Abstract
Transcripts of the myotonic dystrophy protein kinase (DMPK) gene, a member of the Rho kinase family, are subject to cell-type specific alternative splicing. An imbalance in the splice isoform profile of DMPK may play a role in the pathogenesis of DM1, a severe multisystemic disorder. Here, we report how structural subdomains determine biochemical properties and subcellular distribution of DMPK isoforms. A newly developed kinase assay revealed that DMPK is a Lys/Arg-directed kinase. Individual DMPK isoforms displayed comparable transphosphorylation activity and sequence preference for peptide substrates. However, DMPK autophosphorylation and phosphorylation of MYPT1 (as putative in vivo target of DMPK), were dependent on presence of an alternatively spliced VSGGG motif and the nature of the C terminus. In-gel effects of the VSGGG motif on the migration behavior of full-length kinase provide evidence for a model in which this motif mediates 3-D-conformational changes in DMPK isoforms. Finally, different C termini endow DMPK with the ability to bind to either endoplasmic reticulum or mitochondria or to adopt a cytosolic location. Our results suggest that DMPK isoforms have cell-type and location dependent substrate specificities with a role in organellar and cytoarchitectural dynamics.
Myotonic dystrophy (DM1) is the most common form of muscular dystrophy in adults (23), caused by amplification of an unstable (CTG)n repeat in the 3′ untranslated region (3′-UTR) of the DM1 protein kinase (DMPK) gene (10, 20, 29). The severity of the disease is correlated to the length of this repeat expansion, whereas there is an inverse correlation to the age of onset. The favored explanation for the DM1 phenotype is a gain-of-function at the RNA level (reviewed in references 41 and 53), whereby long (CUG)n repeat tracts in DMPK transcripts cause a global perturbation of RNA processing events in the nucleus. Reports of cis-effects on the accumulation of (CUG)n repeat containing DMPK transcripts in the nucleus and trans-effects on alternative splicing of transcripts encoding other muscle or brain proteins support this hypothesis (15, 16, 30, 40, 43, 45).
It is widely accepted, nonetheless, that the highly variable and complex DM1 phenotype is not caused solely by detrimental effects of (CUG)n expansion at the RNA level, but that also direct effects on DMPK gene products and local gene effects are involved in specific disease features. Various studies have shown that expansion of the (CTG)n repeat results in reduced appearance of DMPK in the cytoplasm (53). Studies in knockout mouse and myocyte cell models indicated that lack of DMPK protein may be associated with typical DM1 symptoms like myopathy and heart conduction defects, perhaps via effects on Ca2+ or Na+ ion homeostasis (5, 6, 25, 35, 42).
DMPK is a member of the AGC group of serine/threonine kinases (31) and is most homologous to the p21-activated kinases MRCK (28) and ROCK/rho-kinase/ROK (4). Other mammalian homologues are NDR1 (32), warts/lats (26, 55), and citron kinase (17). DMPK has been shown to modulate skeletal muscle Na+ channels (36). Furthermore, from in vitro studies a number of DMPK substrates have been identified, like the dihydropyridine receptor, CUG-BP, DMAP, MKBP, phospholemman, and myosin phosphatase targeting subunit (reviewed in references 52 and 53), but the candidacy of none of these proteins has yet been firmly established. Besides that, it is not known whether differences in substrate specificity exist between DMPK isoforms.
Study at the mRNA level revealed that six major DMPK isoforms, conserved between humans and mice, are produced by a combination of three different alternative splice events, one of which is cell-type specific (22) (Fig. 1A). All isoforms share an N-terminal domain, a kinase domain and a coiled coil region, while alternative splicing determines presence or absence of a 5-amino-acid (-aa) VSGGG motif and the nature of the C terminus (three cell-type dependent variants). A new human DMPK isoform was recently reported (50). This minor isoform, designated DMPK G here, carries yet another C terminus, but, more importantly, its mRNA lacks the (CUG)n repeat in its 3′-UTR. As a result, unlike DMPK transcripts bearing long (CUG)n repeats, DMPK G transcripts can freely leave the nucleus, thus creating an altered DMPK isoform profile in the cytoplasm of cells of patients where the DMPK gene is expressed.
FIG. 1.
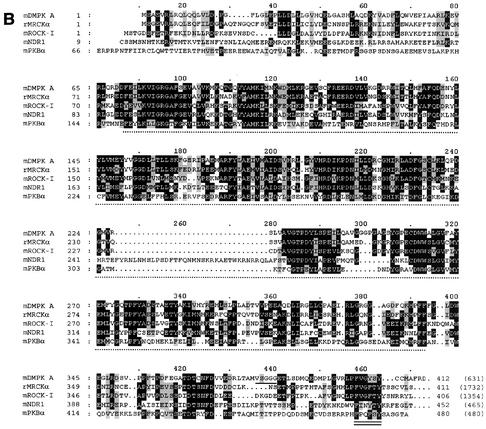
DMPK: domain organization and homology to serine/threonine kinase family members. (A) Major DMPK isoforms A to F have an N-terminal leucine-rich domain (L), a serine/threonine kinase domain, a protein kinase C-terminal domain containing the hydrophobic phosphorylation motif, and a coiled coil region. Differences between isoforms originate from alternative splicing, conserved between humans and mice: (i) a VSGGG-sequence can be present (isoforms A, C, and E) or absent (isoforms B, D, and F) and (ii) three different C-terminal tails occur. Minor splice form DMPK G, only present in humans, carries a fourth type of C terminus, of which the N-terminal half is identical to tail 1. (B) Sequence comparison between mDMPK, rMRCKα, mROCK-I, mNDR1, and mPKBα. Only the first 412 aa of DMPK are shown, since no relevant homology exists with the other kinases beyond this point. Identical amino acids (in at least three of the five kinases) are shown in white on a black background, and similar amino acids are shown in black on a grey background. The kinase domain is indicated with a dotted line below the sequence, the VSGGG sequence is underlined, and the hydrophobic phosphorylation motif is doubly underlined. The total number of amino acids for each full-length protein is indicated in parentheses; note that rMRCKα and mROCK-I are very large proteins compared to mDMPK (accession numbers: P54265, T14039, S74244, AAH09658, and P31750). (C) Sequence identity between mDMPK, rMRCKα, ROCK-I, mNDR1, and mPKBα. The N terminus (aa 1 to 70), the kinase domain (aa 71 to 339), and the protein kinase C-terminal domain (aa 340 to 405) of DMPK were compared with the corresponding parts in rMRCKα, mROCK-I, mNDR1, and mPKBα using ClustalW. The relative sequence identity for each domain is expressed as a percentage relative to mDMPK. Values for the protein kinase C-terminal domain without the VSGGG motif (as in DMPK B, D, and F) are listed in parentheses. Similar results were obtained with rMRCKβ and mROCK-II (data not shown).
In this work, we identify possible cell biological implications of imbalance in the DMPK isoform repertoire. To study activity and substrate specificity of individual DMPK isoforms an in vitro assay was developed which revealed the peptide-substrate specificities of the DMPKs. The internal VSGGG motif appears to simultaneously modulate DMPK autophosphorylation activity and protein conformation, as displayed by protein migration behavior in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) before and after phosphorylation. Furthermore, we demonstrate that the nature of the alternate C terminus of DMPK is a key regulatory factor for substrate specificity, as well as intracellular localization. Proteins with a hydrophobic C terminus target to the endoplasmic reticulum (ER), while proteins with a more hydrophilic C terminus bind to the mitochondrial outer membrane. Proteins with a very short C-terminal tail adopt a cytosolic location. Our data, together with the sequence conservation with respect to other AGC kinases, suggest that the VSGGG and tail domains may provide a critical function in specifying DMPK's role in the regulation of cytoarchitectural-organellar dynamics.
MATERIALS AND METHODS
Cell culture, transfection, and immunofluorescence microscopy.
