staining. Comparison with the mock-transfected negative control and the HDAC1 samples revealed the presence of specific 30- and 32-kDa protein bands in the HDAC4 immunoprecipitate. Peptides derived from these proteins were analyzed by peptide microsequencing and were found to correspond to the β and ɛ isoforms of 14-3-3, respectively (Fig. 1A). Because of the high degree of sequence similarity between HDAC4 and HDAC5 (51% identity), it was hypothesized that HDAC5 associates with 14-3-3 proteins as well. The presence of these two 14-3-3 protein isoforms in both HDAC4 and HDAC5 immunoprecipitates was confirmed by Western blot analysis with isoform-specific antibodies (Fig. 1B). This analysis also confirmed the previously observed association of HDAC3 with both class II HDACs. These immunoprecipitation experiments suggest that HDAC4 and HDAC5 can associate, either directly or indirectly, both with HDAC3, which is nuclear (4), and 14-3-3 proteins, which are generally cytoplasmic (28).
Figure 1.
Association of HDAC4 and HDAC5 with two isoforms of 14-3-3. (A) Recombinant, FLAG-tagged HDAC1 and HDAC4 were transiently expressed in TAg Jurkat cells and were immunoprecipitated by using α-FLAG agarose (Sigma). The immunopurified complexes were separated by SDS/PAGE, and the proteins were visualized by silver stain. (B) The association between HDAC4 and HDAC5 with 14-3-3 was confirmed by Western blot analysis. The recombinant FLAG-tagged proteins were subjected to Western blot analysis using 14-3-3 isoform specific antibodies.
Nuclear-Cytoplasmic Shuttling of HDAC4 and HDAC5 Is Regulated by Binding to 14-3-3.
We and other groups (7) have observed by immunofluorescence that HDAC4 and HDAC5 can be localized exclusively to either the cytoplasm or the nucleus, often aggregating in discrete foci (Fig. 2A). This nuclear and cytoplasmic localization is dynamic, and translocation can occur under normal conditions of cell growth (data not shown). This nuclear-cytoplasmic shuttling process could explain the apparent discrepancy observed in the immunoprecipitation experiments, in which HDAC4 and HDAC5 were found to interact with both nuclear HDAC3 and cytoplasmic 14-3-3.
Figure 2.
Nuclear-cytoplasmic shuttling of HDAC4 and HDAC5 is correlated to 14-3-3 expression levels. (A) Recombinant HDAC4-EGFP and HDAC5-EGFP were transiently expressed in U2OS cells, and the localization of the protein was observed by fluorescence microscopy. (B) Overexpression of 14-3-3 β causes an increased cytoplasmic localization of HDAC4-EGPF. U2OS cells were transiently transfected with HDAC4-EGFP and either a control plasmid (pcDNA3.1, Invitrogen) or myc-tagged 14-3-3 β. The localization of recombinant HDAC4 and 14-3-3 was analyzed by immunofluorescence.
Several cases have been reported in which proteins are sequestered in the cytoplasm by binding to 14-3-3, and disruption of this interaction allows the proteins to translocate into the nucleus and perform their function (29–34). It is possible that binding of HDAC4 and HDAC5 to 14-3-3 sequesters these proteins in the cytoplasm, where they are presumably unable to repress transcription. On loss of 14-3-3 binding, HDAC4 and HDAC5 could translocate into the nucleus, bind to HDAC3 and MEF2, and repress MEF2-dependent gene expression. To study the effect of 14-3-3 binding on HDAC4 localization, HDAC4-EGFP and myc-tagged 14-3-3 β were co-expressed in U2OS cells, and the cellular localization of HDAC4 was analyzed by immunofluorescence. The number of cells with exclusively cytoplasmic localization of HDAC4-EGFP was determined in triplicate experiments (Fig. 2B). Expression of HDAC4-EGFP alone results in an exclusively cytoplasmic localization of HDAC4 in 67 ± 3% of the cells whereas simultaneous overexpression of 14-3-3 β increases this to 97 ± 1% of the cells. This correlation suggests that 14-3-3 may play a role in sequestering HDAC4 in the cytoplasm.
Association of HDAC4 and HDAC5 with 14-3-3 and HDAC3.
