Abstract
HSP27 is an ATP-independent chaperone that confers protection against apoptosis through various mechanisms, including a direct interaction with cytochrome c. Here we show that HSP27 overexpression in various cell types enhances the degradation of ubiquitinated proteins by the 26S proteasome in response to stressful stimuli, such as etoposide or tumor necrosis factor alpha (TNF-α). We demonstrate that HSP27 binds to polyubiquitin chains and to the 26S proteasome in vitro and in vivo. The ubiquitin-proteasome pathway is involved in the activation of transcription factor NF-κB by degrading its main inhibitor, I-κBα. HSP27 overexpression increases NF-κB nuclear relocalization, DNA binding, and transcriptional activity induced by etoposide, ΤNF-α, and interleukin 1β. HSP27 does not affect I-κBα phosphorylation but enhances the degradation of phosphorylated I-κBα by the proteasome. The interaction of HSP27 with the 26S proteasome is required to activate the proteasome and the degradation of phosphorylated I-κBα. A protein complex that includes HSP27, phosphorylated I-κBα, and the 26S proteasome is formed. Based on these observations, we propose that HSP27, under stress conditions, favors the degradation of ubiquitinated proteins, such as phosphorylated I-κBα. This novel function of HSP27 would account for its antiapoptotic properties through the enhancement of NF-κB activity.
Exposure of cells to stressful stimuli induces or enhances the expression of several heat shock proteins (HSPs). One of these proteins is HSP27, a small HSP which is also abundantly expressed in many cancer cells (19). HSP27 has been shown to interact with the actin cytoskeleton, to modulate intracellular reactive oxygen species content, and to prevent apoptotic cell death triggered by a variety of stimuli, including tumor necrosis factor alpha (ΤNF-α) (16, 26) and several commonly used anticancer drugs, such as etoposide (6, 17, 27, 46). Several mechanisms have been proposed to account for the HSP27-mediated negative regulation of programmed cell death. This small HSP specifically interacts with cytochrome c when released from the mitochondria to the cytosol, thus preventing the formation of the apoptosome. A premitochondrial effect at a higher HSP27 expression level has also been described (47, 51). In vitro, this protein behaves as an ATP-independent chaperone that helps in the refolding of denatured proteins (45), a property that could be of key importance for understanding its in vivo functions.
Another mechanism that contributes to cell protection from stressful stimuli through the elimination of unfolded proteins is the extralysosomal, energy-dependent, ubiquitin-proteasome degradation pathway (1, 24, 37, 41) This pathway involves two steps. First, the target protein is conjugated with ubiquitin molecules at lysine residues. Then, the ubiquitin-tagged substrate is transferred to the 26S proteasome, a multisubunit complex consisting of a 20S barrel-shaped proteolytic core and a 19S cap-like regulatory complex (also called PA700). This latter consists of at least 18 distinct subunits with molecular masses of 35 to 110 kDa. It is presumed that ATP hydrolysis by the 19S complex provides the energy needed for the chaperoning and unfolding of substrates degraded within the proteasome cylinder (3, 44). Whereas the enzymatic reactions of ubiquitination are well characterized, the molecular mechanisms underlying the translocation of ubiquitinated substrates to the 26S proteasome remain poorly understood.
Protein degradation by the 26S proteasome, which is present in both the cytoplasm and the nucleus, regulates the expression of a number of intracellular proteins, including cell cycle regulatory proteins, proto-oncogene products, major histocompatibility complex molecules, and NF-κB inhibitors. NF-κB belongs to the Rel/NF-κB family of transcription factors and controls a large variety of processes, such as cell growth and inflammatory and stress responses (48). Unlike many other transcription factors, which are localized in the nucleus, NF-κB is sequestered in the cytoplasm of most cells through binding to a family of inhibitory proteins called I-κB subunits (48). Thus, its regulation is centered around nuclear-cytoplasmic shuttling. I-κB subunits inhibit NF-κB by masking its nuclear localization signal, thereby causing its cytoplasmic retention and blocking both its DNA binding and its transactivation abilities (48). In response to extracellular stimuli, the inhibitors are degraded, thus liberating the transcription factor for translocation to the nucleus. NF-κB is a homo- or heterodimer consisting of various combinations of the family members, including NF-κB1 (p50 and precursor p105), c-Rel, p65 (RelA), NF-κB2 (p52 and precursor p100), and RelB. Most NF-κB inducers act through a common pathway based on the phosphorylation-induced degradation of I-κB proteins, which was first described for the best-studied and major I-κB protein, I-κBα. The induced phosphorylation of I-κBα at N-terminal Ser32 and Ser36 by I-κB kinase provides a signal for rapid I-κBα degradation by the ubiquitin-proteasome pathway. Ubiquitin-dependent degradation of I-κBα is mediated by the ubiquitin ligase protein FWD1 (23, 53, 57).
The present study demonstrates that HSP27 overexpression increases proteasome activation triggered by stressful stimuli, such as inflammatory cytokines (ΤNF-α and interleukin 1β [IL-1β]) and cytotoxic drugs (etoposide). In response to these stimuli, HSP27 enhances NF-κB activity. We show that HSP27 directly associates with the 26S proteasome, with multiubiquitin chains, and with phosphorylated I-κBα. These observations suggest that, when overexpressed in response to various stimuli, HSP27 facilitates phosphorylated I-κBα proteasome-mediated proteolysis, which could account for the protective effect of this small stress protein.
MATERIALS AND METHODS
Culture and reagents.
The U937 human leukemic cell line was grown in RPMI 1640 medium (BioWhittaker, Fontenay-sous-Bois, France) supplemented with 10% (vol/vol) fetal bovine serum and 2 mM l-glutamine. The MEF cell line was grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich, Saint Quentin Fallavier, France) supplemented with 10% (vol/vol) fetal bovine serum, 0.1% sodium pyruvate, and 1% HEPES buffer. Rat colon carcinoma REG cells were grown in Ham's F-10 medium supplemented with 10% (vol/vol) fetal calf serum. Stable transformants of human U937 cells carrying the gene encoding human HSP27 were described previously (17). One of the characterized clones (clone 7) was used in this work. Another clone carrying the hygromycin resistance gene and the empty vector was used as a control. Mutations constructed in HSP27 as well as REG cells transfected with the resulting mutants were described previously (6). Etoposide and acetyl-calpastatin were purchased from Sigma-Aldrich, MG132 was purchased from the Peptide Institute (Osaka, Japan), lactacystin was purchased from Calbiochem (San Diego, Calif.), and IL-1B and ΤNF-α were purchased from R&D Systems (Minneapolis, Minn.).
Immunoblot analysis.
Whole-cell lysates were prepared by lysing cells in 2% sodium dodecyl sulfate-137 mM NaCl-2.7 mM KCl-8 mM Na2HPO4-NaCl-Pi (pH 7.4) at 68°C for 5 min. Protein concentrations were measured by means of a micro-bicinchoninic acid protein assay (Pierce, Asnieres, France). Proteins were separated in sodium dodecyl sulfate-polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes (Bio-Rad, Ivry sur Seine, France). Immunoblot analysis was performed with specific antibodies and enhanced chemoluminescence-based detection (Amersham, Les Ullis, France). The antibodies used were mouse monoclonal human HSP27, HSP60, and HSP90 from StressGen (Victoria, British Columbia, Canada); α-actin from Innogenex (San Ramon, Calif.); ubiquitin and HSC70 from Santa Cruz (Santa Cruz, Calif.); ATPase subunit Rpt4 (PA700) from Affinity Research (Mamhead, United Kingdom); polyclonal human HSP27 from StressGen; I-κBα and the p65 subunit of NF-κB from Santa Cruz (Santa Cruz, Calif.); and phospho-I-κBα from Cell Signaling Technology (Beverly, Mass.). All monoclonal antibodies used were of the immunoglobulin G1 isotype.
Immunoprecipitation.
Cells (107) were lysed in immunoprecipitation buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.1% NP-40). After centrifugation for 10 min at 15,000 × g, the supernatant was incubated with an antibody (1:100) with constant agitation at 4°C. When needed, immunoprecipitations were performed with purified proteins. Five micrograms of the 26S proteasome (Boston Biochem, Cambridge, Mass.) was mixed with 5 μg of recombinant HSP27 (StressGen) in 300 μl of immunoprecipitation buffer. After incubation for 30 min at room temperature, an antibody (1:100) was incubated with the solution with constant agitation at 4°C. The immunocomplexes were precipitated with protein A-Sepharose. The pellet was used for immunoblotting after five successive washings.