COS-1 and N2A cells were cultured as described (22) and transfected using DEAE-dextran or Lipofectamine (Invitrogen), respectively. For immunolocalization, cells were cultured on glass coverslips, transfected and grown for approximately 24 h. Cells were fixed in 2% formaldehyde in phosphate-buffered saline (PBS), permeabilized with PBS containing 0.2% NP-40, and processed for immunofluorescence microscopy using B79, a DMPK-specific antibody (22), or 12CA5 (antihemagglutinin [anti-HA]). ER was visualized using a construct expressing GFP with an N-terminal calreticulin signal sequence and a C-terminal KDEL sequence for ER retention (Clontech), mitochondria were visualized with an antibody against cytochrome c oxidase (47). Preparations were examined and optical sections were recorded on a Bio-Rad MRC1024 confocal scanning laser microscope using LaserSharp software.
DMPK plasmids.
DMPK isoforms N-terminally tagged with a His tag, pSGHismDMPK A through F, were obtained by cloning BglII-BglII DMPK cDNA constructs (22) into the BamHI site of pSG8, i.e., in frame with the His tag present in the vector.
DMPK kinase-dead (kd) mutants were created via a Lys100Ala mutation in the kinase domain, which inactivates the invariant lysine necessary for ATP binding (31). Primers 5′-GGCCAAGTGTATGCCATGGCAATTATGAATAAGTGGGAC-3′ and 5′-GTCCCACTTATTCATAATTGCCATGGCATACACTTGGCC-3′ were used in the QuikChange site-directed mutagenesis kit (Stratagene) with pBlmDMPK C as the template (22). A 1,300-bp AflII-BspEI DMPK fragment containing the Lys100Ala mutation was used to replace the corresponding fragment in pSGHismDMPK A, C, and E. In kd pSGHismDMPK A, C, and E a 937-bp MscI-BspEI fragment was replaced by the corresponding 922-bp MscI-BspEI fragment isolated from pSGmDMPK D to create pSGHismDMPK B, D, and F kd, respectively.
To create plasmids expressing DMPK EVA (Ser379Ala) and EVD (Ser379Asp) mutants two PCRs were done using primers 5′-ACTGCCATGGTGGCCGGGGGCGGGGAGACGCT-3′ and 5′-ACTGCCATGGTCGACGGGGGCGGGGAGACGCT-3′, respectively, as forward primers; 5′-TTTCGGACCTCGGCCTCCTGTA-3′ as the reverse primer; and pSGmDMPK C as the template. PCR products were digested with NcoI and BspEI. The 354-bp NcoI-BspEI fragments were used to replace the corresponding VSGGG-fragment in pSGmDMPK E.
For the HA-tagged DMPK isoforms, untagged pSGmDMPK C was amplified with primers 5′-ATAAGATCTGCCGCCGCCATGTACCCCTACGACGTGCCCGACTATGCTATGTCAGCCGAAGTGCG-3′ and RVI101 5′-ATTCTCGAGTCAAATCTTCATGGCATACACTTG-3′. The 363-bp PCR product was subcloned between the BglII and AflII sites of pSGmDMPK A-F, resulting in pSGHAmDMPK A-F. Subsequently, pSGHAmDMPK A-F kd were obtained by replacing the 1,300-bp (isoforms with VSGGG) or 1,285-bp (isoforms without VSGGG) AflII-BspEI fragment for the corresponding kd fragment.
Isoform DMPK G was expressed as a chimeric protein with a mouse DMPK moiety (aa 1 to 553) and human tail 4 (aa 554 to 623). To clone human tail 4 an RT reaction was done with an oligo(dT) primer on human skeletal muscle RNA followed by a PCR with primers 5′-GCGGATGGAGTTGCTGCAGGC-3′ and 5′-(T)12GGGCAGATGGA-3′. Since the predicted 635-bp E16+ product (50) could not be detected on agarose gel (we therefore estimated DMPK G levels tenfold lower than reported by Tiscornia and Mahadevan), we performed a nested PCR with primers 5′-ACCTGCTGCTCCCTGCCAGGGCTGA-3′ and 5′-CCCAGATCTTTGGGCAGATGGAGGGC-3′. This yielded the expected E16 + 482-bp product. This product was used in a third PCR together with a 244-bp SmaI-SmaI fragment from pSGmDMPK B and primers 5′-CCACGGATCCACCTTCCC-3′ and 5′-CCCAGATCTTTGGGCAGATGGAGGGC-3′. The resulting 572-bp product was digested with XbaI and BglII, resulting in a 543-bp fragment. This fragment was cloned together with the 1.7-kbp BglII-XbaI fragment from pSGmDMPK A in the BglII site of pSG8ΔEco, resulting in pSGDMPK G. The HA tag and kd mutation were incorporated as described above. The sequence of all fragments obtained by PCR was verified by DNA sequencing.
MYPT1.
Myosin phosphatase targeting subunit 1 (MYPT1) was cloned by RT-PCR using mouse stomach RNA with primers 5′-GGAGAATTCATGAAGATGGCGGAC-3′ and 5′-CCTGAATTCCTACTTGGAAAGTTTGCT-3′ (start and stop codons underlined) in a pSG8-based vector in frame with a His tag and a vesicular stomatitis virus (VSV) tag. To purify MYPT1, COS-1 cells were transfected with pSG8HisVSVMYPT1, grown for 24 h and lysed in His lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.05% Tween-20, 1% NP-40) including a protease inhibitor cocktail. Ni-nitrilotriacetic acid beads (Qiagen) were incubated with MYPT1 lysate overnight, washed four times with His lysis buffer, and then used as substrate in the DMPK kinase assay as mentioned below.
Western blot analysis.
Three procedures were followed to prepare and analyze extracts from transfected COS-1 cells: Cells were incubated (i) on ice in RIPA buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium desoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM NaF). Cell extracts were obtained after centrifugation for 10 min at 14,000 × g or (ii) on ice in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.2% NP-40, 1 mM PMSF, 10 mM NaF), and cell extracts were used in an immunoprecipitation with B79 coupled to protein A Sepharose beads, or (iii) to exclude phosphatase activity during lysis, cells were scraped in 10% trichloroacetic acid (TCA)-10 mM dithiothreitol (DTT)-acetone at −80°C (46). Lysate was centrifuged for 10 min at 14,000 × g and the pellet was washed with excess acetone containing 10 mM DTT at 4°C to remove TCA. Cell extracts, immunoprecipitations and pellets were analyzed on an SDS-8 or 10% polyacrylamide gel and blotted to nitrocellulose or polyvinylidene difluoride (PVDF) membrane. Blots were blocked in TBST (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) containing 5% milk powder and incubated with B79, 12CA5, or anti-VSV antibodies in TBST. After washing in TBST blots were incubated with peroxidase-conjugated immunoglobulins (Pierce), washed in TBST-TBS, developed using a chemiluminescence Western blotting reagent and exposed to film.
DMPK in vitro kinase assay.