It is known that 14-3-3 proteins bind to phosphorylated serine or threonine residues in defined consensus sequences (35, 36). It was hypothesized that phosphorylation of HDAC4 and HDAC5 would allow association with 14-3-3 and sequestration in the cytoplasm. Dephosphorylation of these HDACs should result in the loss of interaction with 14-3-3, with subsequent translocation to the nucleus and binding to HDAC3. To test this hypothesis, the effect of phosphorylation of HDAC4 and HDAC5 on their association with 14-3-3 and HDAC3 was analyzed.
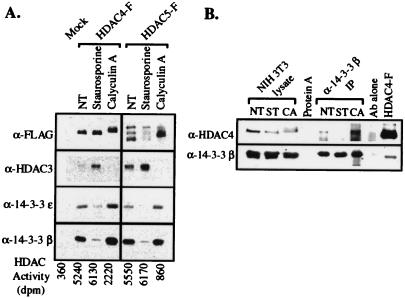
Recombinant FLAG-tagged HDAC4 and HDAC5 were transiently expressed in TAg Jurkat cells that were subsequently treated with the general serine/threonine kinase inhibitor staurosporine or the PP1 and PP2A phosphatase inhibitor calyculin A. The recombinant proteins were immunoprecipitated and analyzed by Western blot analysis (Fig. 3A). Under dephosphorylating conditions caused by staurosporine treatment, there is a decrease in binding of HDAC4 and HDAC5 to both 14-3-3 isoforms, and a corresponding increase in HDAC3 association. In addition, an increase in the overall HDAC activity of the purified complex was observed (Fig. 3A). Similarly, under hyperphosphorylating conditions caused by calyculin A treatment, HDAC4 and HDAC5 undergo a notable electrophoretic mobility shift, probably because of direct phosphorylation. An increase in 14-3-3 binding is observed as well, with a concomitant loss of interaction with HDAC3. This loss of binding to HDAC3 presumably causes the dramatic reduction in immunoprecipitated HDAC activity that is observed, although the activity of isolated HDAC4 and HDAC5 is still above background. These data suggest that binding of 14-3-3 to HDAC4 and HDAC5 requires the presence of phosphorylated serine or threonine residues, and corresponds to a loss of interaction with HDAC3 in the nucleus.
Figure 3.
Phosphorylation-dependent binding of 14-3-3 to HDAC4 and HDAC5. (A) Association of HDAC4 and HDAC5 with 14-3-3 and HDAC3 depends on the phosphorylation state of the proteins. HDAC4-FLAG and HDAC5-FLAG were transiently expressed in TAg Jurkat cells. Forty-eight hours post-transfection, the cells were treated for 1.5 h with staurosporine and calyculin A. The immunopurified complexes were subjected to Western blot analysis and were tested for HDAC activity, as described in Materials and Methods. (B) Association of endogenous HDAC4 with 14-3-3 β in NIH 3T3 cells is phosphorylation-dependent. 14-3-3 β immunoprecipitates from NIH 3T3 cells that had been treated with staurosporine or calyculin A for 1.5 h were analyzed for the presence of endogenous HDAC4 by Western blotting.
To determine whether this mechanism holds true for the endogenous proteins, the association of endogenous HDAC4 with 14-3-3 β in NIH 3T3 cells was assessed (Fig. 3B). Immunoprecipitates of 14-3-3 β from NIH 3T3 cells treated with staurosporine and calyculin A were analyzed by Western blotting for the presence of endogenous HDAC4. An electrophoretic shift of endogenous HDAC4 under calyculin A treatment was observed, suggesting that the protein is directly phosphorylated under these conditions. Furthermore, there is a notable increase in amount of endogenous HDAC4 associated with 14-3-3 β on calyculin A treatment, which is consistent with the data for the recombinant protein. Note, however, that the total amount of endogenous HDAC4 is fairly low, and additional studies suggest that it is expressed in a limited number of cell lines (data not shown).