Determination of proteasome activity.
Proteasome activity was determined by a previously described method (49). Cells (4 × 106 in 200 μl of phosphate-buffered saline [PBS] [pH 7.4]) were incubated for 30 min at 37°C with a 100 μM concentration of the cell-permeating fluorogenic substrate N-succinyl-l-leucyl-l-leucyl-l-tyrosine-7-amido-4-methyl-coumarin (Suc-LLVY-AMC; Bachem, Basel, Switzerland). Fluorescence generated by its cleavage was quantified by using a model SFM 25 spectrofluorometer (Kontron AG, Zurich, Switzerland).
Cell transfection and reporter gene assay.
Cells were cultured at 4 × 105/well in 24-well plates for 24 h before transfection (2 μg of plasmid DNA/well) with the Fugene-6 transfection reagent (Boehringer Mannheim, Mannheim, Germany). To measure NF-κB transcriptional activity, we used plasmid p4NF-κB-luc, which contains four NF-κB sites in tandem coupled to the luciferase reporter gene (kindly provided by A. Brent Carter [7]). Cells were cotransfected with a thymidine kinase-Renilla luciferase plasmid in order to normalize for transfection efficiency. A dual luciferase reporter assay kit from Promega (Charbonnières, France) was used according to the manufacturer's recommendations; 10 μg of total protein was used to measure luciferase and Renilla activities with a Lumat LB 9507 Luminometer (EG&G Berthold).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (20). The NF-κB consensus oligonucleotide sequence was obtained from Promega, labeled with T4 kinase (Boehringer Mannheim), and purified with Sephadex 50 columns (Boehringer Mannheim) to eliminate nonincorporated radioactivity. Antibodies specific to the p65 and p50 transcription factors were purchased from Santa Cruz.
GST in vitro ubiquitination.
Ubiquitin cDNA was cloned in vector pGEX-5X3 (Pharmacia, Uppsala, Sweden) after reverse transcription-PCR amplification. Glutathione S-transferase (GST) and GST fusion protein (GST-ubiquitin) were then purified with gluthatione-Sepharose beads (Pharmacia). Reactions were performed with buffer A, containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.2 mM dithiothreitol. GST-ubiquitin purified with gluthatione-50% Sepharose beads was incubated with cytoplasmic extract from mouse testis. The ubiquitination assay was performed by the addition of 4 mM ATP and incubation for 1 h at room temperature. After incubation, GST-ubiquitin-Sepharose was pelleted and washed three times in buffer A, and 1 volume of buffer A was added to obtain protein fused to 50% Sepharose.
Ubiquitin-binding assays.
GST, GST-monoubiquitin, or GST-polyubiquitin was incubated with cytosolic extracts from U937 cells. After 1 h at room temperature, protein-Sepharose was pelleted and washed three times in buffer A. Bound proteins were eluted and used for Western blot analysis. For ubiquitin-agarose pull-down analysis, 100 μl of cytosolic extract was incubated with 10 μl of ubiquitin (7 mg/ml) or 10 μl of ubiquitin buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2) for 20 min at 4°C. The pull-down analysis was performed by the addition of 20 μl of ubiquitin-50% agarose (Sigma-Aldrich) and incubation for 45 min under rotation. The ubiquitin-agarose matrix was pelleted and washed three times in ubiquitin buffer. Bound proteins were analyzed by Western blotting.
Gel filtration analysis.
Cells (5 × 108) were lysed in 300 μl of a buffer containing 25 mM HEPES (pH 7.5), 0.2 mM dithiothreitol, and 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) by vortexing for 1 min in ice. After centrifugation for 15 min at 14,000 × g, 1 mg of the supernatant was fractionated at room temperature by fast protein liquid chromatography with a Superose-6 column (Pharmacia) at a flow rate of 0.5 ml min−1. Fractions of 1 ml were collected on ice, and 20 μl of each was used to measure proteasome activity or for immunoblot analysis.
Cytometric analysis.
I-κBα levels in whole cells fixed in 3% paraformaldhyde and permeabilized in saponin (0.1% [vol/vol] in PBS-bovine serum albumin) were determined. Nuclear NF-κB levels in cells permeabilized with digitonin (50 μg/ml) were determined. Appropriate concentrations of antibodies were added to cells in 100 μl of PBS containing 0.5% bovine serum albumin. After 1 h of incubation at room temperature and two washes in PBS, cells were incubated for 45 min with fluorescein isothiocyanate-conjugated sheep anti-rabbit immunoglobulin (Jackson ImmunoResearch). Ten thousand cells were analyzed by using a FACScan flow cytometer (Becton Dickinson).
RESULTS
HSP27 potentiates the proteasomal degradation of ubiquitinated proteins.
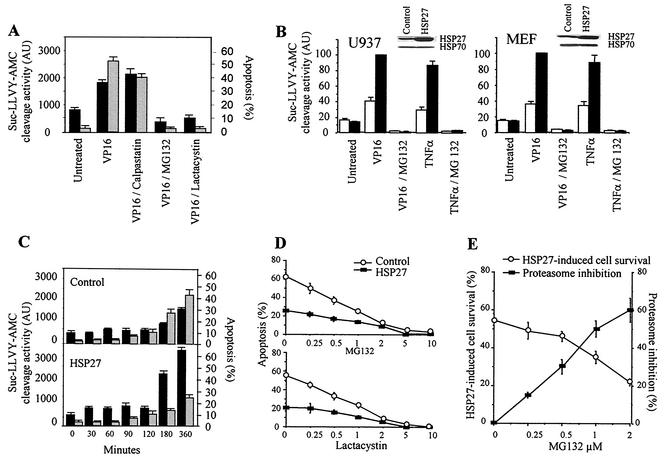
Exposure of U937 human leukemic cells to the topoisomerase II inhibitor etoposide (100 μM) induces rapid cell death by apoptosis, as assessed morphologically by staining of the nuclear chromatin with Hoechst 33342. This treatment also induces the activation of a proteasome proteolytic activity that cleaves Suc-LLVY-AMC. Two proteasome inhibitors, namely, MG132 (25 μM) and lactacystin (25 μM), prevent both etoposide-induced apoptosis and Suc-LLVY-AMC cleavage activity, whereas these events are not influenced by the calpain inhibitor acetyl-calpastatin (Fig. 1A). These results suggested that etoposide activates the proteolytic activity of the 26S proteasome in these cells. To determine whether HSP27 could influence this proteolytic activity in stressed cells, the protein was overexpressed by stable transfection in U937 cells. Exposure of control-transfected U937 cells to either etoposide or ΤNF-α induced a 1.5- to 2-fold increase in Suc-LLVY-AMC cleavage activity. Stable transfection with an hsp27 cDNA-containing vector amplified this cellular response, as demonstrated by the five- to sixfold increase in Suc-LLVY-AMC cleavage activity measured after similar treatments. In all instances, proteasome activation was prevented by MG132 (Fig. 1B) or lactacystin (data not shown). Similar results were obtained for control- and HSP27-transfected MEF (Fig. 1B) and REG (data not shown) cells. In control-transfected cells, the increases in Suc-LLVY-AMC cleavage activity and apoptosis occur simultaneously (Fig. 1A and C). Interestingly, the overexpression of HSP27 prevents etoposide-induced cell death, whereas it amplifies Suc-LLVY-AMC cleavage activity (Fig. 1C). We next determined whether HSP27-induced proteasome activation and apoptosis inhibition were connected events. As shown in Fig. 1A, proteasome inhibitors at 25 μM completely abolished apoptosis induced by etoposide, a result which precluded an analysis of the potential connection. The addition of lower concentrations of proteasome inhibitors to etoposide-treated cells only partially inhibited both proteasome activation and apoptosis induction (Fig. 1D), indicating that the protective effect of HSP27 increased as the proteasome inhibitor concentration decreased (Fig. 1E). Similar results were obtained with both MG132 and lactacystin (Fig. 1D), indicating a correlation between the HSP27 antiapoptotic effect and proteasome activity.
FIG. 1.