A kinase assay for DMPK was developed essentially as described previously (24). COS-1 cells transfected with HA-DMPK constructs or empty pSG8ΔEco vector (as a negative control) were washed with ice-cold PBS and lysed on ice with assay lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 25 mM NaF, 1 mM sodium pyrophosphate, 0.1 mM Na3VO4, 2 μM Microcystin LR [ALEXIS], 1 mM PMSF), supplemented with a protease inhibitor cocktail (Roche). Cell extracts were obtained after centrifuging for 20 min at 14,000 × g. Protein concentrations were determined according to Bradford (7) using bovine serum albumin as a standard. One large or many individual immunoprecipitation(s) were done; typically, 200 μg of cell extract was used for two (duplicate) kinase reactions. Cell extract was used in an immunoprecipitation with monoclonal antibody 12CA5 coupled to protein A-Sepharose for 2 h at 4°C. Beads were washed once in assay lysis buffer supplemented with 0.5 M NaCl, once in assay lysis buffer, and finally once in 50 mM Tris-HCl (pH 7.5)-1 mM DTT-1 mM PMSF-1 mM benzamidine. Beads were aliquoted into the desired number of kinase reactions after the final wash. The final wash was removed, and 40 μl of kinase assay buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 2 mM MnCl2, 1 mM DTT, 1 μM protein kinase A [PKA] inhibitor peptide [Bachem], 1 mM PMSF, 1 mM benzamidine, 2 μM Microcystin LR, 30 μM substrate peptide) was added. For assaying MYPT1 as substrate, kinase buffer (without substrate peptide) and purified MYPT1 bound to Ni-nitrilotriacetic acid beads were added. The reaction was started by adding 5 μl of 0.5 mM [γ-32P]ATP (∼2 μCi) and lasted 60 min at 30°C. Under these conditions phosphate incorporation into peptides was linear. The reaction was quenched by adding 5 μl 0.5 M EDTA. Supernatants were removed and spotted onto P81 phosphocellulose paper (Whatman). Filters were washed 5 times for 5 min in 1% phosphoric acid and once in acetone and were dried and counted in a liquid scintillation counter. In all peptide library experiments duplicate kinase reactions with peptide KKRNRRLSVA were included as a standard. SDS-PAGE sample buffer was added to the beads and one third was used for SDS-PAGE and blotting to PVDF membrane. To verify protein input, the blot was stained with Coomassie brilliant blue or incubated with anti-HA or anti-VSV. To analyze DMPK autophosphorylation and MYPT1 phosphorylation, the blot was exposed to film or used for quantitative phosphorimager analysis.
In vivo [32P]orthophosphate labeling.
For the in vivo [32P]orthophosphate labeling, COS-1 cells transfected with DMPK expression constructs were grown for 20 h, washed with phosphate-free medium and cultured in phosphate-free medium containing [32P]orthophosphate (Amersham Pharmacia Biotech; 0.1 mCi/ml of medium) for 4 h prior to lysis in assay lysis buffer.
RESULTS
DMPK occurs in six major splice isoforms which are multidomain proteins.
To relate DMPK to other AGC kinase family members, we compared the amino acid sequence and domain organization of mDMPK, rMRCKα, mROCK-I, and the more distant, but functionally and structurally well-studied, relatives mNDR1 and mPKBα (also called Akt1) (8). All DMPK isoforms share four protein domains (Fig. 1A): (i) The N terminus (aa 1 to 70) is leucine-rich (23%) and contains a leucine zipper. This domain is also found in MRCKα and in ROCK-I (and in citron kinase; data not shown), but not in NDR1 or PKBα (Fig. 1B and C). (ii) The DMPK kinase domain shows all features of a serine/threonine kinase domain (reference 31 and references therein). The percentage identity with the other kinase domains ranges from 70% for MRCKα to 32% for PKBα (Fig. 1C). Only NDR1 has a characteristic insertion in the activation loop. (iii) Immediately C-terminal to the kinase domain is the “protein kinase C-terminal domain” or “extension to serine/threonine-type protein kinases” (aa 340 to 405). This domain is found in all five kinases investigated and in many other members of the kinase superfamily, such as PKC and p70 S6 kinase, but is, for example, only partly present in PKA and completely absent in myosin light-chain kinase. Its most C-terminal part is known as the hydrophobic phosphorylation motif with the consensus sequence FXX(F/Y)(S/T)(F/Y). (iv) The fourth shared DMPK domain (aa 463 to 530) is an α-helical region with homology to FERM domains and the myosin heavy chain tail region. This domain is thought to be involved in the formation of coiled coil structures. Large coiled coil regions are found in MRCKα/β and ROCK-I/II, but only a low sequence homology between the DMPK coiled coil region and that of MRCKα/β was detected (data not shown). Coiled coil regions are neither found in mNDR1 nor in mPKBα.
The alternatively spliced VSGGG motif in the C-terminal region of the kinase domain is present in approximately 50% of all DMPK isoforms and—based on computer prediction—might serve as a putative phosphorylation site or glycosaminoglycan addition site (Fig. 1B). The VSGGG motif is not present in any of the four kinases investigated and BLAST searches indicate that it is also unique in the entire protein sequence database.
Different C termini exist among the DMPK isoforms. Tail 3 is only 2 aa long and is found in the smooth muscle-specific short isoforms DMPK E and F. The long isoforms, specific for skeletal muscle, heart and brain, have either a hydrophobic (DMPK A and B) or a relatively hydrophilic tail (DMPK C and D). As stated above, minor human isoform DMPK G carries a different, fourth type of tail. C termini comparable to the four tails of DMPK are not found in any of the other DMPK family members. In fact, DMPK tails show no appreciable homology with any other protein in the database.
VSGGG-dependent migration behavior is associated with autophosphorylation.
Previously, we and others have reported that endogenous DMPK from tissues showed a confusingly complex array of signals on Western blots (see reference 22 and references therein). Also the protein products from one single DMPK expression vector in transfected cell lines appeared as multiple bands. Natural occurrence of cell-type specific DMPK splice isoforms in addition to proteolytic processing at the C terminus explained these observations (22; data not shown). Here, we analyzed products of cDNA expression constructs for each of the different DMPKs by use of transfection into COS-1 cells combined with an isolation method with prevention of proteolytic breakdown.
As expected, long DMPK isoforms A-D migrated as products with a higher molecular weight than short isoforms E and F, though all had an apparent molecular weight greater than predicted from their amino acid sequence (Fig. 2A). We observed a correlation between presence of the VSGGG motif and product heterogeneity for each of the isoform pairs A/B, C/D, and E/F. Isoforms with a VSGGG motif appeared as two distinct bands, whereas those without migrated as one, mostly broad band. This conspicuous, VSGGG-dependent mobility indicated that two different DMPK polypeptides with distinct physicochemical properties were present. From here on, we will refer to these as the upper band and lower band, together forming a doublet (Fig. 2A).
FIG. 2.
Analysis of DMPK isoforms and mutants by Western blotting. Individual mouse DMPK cDNA products were analyzed by Western blotting using an anti-DMPK antibody. (A) Expression of isoforms DMPK A to F. The presence or absence of the VSGGG motif, the type of C-terminal tail, and the theoretical molecular weights are summarized below the lanes. The apparent molecular weights of the main bands are indicated (arrows). All isoforms with a VSGGG motif showed two distinct bands (arrowheads), whereas isoforms without a VSGGG motif showed only one (sometimes broad) band. (B) Expression of His-DMPK A through F kd mutants. All kd isoforms showed only one band (i.e., the lower band in A). (C) The effect of in vivo treatment with the phosphatase inhibitor OA (1 μM, 1 h) was tested for DMPK E, His-DMPK E kd, and DMPK EVA. OA treatment changed the doublet ratios of DMPK E and EVA but did not change the migration of His-DMPK E kd. Note that due to the His tag (2.4 kDa) DMPK E kd migrated slightly more slowly than the regular DMPK E isoform. Shown are results obtained by immunoprecipitation. (D) Ser379 mutations alter DMPK E mobility. DMPK E and mutants DMPK EVA and EVD were expressed and treated with OA prior to lysis as in panel C. Lysis was done at −80°C in 10% TCA-10 mM DTT-acetone to preserve intracellular phosphorylation status.