The effect of staurosporine and calyculin A treatment of the localization of HDAC4-EGFP in COS-7 cells was assessed. COS-7 cells transfected with HDAC4-EGFP were treated for 15 min with staurosporine or calyculin A, and the number of cells with exclusively cytoplasmic, nuclear, or general staining of HDAC4-EGFP was determined in triplicate experiments. In the absence of drug treatment, 53 ± 3% of the cells had exclusively cytoplasmic localization of HDAC4 whereas staurosporine treatment decreased this to 31 ± 7%, and calyculin A treatment caused cytoplasmic localization in 73 ± 3% of the cells. Note that the treatment period was short, because of the detachment of the cells from their substrate during prolonged exposure to calyculin A, and hence the localization shifts were not complete. However, this trend is consistent with the observed effect of these treatments on the association of HDAC4 with 14-3-3 and correlates well with a model in which HDAC4 is sequestered in the cytoplasm by 14-3-3.
Binding of 14-3-3 to HDAC4 Blocks Binding of Importin α.
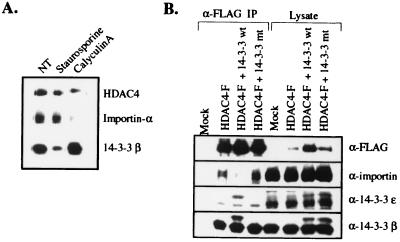
14-3-3 proteins have been shown to sequester Xenopus Cdc25 in the cytoplasm by binding near a bipartite nuclear localization sequence and blocking its interaction with the importin α/β heterodimer (29, 33, 34), which is required for import into the nucleus (37). HDAC4 also contains a nuclear localization sequence (38). To determine whether the interaction with 14-3-3 blocks binding of the importin heterodimer, recombinant FLAG-tagged HDAC4 was immunoprecipitated from untreated, staurosporine-, and calyculin A-treated TAg-Jurkat cells and was analyzed for binding to importin α by Western blotting. On binding to 14-3-3 because of calyculin A treatment, HDAC4 fails to associate with importin α (Fig. 4A).
Figure 4.
Association of 14-3-3 with HDAC4 prevents the binding of importin α. (A) The binding of 14-3-3 to HDAC4 prevents interaction with importin α. Forty-eight hours after transfection with HDAC4-FLAG, TAg Jurkat cells were treated with staurosporine or calyculin A for 1.5 h. HDAC4-FLAG was immunopurified and subjected to Western blot analysis with α-importin α antibodies. (B) Overexpression of 14-3-3 blocks binding of importin α to HDAC4. TAg Jurkat cells were transfected with 1 μg of HDAC4-FLAG and 2 μg each of myc-tagged wild-type or mutant 14-3-3 β and ɛ. pcDNA3.1 (Invitrogen) was used to normalize the amount of DNA transfected. Forty-eight hours post-transfection, HDAC4-FLAG was immunoprecipitated and analyzed for binding to importin α by Western blotting.
To determine whether the loss of importin α binding to HDAC4 is indeed caused by masking of the nuclear localization sequence by 14-3-3, rather than by secondary effects of the calyculin A treatment, the association of importin α and HDAC4 was analyzed under conditions in which the 14-3-3 proteins were overexpressed. Recombinant, myc-tagged 14-3-3 ɛ and β were overexpressed in TAg Jurkat cells, and HDAC4 immunoprecipitates were analyzed for importin α binding by Western blotting. As a control, the effect of overexpression of mutant forms of both 14-3-3 isoforms was determined as well. These mutants are incapable of binding to their phosphorylated consensus sequence because of a substitution of a critical arginine residue in the phosphoserine-binding pocket with alanine (26). The recombinant wild-type 14-3-3 isoforms bound to HDAC4 and caused a significant decrease in importin α association whereas the mutant 14-3-3 isoforms, although expressed at equivalent levels, could not bind to HDAC4 and had no effect on the association of importin α (Fig. 4B). These data are consistent with a model in which the interaction of importin α with HDAC4 is directly blocked by the binding of 14-3-3 to HDAC4, presumably because of the masking of a nuclear localization sequence.
Mutations of the 14-3-3 Binding Sites Cause Increased Nuclear Localization of HDAC4.
14-3-3 proteins bind to well defined consensus sequences containing phosphorylated serine or threonine residues (35, 36). There are four canonical 14-3-3 binding sites in HDAC4, three of which are well conserved in HDAC5. The serine residues in each of these three sites in HDAC4 (S246, S467, S632) were mutated to alanine to prohibit phosphorylation and thus prevent 14-3-3 binding. Mutation of these sites does not abrogate the association of HDAC4 with MEF2D or NCoR, nor does it abrogate HDAC activity (data not shown), and thus protein folding is presumably unaffected. Phosphorylation of these serine residues on calyculin A treatment was confirmed by peptide microsequencing.