HSP27 enhances proteasome activation while inhibiting apoptosis. (A) U937 cells either were left untreated or were treated for 4 h with 100 μM etoposide (VP16) in the absence or presence of acetyl-calpastatin (25 μM), MG132 (25 μM), or lactacystin (25 μM) before measurement of the ability of cell lysates to cleave the substrate Suc-LLVY-AMC (black bars; AU, arbitrary units) and the percentage of apoptotic cells after nuclear chromatin staining with Hoechst 33342 (gray bars). (B) Control-transfected (white bars) and HSP27-transfected (black bars) cells either were left untreated or were treated for 4 h (U937 cells) or 24 h (MEF cells) with 100 μM etoposide (VP16) or 20 ng of ΤNF-α/ml in the absence or presence of 25 μM MG132 before measurement of the ability of cell lysates to cleave the substrate Suc-LLVY-AMC. Results are expressed as percentages (100% is the activity in VP16-treated, HSP27-transfected cells). Insets show Western blot analyses of HSP27 expression in control- and HSP27-transfected cells. (C) Kinetic analysis of Suc-LLVY-AMC cleavage activity (black bars) and apoptosis induction (gray bars) in control- and HSP27-transfected U937 cells treated with 100 μM etoposide for the indicated times. (D and E) Cells were treated with etoposide (100 μM, 4 h) in the presence of decreasing concentrations of MG132 or lactacystin, as indicated. Then, chromatin staining with Hoechst 33342 was used to measure the percentage of control-transfected and HSP27-transfected U937 apoptotic cells (D) or the percentage of cell survival induced by HSP27 overexpression (E). The percentage of proteasome inhibition (Suc-LLVY-AMC cleavage) is also indicated (E). Data are the means and standard deviations for three independent experiments.
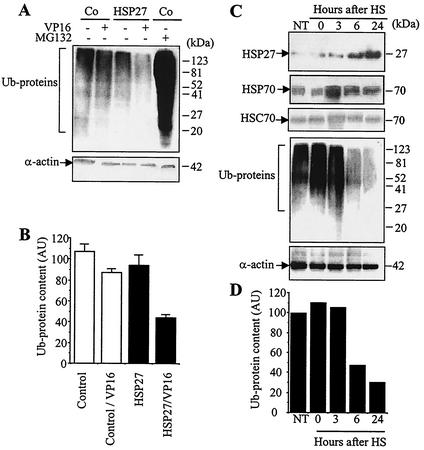
Since the main identified function of the proteasome is the degradation of polyubiquitinated proteins, we analyzed the consequences of HSP27 overexpression on the degradation of ubiquitinated proteins by using a multiubiquitin chain-specific antibody. Multiubiquitinated proteins accumulated in U937 cells exposed for 4 h to 25 μM MG132 (Fig. 2A). The overexpression of HSP27 did not significantly influence the levels of expression of ubiquitinated proteins (Fig. 2B), in accordance with its lack of effect on Suc-LLVY-AMC cleavage activity (Fig. 1B). In contrast, the overexpression of HSP27 amplified the decrease in the ubiquitinated protein content induced by exposure to 100 μM etoposide (Fig. 2A) and to 20 ng of ΤNF-α/ml (data not shown) for 4 h. In order to study the relationship between HSP27 overexpression and proteasome activation in a more physiological model, we exposed U937 cells to 42°C for 1 h and then incubated the cells at 37°C for up to 24 h. This treatment induced a transient increase in HSP70 expression that returned to the basal level when HSP27 expression increased, between 6 and 24 h after heat shock (Fig. 2C). In these heat-shocked cells, we observed a decrease in the expression of ubiquitinated proteins that correlated with an increase in the level of HSP27 (Fig. 2C and D). Altogether, these results suggested that increased HSP27 expression could mediate a decrease in the cellular multiubiquitinated protein content by activating the proteasome.
FIG. 2.
HSP27 enhances the degradation of ubiquitinated proteins in stressed cells. (A) Control-transfected (Co) and HSP27-transfected U937 cells either were left untreated or were treated with 100 μM etoposide (VP16) for 4 h in the absence or presence of 25 μM MG132. Protein ubiquitination (Ub-proteins) was monitored by Western blotting with an antiubiquitin antibody. α-actin served as the loading control. (B) Densitometry analysis of Ub-proteins in three independent experiments (mean and standard deviation) similar to that shown in panel A. AU, arbitrary units. (C) U937 cells either were left untreated (NT) or were exposed for 1 h to 42°C (HS) and then were incubated at 37°C for the indicated times. The indicated proteins were studied by Western blotting. α-Actin served as the loading control. (D) Densitometry analysis of Ub-proteins shown in panel C.
HSP27 is a ubiquitin-binding protein.
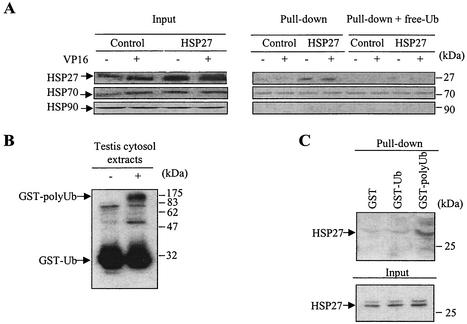
Cytosolic extracts from control- and HSP27-transfected U937 cells were incubated with ubiquitin-agarose to carry out a pull-down experiment. Figure 3 shows that HSP27 efficiently interacts with ubiquitin. Treatment with 100 μM etoposide (Fig. 3A) or 20 ng of ΤNF-α/ml (data not shown) for 4 h did not influence the affinity of HSP27 for ubiquitin. The addition of free ubiquitin (50 μg) significantly decreased HSP27 binding to the ubiquitin-agarose matrix, indicating the specificity of the interaction (Fig. 3A). In contrast, neither HSP70 nor HSP90 demonstrated specific binding to ubiquitin. Although a small amount of HSP70 appeared to be bound to ubiquitin, this amount was not affected by the addition of free ubiquitin, indicating that this association may not be specific (Fig. 3A). To determine whether HSP27 also binds to multiubiquitin chains, GST was polyubiquitinated in vitro (Fig. 3B). Pull-down experiments demonstrated that HSP27 associates more efficiently with GST-polyubiquitin than with GST-monoubiquitin (Fig. 3C).
FIG. 3.
HSP27 associates with ubiquitin. (A) Cytoplasmic extracts obtained from control-transfected and HSP27-transfected U937 cells, either left untreated or treated with 100 μM etoposide (VP16) for 4 h, were incubated with a ubiquitin-agarose matrix. The presence of the indicated proteins in the input material was analyzed by Western blotting (left panels). Pull-down analysis of ubiquitin-agarose was performed in the absence or presence of 50 μg of free ubiquitin (free-Ub) before elution of bound proteins and Western blot analysis of HSPs (right panels). One representative experiment of three independent experiments is shown. (B) Western blot analysis of GST-monoubiquitin (GST-Ub) and GST-polyubiquitin (GST-polyUb) obtained in vitro as described in Material and Methods. (C) Cytosolic extracts from HSP27-overexpressing cells were incubated with GST alone, GST-Ub, or GST-polyUb. The presence of HSP27 in input material was checked by Western blotting after pull-down analysis and elution of bound proteins. One representative experiment of three experiments is shown.
HSP27 binds to the 26S proteasome complex.
Since the exposure of U937 cells to either etoposide or ΤNF-α did not increase the affinity of HSP27 for ubiquitin, we examined whether the increased activity of the proteasome identified in treated cells could result from an interaction of HSP27 with the proteasome under stress conditions. The 26S proteasome results from the association of the PA700 activator complex with the 20S proteasome catalytic unit. The PA700 subunit is needed for ubiquitinated protein degradation (52). Whole-cell lysates from HSP27-transfected U937 cells were immunoprecipitated with an anti-HSP27 polyclonal antibody. As a control for the specificity of the interaction, we used an anti-HSP60 polyclonal antibody (HSP60 is a mitochondrial chaperone that can be released to the cytosol upon exposure to different stimuli to interact with cytosolic proteins) (60). We observed that while HSP60 was not coimmunoprecipitated with PA700, HSP27 was weakly coimmunoprecipitated with PA700, and this interaction was enhanced in HSP27-overexpressing cells by etoposide treatment (Fig. 4A). Figure 4B demonstrates that endogenous HSP27, which accumulates in response to heat shock, associates with PA700 as efficiently as HSP27 overexpressed through stable transfection. Immunoprecipitation experiments performed in vitro with purified proteins indicated that the small stress protein was able to directly interact with the proteolytic complex (Fig. 4C). To determine whether HSP27 binding to the 26S proteasome complex was required for enhancing proteasome activity, we used several hsp27 cDNA mutants stably expressed in the REG colon carcinoma cell line, which does not express endogenous HSP27 (6). Coimmunoprecipitation studies demonstrated that the Δ15-205 and Δ88-205 mutants had lost the capacity to bind to the PA700 subunit, whereas the Δ51-88 and Δ141-175 mutants both retained this binding ability (Fig. 4D). While the two mutants that bound to PA700 retained the capacity to increase proteasome activity upon etoposide (Fig. 4E) or ΤNF-α (data not shown) treatment, the deletion mutants that did not bind to the proteasome lost this property.