To investigate whether intrinsic properties of the VSGGG-segment or other domains of the DMPK protein were involved in the apparent heterogeneity, we analyzed several DMPK mutants. DMPK kd mutants (Lys100Ala; <0.5% residual enzymatic activity, see below) displayed only one band, i.e., the lower band (Fig. 2B). This suggested that the upper band represented an autophosphorylated form of the lower band. Treatment with the serine/threonine phosphatase inhibitor okadaic acid (OA) in vivo resulted in a shift in wt DMPK in favor of higher intensity for the upper band at the expense of that of the lower band (Fig. 2C; from here on we will refer to the signal intensity ratio between the upper and lower band as the doublet ratio). Remarkably, OA treatment did not shift kd mutants. (Note that, due to the presence of the His tag, DMPK E kd migrated slightly more slowly than wild-type [wt] DMPK E.) To exclude in vitro phosphatase activity affecting DMPK (auto)phosphorylation status, we prepared extracts from cells arrested in 10% TCA-acetone at −80°C, an established method to instantly inactivate protease and phosphatase activity during isolation (46). Also under these stringent conditions, only the lower band was observed for kd mutants (data not shown). Taken combined, these findings support the idea that DMPK proteins present in the upper and lower bands differ in the presence of phosphoryl groups positioned somewhere on the polypeptide sequence.
To examine whether the serine residue in the alternatively spliced VSGGG motif itself acts as a site for DMPK autophosphorylation, we made Ser379Ala and Ser379Asp mutants of DMPK E, named DMPK EVA and DMPK EVD, respectively. Under standard isolation conditions, DMPK EVA gave only the lower band. Upon OA preincubation, however, a typical upper band was observed (Fig. 2C). This indicated to us that, despite the mutation at position 379, autophosphorylation was possible and occurred at a residue outside the VSGGG motif. Remarkably, even under stringent TCA and −80°C isolation conditions, and with or without OA, the doublet ratio of these bands was still different from that of wt DMPK E under these conditions (Fig. 2D). Given the altered doublet ratio in DMPK EVA, we assume that the serine to alanine mutation negatively affected DMPK autophosphorylation ability. Since the upper band in DMPK EVA was only observed under conditions that phosphatases were inactivated, either because of OA treatment or stringent buffer conditions, we conclude that the phosphate modification was relatively labile (note that the effect of OA and buffer conditions is less prominent for DMPK E). By inference, the (solely based on computer prediction) putative glycosaminoglycan addition at Ser379 could now be ruled out. Analysis of mutant DMPK EVD, constructed to mimic a phosphoserine residue at position 379, revealed one band with a mobility similar to the DMPK E upper band (Fig. 2D). Upon OA treatment, no additional shifted band was observed, but the band often broadened slightly, suggesting that also this DMPK Ser379 mutant could still be autophosphorylated (see also below). Given the observation that the mere presence of a negative charge at position 379 resulted in a characteristic DMPK shift (probably caused by a conformational change in the protein), but mutation of Ser379 not fully impaired autophosphorylation, we suggest that DMPK is autophosphorylated close to, but not at position 379. Alternatively, Ser379 may be one of two (or more) autophosphorylation sites, each of them contributing independently to the observed migration in SDS-PAGE.
A new in vitro kinase assay shows DMPK to be a Lys/Arg directed kinase.
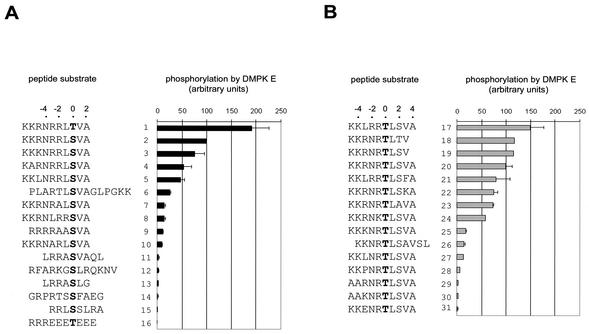
To be able to investigate DMPK auto- and transphosphorylation, we needed a method to measure in vitro enzymatic activity. A simple and sensitive kinase assay based on immunopurified HA-tagged DMPK E was developed and used to screen a library of about 35 synthetic peptides. Peptides tested were assembled in two groups for comparison (Fig. 3A and B) and the activity towards peptide KKRNRRLSVA (peptide 2) was taken as a reference and arbitrarily set at 100% (∼2 pmol Pi/min/μg HA-DMPK E). DMPK E kd was tested to calibrate the specificity of the assay; typically, we found a remaining activity of 0.1 to 0.5%. Among the series of peptides tested, KKRNRRLTVA was preferred over KKRNRRLSVA as substrate, indicating that DMPK E had a preference for threonine over serine. A number of positively charged amino acids were required at positions −1 to −3 (compare peptides 7 and 10 with peptide 2 or peptide 27 with peptides 17 and 18) and to a lesser extent at positions −4 to −6 (peptide 4 versus peptide 2 or peptide 29 versus peptide 20). Furthermore, arginines were preferred over lysines (compare peptides 24 and 25 with 20). The presence of proline and glutamate residues had a negative effect on kinase activity (compare peptides 28 and 31 with peptide 27). Also the aromatic phenylalanine at position +3 was inhibitory (compare peptides 21 and 17). In conclusion, our current best DMPK E substrate sequences comprise three to four arginines (or lysines) at distinct positions N terminal to the phosphoacceptor: (R/K)XRRX(T/S)fX, KKXRRTfX, or KKRXRTfX (where X is any amino acid, preferably not P or E, f is a hydrophobic residue like L or V, and boldface type indicates the phosphoacceptor).
FIG. 3.
Development of an in vitro protein kinase assay for DMPK. Peptides were tested for phosphorylation by HA-DMPK E as described in Materials and Methods. The extent of phosphorylation of each peptide is expressed as the percentage relative to KKRNRRLSVA (peptide 2). The serine or threonine phosphoacceptor (position 0) is shown in boldface type. Bars indicate standard errors. For comparison, peptides are assembled in two groups (A and B).
VSGGG motif and C termini affect DMPK activity.
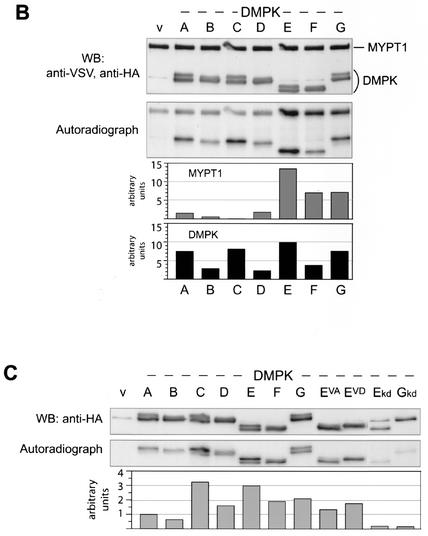
We analyzed the enzymatic activity of DMPK isoforms and mutants in the new kinase assay. Minor isoform DMPK G was also included, which we cloned as a mouse-human chimeric protein since it is not conserved in mouse, to examine possible anomalous functions of its distinct C terminus (Fig. 1A). Expression of HA-tagged DMPK isoforms was verified on Western blot prior to the kinase assay (Fig. 4A, top). DMPK G displayed a doublet, as anticipated from presence of the VSGGG motif. In the kinase assay, irrespective of the peptide tested, DMPK A-G and also mutants EVA and EVD showed rather similar transphosphorylation activity profiles (Fig. 4A).
FIG. 4.