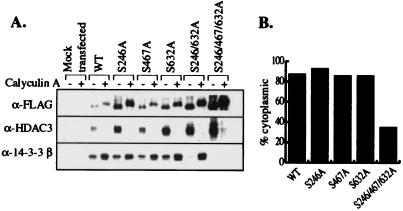
Mutation of individual or two 14-3-3 binding sites is not sufficient to abrogate binding of 14-3-3, but mutation of all three serine residues to alanine (HDAC4 S246/467/632A) abolishes binding to 14-3-3 β and ɛ, even under hyperphosphorylating conditions caused by calyculin A treatment (Fig. 5A; data not shown). Furthermore, localization of the triple mutant to the cytoplasm is dramatically decreased compared with the wild-type and single mutants (Fig. 5B), consistent with a role for 14-3-3 in sequestration of HDAC4 and HDAC5 in the cytoplasm.
Figure 5.
Mutation of 14-3-3 binding sites in HDAC4 causes loss of 14-3-3 binding and increased nuclear localization. (A) Single, double, and triple serine to alanine mutations in the three putative 14-3-3 binding sites of HDAC4 were generated. The recombinant proteins were transiently expressed in TAg Jurkat cells, which were treated with calyculin A for 1.5 h before harvesting. The immunopurified complexes were subjected to Western blot analysis. (B) The single and triple 14-3-3 binding site mutants were fused to EGFP and transiently expressed in U2OS cells. The cellular localization was monitored by fluorescence microscopy. These experiments were repeated multiple times with similar results.
Disruption of HDAC3 and HDAC4 Interaction on Calyculin A Treatment.
Interestingly, despite its inability to bind 14-3-3 and its concomitant nuclear localization, the HDAC4 S246/467/632A triple mutant is still unable to bind to HDAC3 under calyculin A treatment (Fig. 5A). Hence, the inability of HDAC4 to associate with HDAC3 under hyperphosphorylating conditions cannot simply be attributable to its sequestration in the cytoplasm. Other possibilities include changes in HDAC4 or HDAC3 conformations or electrostatic properties on direct phosphorylation that prevents their association, or modifications of additional factors that may be required for mediating the HDAC4-HDAC3 interaction.
To distinguish between these possibilities, immunoprecipitated HDAC4 from untreated, staurosporine-treated, or calyculin A-treated cells was incubated with lysates from untreated or calyculin A-treated TAg Jurkat cells (Fig. 6). Untreated and staurosporine-treated (presumably hypophosphorylated) HDAC4 associated with HDAC3 under all conditions, which is consistent with previous observations. Notably, there is an increase in the amount of associated HDAC3 on incubation with lysate from untreated cells, but not after incubation with lysate from cells treated with calyculin A. This suggests that HDAC3 from untreated cells, but not from calyculin A-treated cells, is still competent to bind HDAC4. Furthermore, there is no factor in calyculin A-treated cells that can disrupt the HDAC4-HDAC3 interaction once it has formed.
Figure 6.
Localization-independent loss of HDAC4 and HDAC3 interaction on calyculin A-treatment. Forty-eight hours after transfection with HDAC4-FLAG, TAg Jurkat cells were treated with staurosporine or calyculin A for 1.5 h. HDAC4-FLAG was immunopurified, and the sample was split into thirds. One-third of the immunopurified protein was prepared for Western blot analysis, one-third was incubated for 1 h with untreated TAg Jurkat lysate, and the remaining one-third of the sample was incubated with calyculin A-treated TAg Jurkat lysate for 1 h. These samples were analyzed for 14-3-3 and HDAC3 binding by Western blotting.
As previously observed, calyculin A-treated (hyperphosphorylated) HDAC4 does not bind to HDAC3 under normal immunoprecipitation conditions. However, on incubation with untreated cell lysates, calyculin A-treated HDAC4 does pull down HDAC3. This interaction is not present on incubation with calyculin A-treated lysate. Hence, calyculin A-treatment of HDAC4 does not cause it to undergo a conformational change that would prevent it from binding to HDAC3. Note that, although there does seem to be a slight decrease in the amount of HDAC3 present in the calyculin A-treated lysate, it is not significant enough to explain the complete lack of binding to HDAC4.