FIG. 4.
HSP27 association with the 26S proteasome is required for HSP27-induced proteasome activity. (A) Immunoprecipitation (IP) performed with control- and HSP27-transfected U937 cells and an anti-HSP27 or an anti-HSP60 antibody was followed by immunodetection of HSP27, HSP70, and the PA700 subunit of the 26S proteasome. ch., chain. (B) Immunodetection of PA700 after immunoprecipitation of HSP27 in control (Co) and heat-shocked (HS; 1 h at 42°C and then 6 h at 37°C) U937 cells. (C) HSP27 was incubated in vitro with the purified 26S proteasome before immunoprecipitation of HSP27 or HSP60 and immunodetection of the indicated proteins. (D) Immunodetection of PA700 after HSP27 immunoprecipitation of REG cells transfected with an empty vector (Control) or a vector containing either wild-type hsp27 or the indicated hsp27 deletion mutations. As a control, cell lysates were immunoprecipitated with nonrelevant immunoglobulin G1. (E) The REG cells shown in panel D were treated with etoposide (VP16, 100 μM, 24 h) before measurement of the ability of cell lysates to cleave the substrate Suc-LLVY-AMC as described in the legend to Fig. 1 (fold induction is relative to that in untreated cells). Results are the means and standard deviations for three independent experiments.
HSP27 enhances NF-κB activation in response to stress.
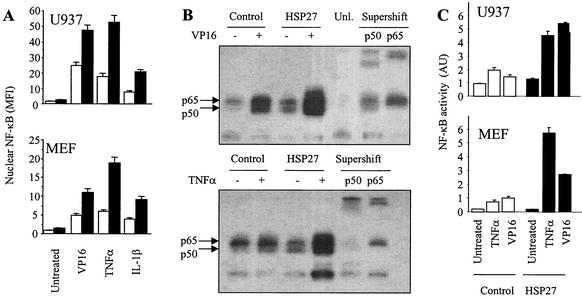
One of the proteins known to be degraded by the 26S proteasome is the NF-κB inhibitor I-κBα. NF-κB is a transcription factor maintained in a latent form in the cytoplasm of unstimulated cells by association with I-κB inhibitory molecules. The degradation of these molecules by the 26S proteasome allows NF-κB migration into the nucleus as an active factor (48). Exposure of U937 human leukemic cells to various stresses, including etoposide (100 μM), ΤNF-α (20 ng/ml), and IL-1β (1 ng/ml), triggers the nuclear localization of NF-κB, as observed by flow cytometry analysis of isolated nuclei, 4 h after the beginning of treatment (Fig. 5A). The overexpression of HSP27 favors the nuclear accumulation of the transcription factor under the three tested conditions (Fig. 5A). Similar observations were made with MEF cells exposed to the same stimuli for 24 h; i.e., the overexpression of HSP27 amplifies NF-κB accumulation in the nuclei of MEF cells exposed to these stimuli (Fig. 5A). In EMSAs, the exposure of U937 cells to either etoposide or ΤNF-α was observed to increase NF-κB DNA-binding activity, and this effect was amplified by the overexpression of HSP27. The specificity of these observations was confirmed by a supershift assay with either an anti-p50 or an anti-p65 antibody (Fig. 5B). This binding was prevented by a 50-fold molar excess of unlabeled double-stranded NF-κB oligonucleotide. Using a luciferase reporter gene under the control of four NF-κB-binding sites in tandem, we observed that NF-κB transcriptional activity in response to either etoposide or ΤNF-α was increased two- to threefold when HSP27 was overexpressed (Fig. 5C). A similar increase in NF-κB transcriptional activity was also measured in the REG rat colon cancer cell line stably transfected with hsp27 cDNA and exposed to the same stimuli (data not shown). Thus, the overexpression of HSP27 increases NF-κB transactivation ability in three different cell lines exposed to two stressful stimuli.
FIG. 5.
HSP27 increases nuclear NF-κB content, DNA binding, and transcriptional activity in stressed cells. (A) Control-transfected (white bars) and HSP27-transfected (black bars) cells either were left untreated or were treated for 4 h (U937 cells) or 24 h (MEF cells) with etoposide (VP16, 100 μM), ΤNF-α (20 ng/ml), or IL-1β (1 ng/ml). The level of nuclear NF-κB expression was determined by flow cytometry (MFI, mean fluorescence index). (B) Nuclear extracts from control-transfected and HSP27-transfected U937 cells either were left untreated or were treated for 4 h with etoposide (VP16, 100 μM) or ΤNF-α (20 ng/ml) and then were subjected to EMSAs with an NF-κB probe. As a control for binding specificity, the extracts were preincubated with a 50-fold molar excess of unlabeled oligonucleotide (Unl.). Supershift analysis was performed by preincubation of the extracts with an anti-p50 or an anti-p65 polyclonal antibody before EMSAs. (C) Control-transfected (white bars) and HSP27-overexpressing (black bars) cells were transiently transfected with an NF-κB luciferase reporter plasmid and then either were left untreated or were treated for 4 h (U937 cells) or 10 h (MEF cells) with 100 μM VP16 or 20 ng of ΤNF-α/ml. Cotransfection of a thymidine kinase-Renilla luciferase plasmid was used to normalize for transfection efficiency. AU, arbitrary units. Results are the means and standard deviations for three independent experiments.
HSP27 potentiates I-κBα degradation by the proteasome.
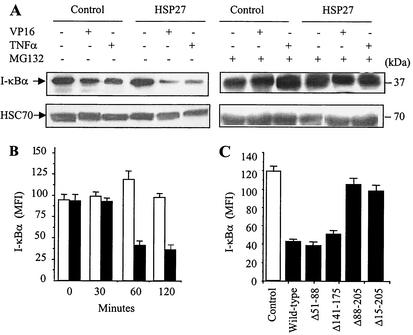
Inactivation of the major NF-κB inhibitor, I-κBα, occurs through its proteasomal degradation. Exposure of either U937 or MEF cells to ΤNF-α or etoposide for 4 h decreased the cellular content of I-κBα, an effect that was amplified by HSP27 overexpression (Fig. 6A, left panels). This decrease in I-κBα content, both in the absence and in the presence of HSP27, was the result of its proteasomal degradation, since it was inhibited by either 25 μM MG132 (Fig. 6A, right panels) or 25 μM lactacystin (data not shown). By using flow cytometry to measure the kinetics of I-κBα degradation induced by etoposide, we observed that HSP27 overexpression accelerated the degradation of I-κBα; i.e., more than 60% of I-κBα was degraded after 1 h of treatment in HSP27-overexpressing cells, while I-κBα content remained unchanged in control cells after 2 h of drug exposure (Fig. 6B). Analysis of REG cells stably transfected with either wild-type HSP27 or HSP27 deletion mutants indicated that Δ15-205 and Δ88-205, the mutants that did not bind to the PA700 subunit of the 26S proteasome (Fig. 4C), lost the capacity to increase I-κBα degradation (Fig. 6C). Altogether, these observations suggested that the proteasome was required for the HSP27-mediated increase in I-κBα degradation in response to etoposide. Similar observations were made with U937 cells exposed to ΤNF-α (data not shown).
FIG. 6.
HSP27 enhances I-κBα degradation by the proteasome. (A) Control-transfected and HSP27-transfected U937 cells were treated as indicated (VP16, 100 μM, 4 h; ΤNF-α, 20 ng/ml, 4 h; MG132, 25 μM) before monitoring of I-κBα expression by Western blotting. HSC70 served as a loading control. (B) Control-transfected (white bars) and HSP27-transfected (black bars) U937 cells were treated with etoposide (100 μM) for the indicated times before measurement of I-κBα cellular content by flow cytometry (MFI, mean fluorescence index). (C) REG cells were transfected with an empty plasmid (Control) or a plasmid encoding wild-type HSP27 or the indicated deletion mutants of HSP27 and then were treated with etoposide (VP16, 100 μM, 24 h) before measurement of I-κBα cellular content by flow cytometry as described for panel B. Results are the means and standard deviations for four independent experiments.
HSP27 interacts with phosphorylated I-κBa.