Differential kinase activity and (auto)phosphorylation of DMPK isoforms and mutants. (A) HA-DMPK isoforms A to G, EVA and EVD were expressed in COS-1 cells. Cell extracts were prepared and tested for HA-DMPK by Western blotting (upper panel). Extracts were used in a kinase assay with substrates KKRNRRLSVA (peptide 2) or KKLRRTLSVA (peptide 17) (middle panel). All DMPK isoforms and mutants showed comparable activities towards these peptides. Immunoprecipitated DMPK (after performing assay with peptide 2) was blotted to PVDF membrane to control input (Coomassie brilliant blue [CBB]staining) and examine DMPK autophosphorylation. All isoforms were autophosphorylated (i.e., upper bands in VSGGG-containing isoforms), but isoforms lacking VSGGG showed lower autophosphorylation activity. (B) HA-DMPK isoforms A to G were used in a kinase assay with full-length MYPT1 protein as substrate. Input of MYPT1 and DMPK was verified by Western blotting (upper panel; note aspecific anti-HA band, unrelated to DMPK, in vector-only lane [lane v]). Phosphorylation of MYPT1 and autophosphorylation of DMPK became evident by autoradiography, which was quantified by phosphorimager analysis (lower two panels; values in arbitrary units, after background [lane v] subtraction). (C) COS-1 cells expressing HA-DMPK isoforms and mutants were cultured in the presence of [32P]orthophosphate to examine in vivo phosphorylation of DMPK. DMPK was immunoprecipitated, separated by SDS-PAGE, and blotted to PVDF membrane. The blot was used for autoradiography (middle panel) and phosphorimager analysis (lower panel). Values are presented in arbitrary units, after background (vector-only lane [lane v]) subtraction. Note that kd mutants E and G were less well expressed (upper panel), which partly explains their low signals on autoradiography.
A full-length protein, MYPT1, which is a known DMPK substrate (37; D. G. Wansink, unpublished data), was also examined in the newly developed kinase assay. Remarkably, DMPK A to D showed only little activity above background towards MYPT1, whereas DMPK E, but also F and G, displayed a strong activity, on average around tenfold higher than DMPK A to D (Fig. 4B). Two reported MYPT1 phosphorylation sites were tested using synthesized peptides mypt1a RQSRRSTQGV and mypt1b RRPGEKRRSTGV. Both peptides were phosphorylated by DMPK E, with an activity of 13 and 7%, measured relative to peptide 2 (Fig. 3), respectively. We also tested full-length myosin light-chain 2a (mlc2a), which we characterized as a possible DMPK interactor using yeast two hybrid screening (21). Mlc2a was not phosphorylated by DMPK E in our in vitro kinase assay, however (data not shown).
Comparing doublet ratios before and after the kinase assay (Fig. 4A, top and bottom panels), we observed a general shift in doublet ratio, in favor of a more intense upper band. This was most obvious for mutant DMPK EVA, which appeared as a singlet before and as a doublet after the assay. This observation demonstrated a precursor-product relationship with the protein in the lower band and predicted that the protein in the upper band was the phosphorylated form. Autoradiography indeed showed unambiguously that only the upper band of each doublet was radioactively labeled (Fig. 4A and B). Isoforms B, D, and F, all lacking the VSGGG motif, were also autophosphorylated, but the signal was threefold less intense than observed for the corresponding VSGGG-containing isoforms. Note that also the transphosphorylation activity towards MYPT1 of DMPK F was twofold lower than that of DMPK E. Irrespective of autophosphorylation, DMPK B, D, and F displayed only one band after the kinase assay, confirming that autophosphorylation in the absence of a VSGGG motif did not result in a mobility shift. As already expected from their migration behavior in SDS-PAGE (Fig. 2D), also DMPK EVA and EVD were autophosphorylated, demonstrating that, if phosphorylated at all, Ser379 cannot be the sole acceptor for autophosphotransfer (Fig. 4A, bottom panel).
Is DMPK phosphorylated in vivo? To investigate this, we cultured transfected COS-1 cells in the presence of [32P]orthophosphate, followed by immunoprecipitation. All isoforms, including both upper and lower bands, contained radioactivity, but not to the same extent. DMPK A/B showed around threefold less signal than DMPK C/D or E/F (Fig. 4C). Interestingly, all VSGGG-containing isoforms incorporated more phosphate than the corresponding isoforms without a VSGGG motif, moreover, the upper band contained more phosphate than the lower band. Phosphorylation of the upper band must, at least in part, be caused by autophosphorylation, but may also be the result of upstream kinases. Evidence for phosphorylation of DMPK by upstream kinases was provided by kd mutants E and G, which, despite being enzymatically inactive themselves, were phosphorylated in vivo. Also both Ser379 mutations were phosphorylated, although they exhibited somewhat lower phosphate incorporation than isoform DMPK E (Fig. 4A).
C termini target DMPK isoforms to ER, mitochondria or cytosol.
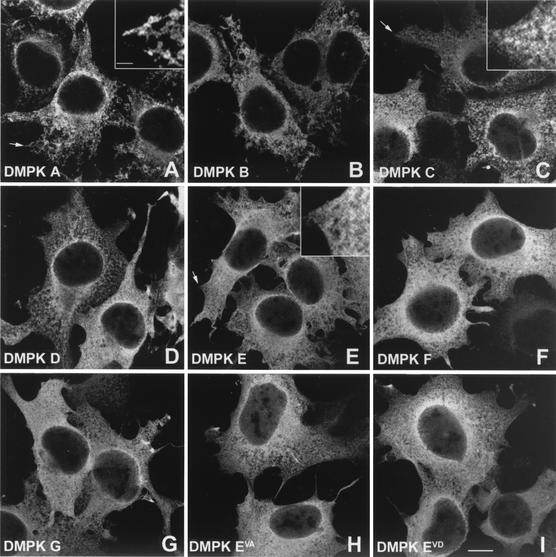
Finally, we investigated the subcellular localization of DMPK isoforms. Judging by immunofluorescent staining in transfected COS-1 cells, all DMPK isoforms were predominantly cytoplasmic, only a very low signal was observed in the nucleus (Fig. 5A to G). No specific effect of presence of the VSGGG motif on DMPK localization was found, this in contrast to the C termini. Isoforms A and B showed a discrete fibrous staining, whereas DMPK C and D displayed a more granular distribution, slightly depending on expression levels. DMPK E and F showed a cytosolic localization, with exclusion of certain vesicular structures. Also DMPK G showed a diffuse staining comparable with that of DMPK E and F. Although their banding pattern on the Western blot was clearly different, DMPK EVA and DMPK EVD showed a similar diffuse cytosolic staining as DMPK E and F (Fig. 5H and I), which agreed with our conclusion that the VSGGG motif was not involved in subcellular localization.
FIG. 5.
Subcellular localization of DMPK isoforms and mutants in COS-1 cells. COS-1 cells were transfected with expression vectors encoding DMPK isoforms and mutants and grown for 24 h. Cells were fixed with 2% formaldehyde and stained with a DMPK-specific antibody. The isoform or mutant shown is indicated in the lower left of each panel. Bar,10 μm. The insets in A, C, and E show a higher magnification of parts indicated with an arrow. The bar in the insets is 2 μm.
To characterize the cellular structures to which DMPK A/B or DMPK C/D were associated, we performed double labeling studies in transfected N2A cells, which display a significantly lower protein expression than COS-1 cells due to lack of SV40-origin based plasmid amplification. Using the appropriate organellar markers, we found that DMPK A and B associated with the ER (Fig. 6A to C), whereas DMPK C and D were located at the outer membrane of mitochondria (Fig. 6D to F; EM ultrastructural analysis not shown). DMPK E and F were found in the cytosol (data not shown). Again, this confirmed that partitioning occurred irrespective of presence of the VSGGG motif. Together with the differential enzymatic activity shown above, our observations suggest that DMPK splice isoforms phosphorylate different substrates at distinct sites in the cell and may have a role in ER or mitochondrial dynamics.
FIG. 6.
Tail 1 and tail 2 target DMPK isoforms to the ER or mitochondria, respectively. HA-DMPK isoforms were transfected to N2A cells to examine their subcellular localization in more detail. Colocalization with an ER-resident GFP mutant demonstrated localization of DMPK A to the ER (A to C). Similarly, colocalization with cytochrome c oxidase, a mitochondrial protein, showed that DMPK C is confined to mitochondria (D to F). Bars, 10 μm.
DISCUSSION
Alternative splicing confers distinct properties to DMPK.