These experiments suggest that, although sequestration of HDAC4 in the cytoplasm by 14-3-3 may serve to prevent its interaction with HDAC3, this association is abrogated by an additional mechanism when the cell is exposed to hyperphosphorylating conditions. This loss of interaction may be attributable to the phosphorylation of HDAC3 or to the modification of additional proteins required for mediating the HDAC3-HDAC4 association.
Increased Nuclear Localization of HDAC4 Enhances MEF2-Dependent Repression.
The sequestration of HDAC4 and HDAC5 in the cytoplasm by 14-3-3 presumably prevents these proteins from repressing gene transcription, and therefore represents a mechanism for controlling HDAC activity. To determine whether cellular localization affects HDAC4-mediated transcriptional repression by MEF2, the following series of reporter gene assays was performed. TAg-Jurkat cells were transfected with a MEF2-luciferase reporter, the MEF2D transcription factor, and either wild-type HDAC4 or the HDAC4 S246/467/632A triple mutant, which no longer binds to 14-3-3 and displays enhanced nuclear localization. Equivalent expression levels of the wild-type and triple mutant HDAC4 recombinant proteins was confirmed by Western blot analysis (data not shown). Expression of wild-type HDAC4 decreases MEF2-dependent transcription slightly whereas expression of the HDAC4 triple mutant completely represses transcription (Fig. 7). Furthermore, in the absence of HDAC3, wild-type HDAC4 associated with 14-3-3 displays slightly higher HDAC activity than the triple mutant HDAC4 (data not shown), and hence the decreased ability of the wild-type HDAC4 to repress transcription is not attributable to inherently lower activity of the 14-3-3-bound protein. These data are consistent with a model in which loss of association with 14-3-3 results in nuclear localization of HDAC4 and association with MEF2 and HDAC3, causing increased transcriptional repression.
Discussion
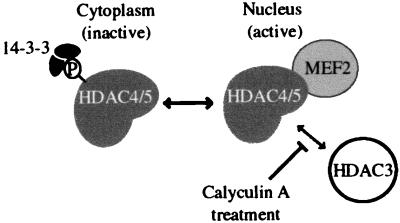
Analysis of mammalian HDAC regulation has focused primarily on the recruitment of HDACs to specific gene promoters or general chromosomal domains through interactions with DNA-binding proteins. In this paper, we present evidence for the regulation of two mammalian class II HDACs by phosphorylation-dependent cellular localization. HDAC4 and HDAC5 contain three canonical 14-3-3 binding sites, all of which are involved in mediating the interaction with 14-3-3. This binding to 14-3-3 is correlated with the cytoplasmic localization of HDAC4. This sequestration in the cytoplasm appears to be caused by the occlusion of a nuclear localization sequence by the bound 14-3-3 proteins, thereby preventing association with the importin α/β heterodimer, which is required for nuclear transport. Cytoplasmic HDAC4 and HDAC5 cannot interact with nuclear proteins, such as MEF2 and HDAC3, and hence cannot be involved in directly repressing transcription via deacetylation of nuclear histones. Although it is a formal possibility that these class II HDACs are performing functions in the cytoplasm, the association of HDAC4 with the MEF2 transcription factors suggests that a primary role of these HDACs is to silence gene expression. Hence, it is more likely that HDAC4 and HDAC5 are being held in a “inactive” state when they are present in the cytoplasm. On loss of 14-3-3 binding, HDAC4 and HDAC5 can translocate into the nucleus and associate with HDAC3 and transcription factors such as MEF2. This association targets the HDACs to specific promoters and silences gene expression (Fig. 8).
Figure 8.
Model for regulation of HDAC4 activity by cellular localization. Binding of phosphorylated HDAC4 or HDAC5 to 14-3-3 sequesters these proteins in the cytoplasm, where they cannot function in repression of transcription. Dephosphorylation allows HDAC4 and HDAC5 to shuttle to the nucleus, where they associate with HDAC3 and the MEF2 transcription factor, and silence transcription of MEF2-dependent genes. Treatment of cells with the phosphatase inhibitor calyculin A also disrupts the interaction of HDAC3 with HDAC4 and HDAC5 by an unknown mechanism, possibly involving additional protein factors.