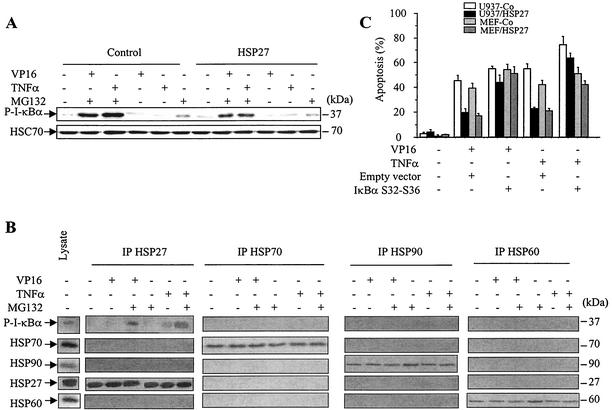
The phosphorylation of I-κBα is necessary for its ubiquitination and proteasomal degradation. We therefore analyzed whether HSP27 could also affect I-κBα phosphorylation. Control- and HSP27-transfected U937 cells were treated with etoposide (100 μM) or ΤNF-α (20 ng/ml) in the absence and in the presence of the proteasome inhibitor MG132 (25 μM). The expression of phosphorylated I-κBα, studied by using an antibody that specifically recognizes the phosphorylated protein, was hardly detected in cells exposed to either etoposide or ΤNF-α in the absence of the proteasome inhibitor, while the protein accumulated when proteasomal function was inhibited by MG132 (Fig. 7A). In any event, the overexpression of HSP27 did not significantly influence the level of phosphorylated I-κBα (Fig. 7A), suggesting that HSP27 does not affect the process of I-κBα phosphorylation in response to these stimuli.
FIG. 7.
HSP27 associates with phosphorylated I-κBα. (A) Control-transfected and HSP27-transfected U937 cells were treated as indicated (VP16, 100 μM, 4 h; ΤNF-α, 20 ng/ml, 4 h; MG132, 25 μM) before monitoring of phosphorylated I-κBα (P-I-κBα) expression by Western blotting. HSC70 served as a loading control. (B) Total cell lysates were prepared from HSP27-transfected U937 cells either left untreated or treated for 4 h with etoposide (VP16, 100 μM), ΤNF-α (20 ng/ml), and/or MG132 (25 μM) and then immunoprecipitated (IP) with the indicated antibodies. The indicated proteins were detected by Western blotting. (C) Control-transfected and HSP27-transfected U937 cells and control-transfected and HSP27-transfected MEF cells were transiently transfected with an empty vector or a plasmid containing a nonphosphorylatable, nondegradable mutant form of I-κBα (I-κBα S32A/S36A) (transfection efficiency measured with a β-galactosidase-expressing plasmid, 35 to 40%). At 36 h later, cells were treated with 100 μM etoposide (VP16) or 20 ng of TNF-α/ml for 4 h (U937 cells) or 24 h (MEF cells) before measurement of the percentage of apoptotic cells. Error bars indicate standard deviations (n = 4).
Next, we determined whether HSP27 could interact with phosphorylated I-κBα (Fig. 7B). Immunoprecipitation experiments indicated that, in response to etoposide and in the presence of MG132 to allow the accumulation of the phosphorylated and ubiquitinated protein, HSP27 strongly interacted with the inhibitory protein. The interaction of phosphorylated I-κBα with HSP27 appeared to be specific for this stress protein, since no interaction could be detected with either HSP60, HSP70, or HSP90 (Fig. 7B). Similar results were obtained with ΤNF-α-treated cells (Fig. 7B).
To determine whether the effect of HSP27 on I-κBα degradation or NF-κB activation was involved in the antiapoptotic function of HSP27, we used a nonphosphorylatable, nondegradable I-κBα mutant in which serines 32 and 36 have been replaced by alanines. This “superrepressor” mutant has been shown to sensitize prostate cancer cells to TNF-α (42). Control and HSP27-overexpressing U937 and MEF cells were transiently transfected with either the mutant plasmid or an empty vector (the efficiency of the transient transfection, measured with a β-galactosidase control plasmid, was 35 to 40%) before measurement of apoptosis induced by etoposide (100 μM) or TNF-α (20 ng/ml) in the transfected cells. We observed that the I-κBα superrepressor slightly sensitizes U937 and MEF cells to both etoposide and TNF-α. Interestingly, the protective effect of HSP27 toward drug-induced apoptosis was significantly and consistently reduced in the presence of the I-κBα mutant plasmid; in the two cell lines, HSP27 overexpression induced a 60% decrease in etoposide- or TNF-α-induced apoptosis in control cells compared to only a 15% decrease in cells expressing the superrepressor (Fig. 7C).
HSP27, phosphorylated I-κBα, and PA700 colocalize in a same cell fraction.
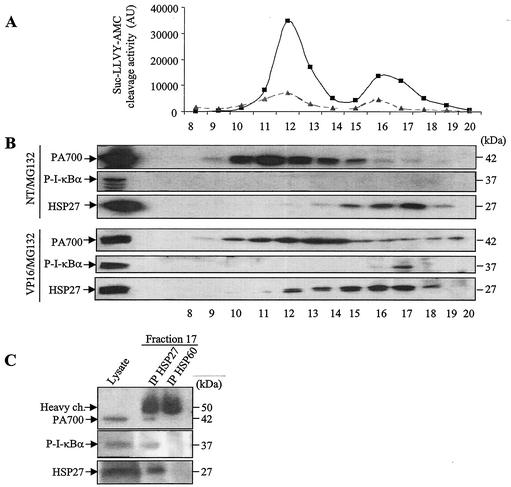
To determine whether an interaction among HSP27, phosphorylated I-κBα, and the 26S proteasome could exist in vivo, lysates from HSP27-overexpressing U937 cells treated with etoposide in the absence or presence of MG132 were centrifuged and fractionated with a Superose-6 column. The activities of the fractions against the proteasome substrate Suc-LLVY-AMC were tested. As shown in Fig. 8A, two peaks of activity were detected. These activities were inhibited by MG132.
FIG. 8.
HSP27, phosphorylated I-κBα, and PA700 can be found in the same cellular protein fraction. (A) Lysates from HSP27-transfected U937 cells not treated (triangles) or treated (squares) with etoposide (100 μM, 4 h) were centrifuged and fractionated with a Superose-6 column. Fractions were tested for hydrolysis of the substrate Suc-LLVY-AMC. AU, arbitrary units. (B) The presence of PA700, HSP27, and phosphorylated I-κBα (P-I-κBα) in fractions of cell extracts from nontreated (NT) and etoposide-treated (VP16) cells in the presence of MG132 (25 μM) to stabilize phosphorylated I-κBα was determined by Western blotting. (C) Immunodetection of PA700 and P-I-κBα after immunoprecipitation (IP) of HSP27 in fraction 17 of cells exposed to etoposide in the presence of MG132. ch., chain.
Western blot analysis of the different fractions indicated that phosphorylated I-κBα could be detected only when the treated cells were cultured in the presence of MG132, i.e., when the degradation of the phosphorylated protein was inhibited. Phosphorylated I-κBα could not be detected in extracts from untreated cells cultured in the presence of MG132 alone, whereas the protein was easily detected in etoposide-treated cells cultured under the same conditions (Fig. 8B). In this latter situation, phosphorylated I-κBα, HSP27, and PA700 were found together in fraction 17, which corresponds to the second peak of proteasome activity (Fig. 8A). Coimmunoprecipitation analysis of HSP27 and phosphorylated I-κBα or PA700 in that fraction confirmed the interaction among the three components (Fig. 8C). These results suggested that the formation of a ternary complex involving phosphorylated I-κBα, HSP27, and the proteasome occurs mainly in etoposide-treated cells. Together with other results described above, these observations suggested that stress conditions are required for HSP27 to enforce the proteasomal degradation of specific proteins.
DISCUSSION
One of the functions of small heat shock protein HSP27 is to protect stressed cells from undergoing death by apoptosis. Various mechanisms have been proposed to account for this protective effect, including a decreased content of radical oxygen species (40) and a direct interaction with cytochrome c when released from mitochondria (6). The present study identifies another role for HSP27, i.e., facilitating the activation of the ubiquitin-proteasome pathway. One of the consequences of this effect is an increase in NF-κB activity through enhanced degradation of its main inhibitor, I-κBα. Since the suppression of apoptosis is an important function of NF-κB (32), increased NF-κB activation may contribute to the antiapoptotic effect of HSP27.