The functions of well-known kinases, like PKCβ (14), PKBγ (9), and doublecortin-like kinase (11) are controlled by alternative splicing. In the present work, we provide evidence that also the alternative splice products A to G from the DMPK gene are structurally different, and that each has a distinct substrate specificity and subcellular localization. If we may extrapolate our findings to the situation in human patients, and based on the observation that the DMPK mRNA profile may be changed in patients with DM1 (50), our findings suggest that shifts in the type or concentration of DMPK variants may not only have cell biological consequences, but may also well be involved in the pathological aspects of DM1 disease manifestation (summarized in Fig. 7A). This would place DM1 into the large category of alternative splicing disorders (reviewed in references 13 and 38). Previously, already strong evidence was provided that a different mechanism, involving titration of RNA processing factors by the abnormal long expanded (CUG)n repeats in the 3′-UTR of DMPK mRNA, has trans-effects on the processing of other transcripts, like that of the chloride channel, tau protein, and insulin receptor in patients with DM1 (53).
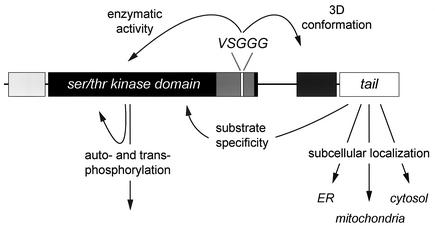
FIG. 7.
Alternatively spliced protein domains. The VSGGG motif and the C terminus determine differential functioning of DMPK isoforms.
Comparison of DMPK with its homologues MRCKα/β and ROCK-I/-II revealed four common protein domains, but also additional, different domains. The leucine-rich domain and the coiled coil region determine multimerization of DMPK (12; Van Herpen and Wansink, unpublished data) and MRCK (48). Given the high degree of homology with the kinase domains of MRCK and ROCK, one might suggest DMPK to function in actin dynamics (4, 28). However, DMPK apparently lacks domains that are crucial for this activity (e.g., a p21 GTPase-binding domain or a pleckstrin homology domain). Studies on PKB/Akt, NDR and MRCK found that the hydrophobic phosphorylation motif is involved in modulation of kinase activity via phosphorylation of its serine/threonine residue (1, 34, 48, 56), which suggests that also DMPK may underlie a similar regulatory regime. Clear distinct features such as the alternatively spliced VSGGG motif and C termini, which are found in no protein other than DMPK, form unique regulatory domains.
VSGGG motif and DMPK (auto)phosphorylation.
Based on indirect evidence, inferred from the characteristic gel migration behavior of VSGGG-containing DMPK isoforms and the in vitro kinase assays, we suggest that the VSGGG motif has a role in determining the three-dimensional conformation of DMPK, and therewith in the regulation of DMPK (auto)phosphorylation activity. Close proximity of the VSGGG-segment to the hydrophobic phosphorylation motif and the active site of the kinase domain (results from molecular modeling studies, data not shown) may explain its modulating effect on autophosphorylation.
Concerning the actual location of DMPK autophosphorylation site(s), different models can be envisaged. First, we consider it unlikely that Ser379 is the unique autophosphorylation site, since DMPK autophosphorylation still occurred in Ser379 mutants. In addition, DMPK isoforms B, D, and F are also autophosphorylated, in spite of the fact that they contain a different amino acid sequence around position 379, due to absence of a VSGGG motif, although now a phosphorylatable threonine is at that position. Since placement of a negative charge at (in DMPK EVD) or near Ser379 (data not shown) resulted in a very similar shifted gel migration behavior, we assume that the autophosphorylation site is located close to Ser379. This could be any one of four potential phosphoacceptor sites (one serine and three threonines) located within stretches of 15 aa flanking the Ser379 site. Based on the evidence provided, we cannot, however, rule out that an amino acid residue elsewhere in the protein is the autophosphorylation acceptor. Secondly, Ser379 could be one of several autophosphorylation sites. In this case, mutation of Ser379 would still allow phosphorylation at one or more other sites, explaining the radioactive phosphate incorporation in the DMPK EVA and EVD mutants in the in vitro kinase assay. In both scenarios, we assume that the altered doublet ratio of DMPK EVA is related to an effect of the Ser->Ala mutation on the in vivo autophosphorylation ability of DMPK. Since DMPK does not contain a DMPK consensus phosphorylation site (see below), we assume that autophosphorylation is mainly conformationally driven, whereas the exact amino acid context may be less important.
Finally, based on our radioactive in vivo phosphorylation assay, we found that not only the upper but also the lower gel bands represent phosphorylated DMPK isoforms. Since also DMPK kd mutants were phosphorylated, we conclude that this was caused by kinases other than DMPK, so called upstream kinases. Thus, the total number of (auto)phosphorylation sites in DMPK remains unclear, but equals at least two.
DMPK kinase activity.
Reports on the (auto)phosphorylation activity of DMPK are scarce and mostly based on use of (improperly folded) bacterially expressed protein and/or standard, inefficient substrates like histone H1 or MBP (18, 49, 54). We demonstrate here that DMPK is a highly active kinase with a preferred substrate sequence consisting of a threonine (or serine) N-terminally preceded by three to four arginines (or lysines) at distinct positions. Our consensus motif is different from the one defined by Bush and coworkers, perhaps because only five peptides were tested in that report (12). AGC kinase family member NDR1 has a comparable consensus as DMPK, but does not accept threonine as phosphoacceptor (33). PKB, on the other hand, requires fewer positively charged amino acids N-terminally, i.e., on the −3 and −5 positions, and strongly prefers arginine over lysine (2, 8, 39). Typically, the classical PKB substrate crosstide (GRPRTSSFAEG) was not phosphorylated by DMPK (Fig. 3A). Although its precise consensus sequence still needs to be determined, ROCK-II was also found to be a Lys/Arg-directed kinase, with a preference for positively charged residues at the −2 and −3 positions (51).
Using peptides as substrate, no fundamental differences in activity between DMPK isoforms were observed. Full-length MYPT1 protein, however, was preferentially phosphorylated by DMPK E, F, and G. As E and F can be regarded as clipped forms of DMPK A-D, this is in accordance with an autoregulatory function for C-terminal tails 1 and 2, similar to what has been reported for MRCK (48), ROCK (3), and human DMPK A/B (12). It is of note though that the putative pseudosubstrate autoinhibitory sequence in human DMPK A (TAVWRRPGAARAP) as defined by Bush et al. (12) is not conserved in mouse tail 1 (TPVWCFPGATFAP) (critical amino acids that are not conserved between humans and mice are underlined). Also in tail 2 no pseudosubstrate site could be identified. We have no experimental evidence for a MYPT1 docking site in any of the tails, since it seems that MYPT1 binds to all DMPK isoforms, independent of the nature of the C terminus (data not shown). We propose, therefore, that tails 1 and 2 are not involved in specific substrate binding or exclusion, but exert their effect via a different mechanism, possibly steric hindrance, which may be relieved after DMPK activation. We cannot exclude the possibility, however, that substrates other than MYPT1 do bind to (specific) DMPK C termini. Since DMPK G does phosphorylate MYPT1, tail type 4 apparently serves another role, and this isoform may be constitutively active. With well-chosen peptides, the now developed DMPK kinase assay offers us better possibilities to investigate the regulatory function of the various C termini in more detail.