There is evidence for a second level of phosphorylation-dependent regulation of HDAC4 activity. HDAC3 from cells treated with a phosphatase inhibitor cannot bind HDAC4, even in the absence of differential cellular compartmentalization. Hence, HDAC3 may alter its conformation or electrostatic properties on direct phosphorylation, or it may acquire or lose binding to another factor involved in mediating its interaction with the class II HDACs. Preliminary experiments have not revealed any alteration in the phosphorylation state of HDAC3, or the calyculin A-regulated binding of any proteins to HDAC3 (data not shown). Because the association of HDAC family members is a common phenomenon (6), this loss of HDAC3/HDAC4 interaction may result in the inactivation of HDAC4 in the nucleus, even before its sequestration in the cytoplasm.
Recent studies suggest that 14-3-3 also may be involved in regulation of RbAp48 function. RbAp48 has been shown to interact directly with histones, along with its homologue RbAp46 (39). Xenopus RbAp48 was found to form a complex with Hat1 and three isoforms of 14-3-3. Because RbAp48 also contains a putative 14-3-3 binding motif, it is possible that the 14-3-3 proteins bind to RbAp48 directly and modify its function (40). Thus, 14-3-3 mediated cellular localization may be a common mechanism for regulating acetylation and deacetylation of histones.
Mediation of the cellular localization of HDAC4 and HDAC5 by phosphorylation-dependent 14-3-3 binding represents a general mechanism for modulating HDAC function. This process is most likely controlled by signal transduction pathways of the cell. Preliminary experiments using cell cycle inhibitors, specific kinase and phosphatase inhibitors, and various cell culture conditions, such as oxidative or osmotic stress, have not revealed any obvious connections to common signaling pathways (data not shown). However, the interaction of HDAC4 with MEF2, coupled with its high expression levels in muscle tissues, suggests that this regulatory process may be involved in muscle cell differentiation.
Acknowledgments
We thank Gregory Verdine and Christian Hassig for critical reading of the manuscript, Mary Kay Pflum, Jeff Tong, and Randy Peterson for helpful discussions, and William S. Lane (Harvard Micro-chemistry Facility) for peptide microsequencing analysis. This work was supported by a National Institute of General Medical Sciences grant (GM38627) to S.L.S. C.M.G. is supported by a National Science Foundation predoctoral training grant. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- HDAC
histone deacetylase
- MEF
myocyte enhancer factor
- EGFP
enhanced green fluorescent protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140199597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140199597
References
- 1.Ayer D E. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 2.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 4.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 6.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 9.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 10.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X J. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 12.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 13.Wade P A, Jones P L, Vermaak D, Wolffe A P. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 14.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 16.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 18.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 21.Black B L, Olson E N. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 22.Kao H Y, Downes M, Ordentlich P, Evans R M. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 23.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 24.Kolle D, Brosch G, Lechner T, Pipal A, Helliger W, Taplick J, Loidl P. Biochemistry. 1999;38:6769–6773. doi: 10.1021/bi982702v. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Chen H Y, Davie J R. Biochem J. 1996;314:631–637. doi: 10.1042/bj3140631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorson J A, Yu L W, Hsu A L, Shih N Y, Graves P R, Tanner J W, Allen P M, Piwnica-Worms H, Shaw A S. Mol Cell Biol. 1998;18:5229–5238. doi: 10.1128/mcb.18.9.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burbelo P D, Hall A. Curr Biol. 1995;5:95–96. doi: 10.1016/s0960-9822(95)00022-4. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Winkler K, Yoshida M, Kornbluth S. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nature (London) 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 31.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang H G, Pathan N, Ethell I M, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke T F, Reed J C. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 33.Kumagai A, Dunphy W G. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Y, Piwnica-Worms H. Mol Cell Biol. 1999;19:7410–7419. doi: 10.1128/mcb.19.11.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 36.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 37.Gorlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 38.Hicks G R, Raikhel N V. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 39.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;9:6–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 40.Imhof A, Wolffe A P. Biochemistry. 1999;38:13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]