HSP27 shares several properties with HSP70, another inducible HSP. When overexpressed, both stress proteins inhibit apoptosis in vitro and in vivo, induce resistance to most chemotherapeutic agents, and enhance tumorigenesis in rodents (18). However, several differences between the two chaperones have been identified: (i) the function of HSP70 depends on ATP hydrolysis, whereas that of HSP27 does not; (ii) hsp70 is an early responsive gene, whereas hsp27 is a late responsive one; and (iii) their antiapoptotic effects involve distinct molecular mechanisms (16). Consequently, these proteins have been referred to as complementary protective proteins (29). The present study suggests another difference between them, based on their interaction with the proteasomal machinery. Although HSP70 overexpression also enhances proteasome activity in response to various stresses (unpublished data), we have shown here that this effect does not depend on a direct interaction with ubiquitin chains, as was observed with HSP27. Furthermore, HSP70 does not bind to phosphorylated I-κBα. This finding is consistent with recent studies showing that HSP70-induced stimulation of proteasome activity could be mediated by interactions with its cochaperones, BAG-1 and CHIP (see below).
HSP27 activates the ubiquitin-proteasome pathway.
When proteins unfold as a result of cellular stress, molecular chaperones recognize them, prevent their aggregation, and orchestrate cellular folding processes, in conjunction with regulatory cofactors. However, not every attempt to fold a protein is successful, and misfolded proteins can be targeted to the cellular degradation machinery for destruction (25). The molecular mechanisms underlying the cooperation of molecular chaperones with the degradation machinery remain largely unknown. ATP-dependent HSP90 was shown to protect the trypsin-like activity of the proteasome system from oxidative inactivation (9) and to inhibit the activity of the proteasome (4, 15, 33). Proteasome inhibitors activate heat shock transcription factors, with the subsequent induction of HSPs (55, 56); e.g., the exposure of U373 MG human glioma cells to proteasome inhibitors induces HSF2-mediated accumulation of HSP27 (38). In the present study, we identified another relationship between the ubiquitin-proteasome pathway and HSP27. The overexpression of HSP27 in different cell types enhanced the catalytic activity of the proteasome and the degradation of ubiquitinated proteins in response to stressful stimuli.
HSP27 interacts with polyubiquitin chains.
Cooperation between the ATP-dependent molecular chaperones HSP70 and HSP90 and the ubiquitination machinery has been described. HSP70 and HSP90 cooperate to promote the degradation of endoplasmic reticulum-associated apoprotein B by the ubiquitin-proteasome system (22). It was demonstrated that HSP90 and HSP70 could capture unfolded substrates and then interact with an E3 ubiquitin ligase, such as CHIP (C terminus of Hsc70-interacting protein), a tetratricopeptide domain-containing protein that efficiently ubiquitinates the captured unfolded protein (10, 39, 43). The ubiquitin-like domain-containing protein BAG-1 cooperates with CHIP to shift the activity of the chaperone systems from protein folding to degradation (13). In the present study, neither HSP70 nor HSP90 was observed to directly interact with ubiquitin, whereas HSP27 associated with multiubiquitin chains, a result further suggesting a distinct mode of connection with the proteolytic system. Few interactions between small stress proteins and the ubiquitin-proteasome machinery have been described so far. A two-hybrid approach has revealed that Drosophila small HSPs (HSP23 and HSP27) can associate in vivo with the nuclear ubiquitin-conjugating enzyme DmUbc9, a protein involved in conjugating ubiquitin-related protein SUMO-1 to substrate proteins, but the functional consequences of this interaction are unknown (31). The ability of HSP27 to directly interact with multiubiquitin chains could account for its recently described colocalization with ubiquitinated proteins in cytoplasmic inclusions, called “aggregosomes,” that characterize a variety of degenerative diseases (28, 61). These inclusions contain misfolded, ubiquitinated proteins sheathed in a cage of intermediate filaments. HSP27 and the 20S proteasome are recruited to these aggregosomes, a fact which led to the hypothesis that HSP27 may interact with ubiquitinated proteins for the proper folding or disruption of cellular proteins.
HSP27 interacts with the 26S proteasome.
Various interactions of HSPs with the proteasome complex have been reported. The HSC70 and HSP70 cofactor BAG-1 binds to the proteolytic machinery in an ATP-dependent manner and provides a link between the HSC70/HSP70 chaperone system and the 26S proteasome (36). More recently, a yeast two-hybrid approach identified a direct and specific interaction between the small HSP αβ-crystallin (but not HSP27) and one of the 14 subunits of the 20S proteasome (5). An enzyme-linked immunosorbent assay-based method suggested that αB-crystallin and HSP90 could interact weakly with the 20S proteasome and inhibit its catalytic activity (8, 59). Again, the functional consequences of these interactions are unknown. Here, we show that HSP27 coimmunoprecipitates with the 26S proteasome, whereas HSP60 does not. The HSP27 domain involved in this interaction is distinct from that involved in the interaction of HSP27 with cytochrome c, since the deletion of amino acids 55 to 88 suppresses cytochrome c binding (6) without affecting the association of HSP27 with the 26S proteasome (this study). Interestingly, this region also was shown to be dispensable for HSP27 chaperone function in vitro (21). These results suggest that the chaperone function of HSP27 is not required for its interaction with cytochrome c and the inhibition of caspase-dependent apoptosis but is necessary for its interaction with the 26S proteasome and the ability to enhance the proteasomal degradation of ubiquitinated proteins.
HSP27 enhances NF-κB activity.
One of the well-identified functions of the ubiquitin-proteasome system is regulation of the shuttling of the Rel family of transcription factors from the cytoplasm to the nucleus in response to cell stimulation (2). The ability of HSPs to influence NF-κB activation is a complex issue. Overexpressed HSP70 was shown to have no significant influence on NF-κB activation in response to ΤNF-α or UV light (30, 54). Accordingly, NF-κB dissociation from its complexing partners during recovery from heat shock could be due to the thermolability of NF-κB/I-κBα complexes rather than to the influence of HSPs (35). On the other hand, it has been shown that heat shock can prevent or delay subsequent NF-κB activation by a stressful stimulus (14). The present study introduces a new role for HSPs in the regulation of NF-κB activation, showing that this activation is enhanced by HSP27 in response to stressful stimuli.
HSP27 interacts with phosphorylated I-κBα as well as with the 26S proteasome.
Ubiquitination-mediated proteolysis of I-κBα by the 26S proteasome is a critical step in most pathways leading to NF-κB activation (32). NF-κB activation can be inhibited by antioxidant drugs or overexpression of detoxifying enzymes, suggesting that crucial redox events or intracellular reactive oxygen species are involved (34). Despite the ability of HSP27 overexpression to decrease reactive oxygen species content (40), we show here that increased expression of the protein enhances the degradation of I-κBα in response to etoposide, ΤNF-α, and IL-1β in different cell lines. HSP27 interacts with phosphorylated I-κBα, mainly in the presence of a proteasome inhibitor that prevents the rapid degradation of the phosphorylated protein. In contrast, HSP27 does not affect I-κBα phosphorylation, nor does it interact with NF-κB (data not shown). The interaction between HSP27 and phosphorylated I-κBα appears to be specific for HSP27, since neither HSP70, HSP60, nor HSP90 could interact with the inhibitory protein. Phosphorylated I-κBα was identified in combination with the 26S proteasome and HSP27 in a high-molecular-weight fraction where the three components interact, demonstrating the existence of a ternary complex in vivo.
Concluding remarks.
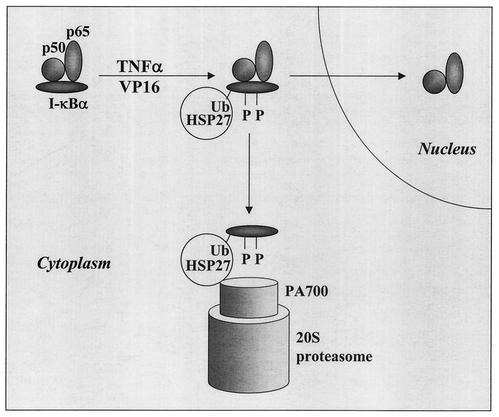
Based on these observations, we propose the hypothetical model shown in Fig. 9. In response to stimulation, I-κBα is phosphorylated and ubiquitinated, favoring its association with HSP27, and disrupts the NF-κB dimer. Supporting this hypothesis, ubiquitinated I-κBα was shown to bind to NF-κB in vitro (11), while it dissociated from p50-containing NF-κB in vivo (50), arguing for an in vivo molecular chaperone (HSP27?) that disrupts the complex. NF-κB then migrates to the nucleus, whereas HSP27 drives I-κBα to the 26S proteasome, where the protein is degraded. It is likely that HSP27 promotes efficient degradation of other ubiquitinated proteins as well, since HSP27 binds to ubiquitin and strongly decreases ubiquitinated protein cellular contents under different stressful conditions. A similar function was recently described for the valosin-containing protein, an AAA (ATPases associated with a variety of cellular activities) family member that associates with ubiquitinated proteins (12). However, it was shown that the valosin-containing protein alone was not sufficient to promote the degradation of a model substrate (58), suggesting that other proteins may be involved in the targeting of multiubiquitinated proteins to the 26S proteasome. Based on the results of the present study, we propose that HSP27 may be one of these proteins, acting as a chaperone to target different ubiquitinated substrates to the proteolytic machinery.