What more can we learn from the identified DMPK consensus phosphoacceptor site? Of the putative DMPK substrates reported in literature only the dihydropyridine-sensitive L-type, calcium channel beta-3 subunit (EQKARRSGN; note that this is not the site suggested by Timchenko et al. [49]) and MYPT1 (37) share the consensus sequence. Two MYPT1 phosphoacceptor sites have been described in literature, which are both phosphorylated by ROCK-I (19, 27). One of these, RQSRRSTQG, meets our DMPK consensus sequence precisely, whereas the other, GEKRRSTGV, is very similar. Remarkably, synthetic peptides covering these putative sites were both phosphorylated by DMPK E in an in vitro kinase assay, although none of them was absolutely preferred. Together with the identification of MYPT1 as a DMPK interactor in Y2H, this strongly suggests that DMPK phosphorylates one or both of these MYPT1 sites. We cannot exclude, however, that it occurs on one of the other putative DMPK sites that we identified in MYPT1. Interestingly, we have thus far not been able to demonstrate phosphorylation of putative DMPK binding protein mlc2a (21). This agrees with absence of a true DMPK consensus sequence in mlc2a, the sequence coming closest to the consensus being ASRKAGTRG. Conversely, although the presumed site in the Na+ channel (AMKKLGSKK) does not fit our consensus, there is electrophysiological evidence that it may very well be a DMPK phosphorylation site (36). Alternatively, the observed modulating effect by DMPK may function via another site found in the Na+ channel (PERKPRSDL), or may be indirect. Our database searches using the DMPK phosphorylation motif also revealed a number of new potential DMPK substrates, such as (i) ryanodine receptor 2 (NRTRRISQT) and (ii) α-2C adrenergic receptor (RRRRARSSV).
Similarity to consensus may not be a very reliable predictive index. As was also found for PKB (39), amino acids surrounding the arginine and/or lysine residues may affect the kinases phosphorylation ability, and even not all reported PKB substrates contain the RXRXX(S/T) consensus motif (8). Although DMPK clearly exhibits autophosphorylation ability, no clear match to the consensus motif can be found in the protein. We therefore suggest that additional criteria be used like evolutionary conservation of predicted sites. Also cell and tissue distribution and subcellular localization of the putative substrates should match the requirements known for DMPK (see below). Knowing the subcellular localization of individual DMPK isoforms (e.g., ER and mitochondria in skeletal muscle versus cytosol in smooth muscle cells) will help us to identify new (potential) substrates.
DMPK isoforms localize at ER, mitochondria or in the cytosol.
The subcellular location of DMPK has been a matter of debate ever since its discovery. Partly this was due to a general lack of highly specific DMPK antibodies, but the main reason being that microscopic images were blurred by the simultaneous expression of multiple isoforms in myoblasts, neuronal cells or other cell types in which DMPK is expressed (reviewed in references 21 and 52). We report here that alternative splicing endows DMPK with the ability to locate at distinct subcellular distributions through the formation of different C termini. The alternatively spliced VSGGG motif seems to have no role in DMPK distribution. Pilot experiments suggest that both tail 1 and tail 2 interact with membranes via long stretches of hydrophobic amino acids (data not shown), although no evidence exists for a known membrane targeting signal. Whether DMPK is a membrane-associated or transmembrane protein is as yet unclear. Protein-protein interactions may also play a role in DMPK localization, especially in the case of DMPK C and D, since binding to mitochondria displayed saturable binding characteristics.
In a situation where ectopic location due to overexpression is prevented, DMPK A/B occur attached to the ER, DMPK C/D are located at mitochondria, and DMPK E/F and G have a cytosolic location. These localizations are consistent with previous studies on the location of a human variant homologous to DMPK B, and a C-terminally truncated form (12, 54). In cell types where different variants occur simultaneously (e.g., muscle and heart), DMPK may be involved in the regulation of ER-mitochondrial positioning or dynamics. Presumably, the functional spectrum of DMPK may be even further enlarged by its ability to form dimeric or multimeric structures via coil-coil associations (Van Herpen and Wansink, unpublished data). Interestingly, strict regulation of mitochondrial positioning with respect to the ER Ca2+ release sites is necessary to activate and control mitochondrial calcium uptake in muscles, and recent evidence also points to a role in apoptosis control via Ca2+ homeostasis regulation (44). Ca2+ homeostasis is indeed disturbed in myotubes lacking DMPK (5). Selective targeting to either ER, mitochondria or cytosol may enable DMPK to phosphorylate distinct substrates in different cell types, thereby broadening its scope of action. Likewise, cytosolic human isoform DMPK G, found at relatively high levels in skeletal muscle and brain cells of DM1 patients, may erroneously phosphorylate proteins which are normally only targets for cytosolic DMPK E and F in smooth muscle cells. As distinctly localized DMPK isoforms incorporated different amounts of phosphate in vivo, future studies will therefore have to focus on deciphering signal transduction pathways in which DMPK isoforms are active, perhaps in location (i.e., ER-mitochondria-cytosol), cell type- and isoform-specific regulation schemes. Knowing such schemes would perhaps open up the possibility to manipulate DMPK activity pharmacologically.
Acknowledgments
D.G.W. and R.E.M.A.V.H. contributed equally to this work.
We thank Michelle Hill for help in developing the DMPK kinase assay and Susan Mulders for MYPT1 expression.
Part of this work was performed at the Friedrich Miescher Institute by D.G.W., who was supported by an EMBO fellowship. B.W. is supported by the Dutch Beatrixfonds, the American Muscular Dystrophy Association, and the Association Française contre les Myopathies.
REFERENCES
- 1.Alessi, D. R., M. Andjelkovic, B. Caudwell, P. Cron, N. Morrice, P. Cohen, and B. A. Hemmings. 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15:6541-6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi, D. R., F. B. Caudwell, M. Andjelkovic, B. A. Hemmings, and P. Cohen. 1996. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399:333-338. [DOI] [PubMed] [Google Scholar]
- 3.Amano, M., K. Chihara, N. Nakamura, T. Kaneko, Y. Matsuura, and K. Kaibuchi. 1999. The COOH terminus of Rho-kinase negatively regulates Rho-kinase activity. J. Biol. Chem. 274:32418-32424. [DOI] [PubMed] [Google Scholar]
- 4.Amano, M., Y. Fukata, and K. Kaibuchi. 2000. Regulation and functions of Rho-associated kinase. Exp. Cell Res. 261:44-51. [DOI] [PubMed] [Google Scholar]
- 5.Benders, A. G. M., P. J. T. A. Groenen, F. T. J. J. Oerlemans, J. H. Veerkamp, and B. Wieringa. 1997. Myotonic dystrophy protein kinase is involved in the modulation of the Ca2+ homeostasis in skeletal muscle cells. J. Clin. Investig. 100:1440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berul, C. I., C. T. Maguire, M. J. Aronovitz, J. Greenwood, C. Miller, J. Gehrmann, D. Housman, M. E. Mendelsohn, and S. Reddy. 1999. DMPK dosage alterations result in atrioventricular conduction abnormalities in a mouse myotonic dystrophy model. J. Clin. Investig. 103:R1-R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brazil, D. P., and B. A. Hemmings. 2001. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem. Sci. 26:657-664. [DOI] [PubMed] [Google Scholar]
- 9.Brodbeck, D., M. M. Hill, and B. A. Hemmings. 2001. Two splice variants of protein kinase Bγ have different regulatory capacity depending on the presence or absence of the regulatory phosphorylation site serine 472 in the carboxy-terminal hydrophobic domain. J. Biol. Chem. 276:29550-29558. [DOI] [PubMed] [Google Scholar]