FIG. 9.
Hypothetical model for the influence of HSP27 on proteasome-mediated I-κBα degradation. In response to stimulation, I-κBα is phosphorylated and ubiquitinated (Ub), which could enhance its association with HSP27 and its targeting to the 26S proteasome, thus permitting the release of NF-κB dimers that translocate to the nucleus. When associated with ubiquitinated and phosphorylated I-κBα, HSP27 may favor its transfer, recognition, and/or unfolding by the 26S proteasome.
Acknowledgments
We thank A. P. Arrigo, C. Ducasse, and C. Diaz-Latoud (CNRS UMR 5534, Villeurbanne, France) for the HSP27-derived plasmids and helpful discussions. We thank A. Brent Carter (University of Iowa Health Care) for plasmid p4NF-κB-luc. We thank A. Hammann, F. Ghiringhelli, and C. Rébé for technical assistance.
This work was supported by the Institut National pour la Santé et la Recherche Medicale (INSERM) and by grants from the Ligue Nationale contre le Cancer and the Association pour la Recherche contre le Cancer (ARC grants 9567 and 5204). A.P. was supported by a fellowship from the Ministère de l'Education Nationale, and S.G. was supported by a poste vert from the INSERM.
REFERENCES
- 1.Adams, J. 2002. Proteasome inhibitors as new anticancer drugs. Curr. Opin. Oncol. 14:628-634. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A. S., Jr. 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 3.Benaroudj, N., and A. L. Goldberg. 2000. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat. Cell Biol. 2:833-839. [DOI] [PubMed] [Google Scholar]
- 4.Bender, A. T., D. R. Demady, and Y. Osawa. 2000. Ubiquitination of neuronal nitric-oxide synthase in vitro and in vivo. J. Biol. Chem. 275:17407-17411. [DOI] [PubMed] [Google Scholar]
- 5.Boelens, W. C., Y. Croes, and W. W. de Jong. 2001. Interaction between alphaB-crystallin and the human 20S proteasomal subunit C8/alpha7. Biochim. Biophys. Acta 1544:311-319. [DOI] [PubMed] [Google Scholar]
- 6.Bruey, J. M., C. Ducasse, P. Bonniaud, L. Ravagnan, S. A. Susin, C. Diaz-Latoud, S. Gurbuxani, A. P. Arrigo, G. Kroemer, E. Solary, and C. Garrido. 2000. Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat. Cell Biol. 2:645-652. [DOI] [PubMed] [Google Scholar]
- 7.Carter, A. B., K. L. Knudtson, M. M. Monick, and G. W. Hunninghake. 1999. The p38 mitogen-activated protein kinase is required for NF-kappaB-dependent gene expression. The role of TATA-binding protein (TBP). J. Biol. Chem. 274:30858-30863. [DOI] [PubMed] [Google Scholar]
- 8.Conconi, M., L. Djavadi-Ohaniance, W. Uerkvitz, K. B. Hendil, and B. Friguet. 1999. Conformational changes in the 20S proteasome upon macromolecular ligand binding analyzed with monoclonal antibodies. Arch. Biochem. Biophys. 362:325-328. [DOI] [PubMed] [Google Scholar]
- 9.Conconi, M., I. Petropoulos, I. Emod, E. Turlin, F. Biville, and B. Friguet. 1998. Protection from oxidative inactivation of the 20S proteasome by heat-shock protein 90. Biochem. J. 333:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 11.Dai, R. M., E. Chen, D. L. Longo, C. M. Gorbea, and C. C. Li. 1998. Involvement of valosin-containing protein, an ATPase co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J. Biol. Chem. 273:3562-3573. [DOI] [PubMed] [Google Scholar]
- 12.Dai, R. M., and C. C. Li. 2001. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3:740-744. [DOI] [PubMed] [Google Scholar]
- 13.Demand, J., S. Alberti, C. Patterson, and J. Hohfeld. 2001. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11:1569-1577. [DOI] [PubMed] [Google Scholar]
- 14.DeMeester, S. L., T. G. Buchman, and J. P. Cobb. 2001. The heat shock paradox: does NF-kappaB determine cell fate? FASEB J. 15:270-274. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, N., S. Sato, A. Ishida, and T. Tsuruo. 2002. Involvement of Hsp90 in signaling and stability of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 277:10346-10353. [DOI] [PubMed] [Google Scholar]
- 16.Garrido, C. 2002. Size matters: of the small HSP27 and its large oligomers. Cell Death Differ. 9:483-485. [DOI] [PubMed] [Google Scholar]
- 17.Garrido, C., J. M. Bruey, A. Fromentin, A. Hammann, A. P. Arrigo, and E. Solary. 1999. HSP27 inhibits cytochrome c-dependent activation of procaspase-9. FASEB J. 13:2061-2070. [DOI] [PubMed] [Google Scholar]
- 18.Garrido, C., S. Gurbuxani, L. Ravagnan, and G. Kroemer. 2001. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem. Biophys. Res. Commun. 286:433-442. [DOI] [PubMed] [Google Scholar]
- 19.Garrido, C., P. Ottavi, A. Fromentin, A. Hammann, A. P. Arrigo, B. Chauffert, and P. Mehlen. 1997. HSP27 as a mediator of confluence-dependent resistance to cell death induced by anticancer drugs. Cancer Res. 57:2661-2667. [PubMed] [Google Scholar]
- 20.Goldring, C. E., S. Reveneau, A. Chantome, A. Pance, C. Fleury, D. A. Hume, D. Sester, B. Mignotte, and J. F. Jeannin. 2000. Heat shock enhances transcriptional activation of the murine-inducible nitric oxide synthase gene. FASEB J. 14:2393-2395. [DOI] [PubMed] [Google Scholar]
- 21.Guo, Z., and L. F. Cooper. 2000. An N-terminal 33-amino-acid-deletion variant of hsp25 retains oligomerization and functional properties. Biochem. Biophys. Res. Commun. 270:183-189. [DOI] [PubMed] [Google Scholar]
- 22.Gusarova, V., A. J. Caplan, J. L. Brodsky, and E. A. Fisher. 2001. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J. Biol. Chem. 276:24891-24900. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama, S., M. Kitagawa, K. Nakayama, M. Shirane, M. Matsumoto, K. Hattori, H. Higashi, H. Nakano, K. Okumura, K. Onoe, and R. A. Good. 1999. Ubiquitin-dependent degradation of IkappaBalpha is mediated by a ubiquitin ligase Skp1/Cul1/F-box protein FWD1. Proc. Natl. Acad. Sci. USA 96:3859-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hideshima, T., C. Mitsiades, M. Akiyama, T. Hayashi, D. Chauhan, P. Richardson, R. Schlossman, K. Podar, N. C. Munshi, N. Mitsiades, and K. C. Anderson. 2002. Molecular mechanisms mediating anti-myeloma activity of proteasome inhibitor PS-341. Blood 26:26.. [DOI] [PubMed] [Google Scholar]
- 25.Hohfeld, J., D. M. Cyr, and C. Patterson. 2001. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2:885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horman, S., P. Galand, R. Mosselmans, N. Legros, G. Leclercq, and N. Mairesse. 1997. Changes in the phosphorylation status of the 27 kDa heat shock protein (HSP27) associated with the modulation of growth and/or differentiation in MCF-7 cells. Cell Prolif. 30:21-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huot, J., G. Roy, H. Lambert, P. Chretien, and J. Landry. 1991. Increased survival after treatments with anticancer agents of Chinese hamster cells expressing the human Mr 27, 000 heat shock protein. Cancer Res. 51:5245-5252. [PubMed] [Google Scholar]
- 28.Ito, H., K. Kamei, I. Iwamoto, Y. Inaguma, R. Garcia-Mata, E. Sztul, and K. Kato. 2002. Inhibition of proteasomes induces accumulation, phosphorylation, and recruitment of HSP27 and alphaB-crystallin to aggresomes. J. Biochem. (Tokyo) 131:593-603. [DOI] [PubMed] [Google Scholar]
- 29.Jaattela, M. 1999. Heat shock proteins as cellular lifeguards. Ann. Med. 31:261-271. [DOI] [PubMed] [Google Scholar]
- 30.Jaattela, M., and D. Wissing. 1993. Heat-shock proteins protect cells from monocyte cytotoxicity: possible mechanism of self-protection. J. Exp. Med. 177:231-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joanisse, D. R., Y. Inaguma, and R. M. Tanguay. 1998. Cloning and developmental expression of a nuclear ubiquitin-conjugating enzyme (DmUbc9) that interacts with small heat shock proteins in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 244:102-109. [DOI] [PubMed] [Google Scholar]
- 32.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 33.Katschinski, D. M., L. Le, D. Heinrich, K. F. Wagner, T. Hofer, S. G. Schindler, and R. H. Wenger. 2002. Heat induction of the unphosphorylated form of hypoxia-inducible factor-1alpha is dependent on heat shock protein-90 activity. J. Biol. Chem. 277:9262-9267. [DOI] [PubMed] [Google Scholar]
- 34.Kretz-Remy, C., P. Mehlen, M. E. Mirault, and A. P. Arrigo. 1996. Inhibition of I kappa B-alpha phosphorylation and degradation and subsequent NF-kappa B activation by glutathione peroxidase overexpression. J. Cell Biol. 133:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretz-Remy, C., B. Munsch, and A. P. Arrigo. 2001. NFkappa B-dependent transcriptional activation during heat shock recovery. Thermolability of the NF-kappaB.Ikappa B complex. J. Biol. Chem. 276:43723-43733. [DOI] [PubMed] [Google Scholar]
- 36.Luders, J., J. Demand, and J. Hohfeld. 2000. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275:4613-4617. [DOI] [PubMed] [Google Scholar]
- 37.MacFarlane, M., W. Merrison, S. B. Bratton, and G. M. Cohen. 2002. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 277:36611-36616. [DOI] [PubMed] [Google Scholar]
- 38.Mathew, A., S. K. Mathur, and R. I. Morimoto. 1998. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 18:5091-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 40.Mehlen, P., E. Hickey, L. A. Weber, and A. P. Arrigo. 1997. Large unphosphorylated aggregates as the active form of hsp27 which controls intracellular reactive oxygen species and glutathione levels and generates a protection against TNFalpha in NIH-3T3-ras cells. Biochem. Biophys. Res. Commun. 241:187-192. [DOI] [PubMed] [Google Scholar]
- 41.Mitsiades, N., C. S. Mitsiades, V. Poulaki, D. Chauhan, G. Fanourakis, X. Gu, C. Bailey, M. Joseph, T. A. Libermann, S. P. Treon, N. C. Munshi, P. G. Richardson, T. Hideshima, and K. C. Anderson. 2002. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 99:14374-14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muenchen, H. J., D. L. Lin, M. A. Walsh, E. T. Keller, and K. J. Pienta. 2000. Tumor necrosis factor-alpha-induced apoptosis in prostate cancer cells through inhibition of nuclear factor-kappaB by an IkappaBalpha “super-repressor.” Clin. Cancer Res. 6:1969-1977. [PubMed] [Google Scholar]
- 43.Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navon, A., and A. L. Goldberg. 2001. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell 8:1339-1349. [DOI] [PubMed] [Google Scholar]
- 45.Nicholl, I. D., and R. A. Quinlan. 1994. Chaperone activity of alpha-crystallins modulates intermediate filament assembly. EMBO J. 13:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oesterreich, S., S. G. Hilsenbeck, D. R. Ciocca, D. C. Allred, G. M. Clark, G. C. Chamness, C. K. Osborne, and S. A. Fuqua. 1996. The small heat shock protein HSP27 is not an independent prognostic marker in axillary lymph node-negative breast cancer patients. Clin. Cancer Res. 2:1199-1206. [PubMed] [Google Scholar]
- 47.Paul, C., F. Manero, S. Gonin, C. Kretz-Remy, S. Virot, and A. P. Arrigo. 2002. Hsp27 as a negative regulator of cytochrome c release. Mol. Cell. Biol. 22:816-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 49.Reidlinger, J., A. M. Pike, P. J. Savory, R. Z. Murray, and A. J. Rivett. 1997. Catalytic properties of 26 S and 20 S proteasomes and radiolabeling of MB1, LMP7, and C7 subunits associated with trypsin-like and chymotrypsin-like activities. J. Biol. Chem. 272:24899-24905. [DOI] [PubMed] [Google Scholar]
- 50.Roff, M., J. Thompson, M. S. Rodriguez, J. M. Jacque, F. Baleux, F. Arenzana-Seisdedos, and R. T. Hay. 1996. Role of IkappaBalpha ubiquitination in signal-induced activation of NFkappaB in vivo. J. Biol. Chem. 271:7844-7850. [DOI] [PubMed] [Google Scholar]
- 51.Samali, A., J. D. Robertson, E. Peterson, F. Manero, L. van Zeijl, C. Paul, I. A. Cotgreave, A. P. Arrigo, and S. Orrenius. 2001. Hsp27 protects mitochondria of thermotolerant cells against apoptotic stimuli. Cell Stress Chaperones 6:49-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawada, H., T. Akaishi, M. Katsu, and H. Yokosawa. 1997. Difference between PA700-like proteasome activator complex and the regulatory complex dissociated from the 26S proteasome implies the involvement of modulating factors in the 26S proteasome assembly. FEBS Lett. 412:521-525. [DOI] [PubMed] [Google Scholar]
- 53.Shirane, M., S. Hatakeyama, K. Hattori, and K. Nakayama. 1999. Common pathway for the ubiquitination of IkappaBalpha, IkappaBbeta, and IkappaBepsilon mediated by the F-box protein FWD1. J. Biol. Chem. 274:28169-28174. [DOI] [PubMed] [Google Scholar]
- 54.Simon, M. M., A. Reikerstorfer, A. Schwarz, C. Krone, T. A. Luger, M. Jaattela, and T. Schwarz. 1995. Heat shock protein 70 overexpression affects the response to ultraviolet light in murine fibroblasts. Evidence for increased cell viability and suppression of cytokine release. J. Clin. Investig. 95:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stangl, K., C. Gunther, T. Frank, M. Lorenz, S. Meiners, T. Ropke, L. Stelter, M. Moobed, G. Baumann, P. M. Kloetzel, and V. Stangl. 2002. Inhibition of the ubiquitin-proteasome pathway induces differential heat-shock protein response in cardiomyocytes and renders early cardiac protection. Biochem. Biophys. Res. Commun. 291:542-549. [DOI] [PubMed] [Google Scholar]
- 56.Tacchini, L., P. Dansi, E. Matteucci, A. Bernelli-Zazzera, and M. A. Desiderio. 2001. Influence of proteasome and redox state on heat shock-induced activation of stress kinases, AP-1 and HSF. Biochim. Biophys. Acta 1538:76-89. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka, K., T. Kawakami, K. Tateishi, H. Yashiroda, and T. Chiba. 2001. Control of IkappaBalpha proteolysis by the ubiquitin-proteasome pathway. Biochimie 83:351-356. [DOI] [PubMed] [Google Scholar]
- 58.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, B. J., and J. W. Margolis. 1995. Age-dependent association of isolated bovine lens multicatalytic proteinase complex (proteasome) with heat-shock protein 90, an endogenous inhibitor. Arch. Biochem. Biophys. 323:455-462. [DOI] [PubMed] [Google Scholar]
- 60.Xanthoudakis, S., S. Roy, D. Rasper, T. Hennessey, Y. Aubin, R. Cassady, P. Tawa, R. Ruel, A. Rosen, and D. W. Nicholson. 1999. Hsp60 accelerates the maturation of pro-caspase-3 by upstream activator proteases during apoptosis. EMBO J. 18:2049-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zatloukal, K., C. Stumptner, A. Fuchsbichler, H. Heid, M. Schnoelzer, L. Kenner, R. Kleinert, M. Prinz, A. Aguzzi, and H. Denk. 2002. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160:255-263. [DOI] [PMC free article] [PubMed] [Google Scholar]