- 10.Brook, J. D., M. E. McCurrach, H. G. Harley, A. J. Buckler, D. Church, H. Aburatani, K. Hunter, V. P. Stanton, J.-P. Thirion, T. Hudson, R. Sohn, B. Zemelman, R. G. Snell, S. A. Rundle, S. Crow, J. Davies, P. Shelbourne, J. Buxton, C. Jones, V. Juvonen, K. Johnson, P. S. Harper, D. J. Shaw, and D. E. Housman. 1992. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68:799-808. [DOI] [PubMed] [Google Scholar]
- 11.Burgess, H. A., and O. Reiner. 2002. Alternative splice variants of doublecortin-like kinase are differentially expressed and have different kinase activities. J. Biol. Chem. 277:17696-17705. [DOI] [PubMed] [Google Scholar]
- 12.Bush, E. W., S. M. Helmke, R. A. Birnbaum, and M. B. Perryman. 2000. Myotonic dystrophy protein kinase domains mediate localization, oligomerization, novel catalytic activity, and autoinhibition. Biochemistry 39:8480-8490. [DOI] [PubMed] [Google Scholar]
- 13.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 14.Chalfant, C. E., H. Mischak, J. E. Watson, B. C. Winkler, J. Goodnight, R. V. Farese, and D. R. Cooper. 1995. Regulation of alternative splicing of protein kinase C beta by insulin. J. Biol. Chem. 270:13326-13332. [DOI] [PubMed] [Google Scholar]
- 15.Charlet-B., N., R. S. Savkur, G. Singh, A. V. Philips, E. A. Grice, and T. A. Cooper. 2002. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10:45-53. [DOI] [PubMed] [Google Scholar]
- 16.Davis, B. M., M. E. McCurrach, K. L. Taneja, R. H. Singer, and D. E. Housman. 1997. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA 94:7388-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Cunto, F., E. Calautti, J. Hsiao, L. Ong, G. Topley, E. Turco, and G. P. Dotto. 1998. Citron Rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J. Biol. Chem. 273:29706-29711. [DOI] [PubMed] [Google Scholar]
- 18.Dunne, P. W., E. T. Walch, and H. F. Epstein. 1994. Phosphorylation reactions of recombinant human myotonic dystrophy protein kinase and their inhibition. Biochemistry. 33:10809-10814. [DOI] [PubMed] [Google Scholar]
- 19.Feng, J., M. Ito, K. Ichikawa, N. Isaka, M. Nishikawa, D. J. Hartshorne, and T. Nakano. 1999. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J. Biol. Chem. 274:37385-37390. [DOI] [PubMed] [Google Scholar]
- 20.Fu, Y.-H., A. Pizzuti, R. G. Fenwick Jr., J. King, S. Rajnarayan, P. W. Dunne, J. Dubel, G. A. Nasser, T. Ashizawa, P. de Jong, B. Wieringa, R. G. Korneluk, M. B. Perryman, H. F. Epstein, and C. T. Caskey. 1992. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255:1256-1258. [DOI] [PubMed] [Google Scholar]
- 21.Groenen, P., and B. Wieringa. 1998. Expanding complexity in myotonic dystrophy. BioEssays 20:901-912. [DOI] [PubMed] [Google Scholar]
- 22.Groenen, P. J. T. A., D. G. Wansink, M. Coerwinkel, W. van den Broek, G. Jansen, and B. Wieringa. 2000. Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum. Mol. Genet. 9:605-616. [DOI] [PubMed] [Google Scholar]
- 23.Harper, P. S. 2001. Myotonic dystrophy, 3rd ed., vol. 37. W. B. Saunders, London, United Kingdom.
- 24.Hill, M. M., and B. A. Hemmings. 2002. Analysis of protein kinase B/Akt. Methods Enzymol. 345:448-463. [DOI] [PubMed] [Google Scholar]
- 25.Jansen, G., P. J. Groenen, D. Bachner, P. H. Jap, M. Coerwinkel, F. Oerlemans, W. van den Broek, B. Gohlsch, D. Pette, J. J. Plomp, P. C. Molenaar, M. G. Nederhoff, C. J. van Echteld, M. Dekker, A. Berns, H. Hameister, and B. Wieringa. 1996. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 13:316-324. [DOI] [PubMed] [Google Scholar]
- 26.Justice, R. W., O. Zilian, D. F. Woods, M. Noll, and P. J. Bryant. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9:534-546. [DOI] [PubMed] [Google Scholar]
- 27.Kawano, Y., Y. Fukata, N. Oshiro, M. Amano, T. Nakamura, M. Ito, F. Matsumura, M. Inagaki, and K. Kaibuchi. 1999. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147:1023-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung, T., X.-Q. Chen, I. Tan, E. Manser, and L. Lim. 1998. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 18:130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahadevan, M., C. Amemiya, G. Jansen, L. Sabourin, S. Baird, C. Neville, N. Wormskamp, B. Segers, M. Batzer, J. Lamerdin, P. de Jong, B. Wieringa, and R. Korneluk. 1993. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum. Mol. Genet. 2:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Mankodi, A., M. P. Takahashi, H. Jiang, C. L. Beck, W. J. Bowers, R. T. Moxley, S. C. Cannon, and C. A. Thornton. 2002. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10:35-44. [DOI] [PubMed] [Google Scholar]
- 31.Manning, G., G. D. Plowman, T. Hunter, and S. Sudarsanam. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27:514-520. [DOI] [PubMed] [Google Scholar]
- 32.Millward, T., P. Cron, and B. A. Hemmings. 1995. Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc. Natl. Acad. Sci. 92:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millward, T. A., C. W. Heizmann, B. W. Schafer, and B. A. Hemmings. 1998. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 17:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millward, T. A., D. Hess, and B. A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847-33850. [DOI] [PubMed] [Google Scholar]
- 35.Mounsey, J. P., D. J. Mistry, C. W. Ai, S. Reddy, and J. R. Moorman. 2000. Skeletal muscle sodium channel gating in mice deficient in myotonic dystrophy protein kinase. Hum. Mol. Genet. 9:2313-2320. [DOI] [PubMed] [Google Scholar]
- 36.Mounsey, J. P., P. Xu, J. E. John, 3rd, L. T. Horne, J. Gilbert, A. D. Roses, and J. R. Moorman. 1995. Modulation of skeletal muscle sodium channels by human myotonin protein kinase. J. Clin. Investig. 95:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muranyi, A., R. Zhang, F. Liu, K. Hirano, M. Ito, H. F. Epstein, and D. J. Hartshorne. 2001. Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett. 493:80-84. [DOI] [PubMed] [Google Scholar]
- 38.Nissim-Rafinia, M., and B. Kerem. 2002. Splicing regulation as a potential genetic modifier. Trends Genet. 18:123-127. [DOI] [PubMed] [Google Scholar]
- 39.Obata, T., M. B. Yaffe, G. G. Leparc, E. T. Piro, H. Maegawa, A. Kashiwagi, R. Kikkawa, and L. C. Cantley. 2000. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275:36108-36115. [DOI] [PubMed] [Google Scholar]
- 40.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737-740. [DOI] [PubMed] [Google Scholar]
- 41.Ranum, L. P. W., and J. W. Day. 2002. Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev. 12:266-271. [DOI] [PubMed] [Google Scholar]
- 42.Reddy, S., D. B. J. Smith, M. M. Rich, J. M. Leferovich, P. Reilly, B. M. Davis, K. Tran, H. Rayburn, R. Bronson, D. Cros, R. J. Balice-Gordon, and D. Housman. 1996. Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat. Genet. 13:325-335. [DOI] [PubMed] [Google Scholar]
- 43.Savkur, R. S., A. Philips, and T. A. Cooper. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29:40-47. [DOI] [PubMed] [Google Scholar]
- 44.Scorrano, L., S. A. Oakes, J. T. Opferman, E. H. Cheng, M. D. Sorcinelli, T. Pozzan, and S. J. Korsmeyer. 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300:135-139. [DOI] [PubMed] [Google Scholar]
- 45.Sergeant, N., B. Sablonniere, S. Schraen-Maschke, A. Ghestem, C. Maurage, A. Wattez, P. Vermersch, and A. Delacourte. 2001. Dysregulation of human brain microtubule-associated tau mRNA maturation in myotonic dystrophy type 1. Hum. Mol. Genet. 10:2143-2155. [DOI] [PubMed] [Google Scholar]
- 46.Seto, M., Y. Sasaki, and Y. Sasaki. 1990. Alteration in the myosin phosphorylation pattern of smooth muscle by phorbol ester. Am. J. Physiol. 259:C769-C774. [DOI] [PubMed] [Google Scholar]
- 47.Stadhouders, A. M., P. H. Jap, H. P. Winkler, H. M. Eppenberger, and T. Wallimann. 1994. Mitochondrial creatine kinase: a major constituent of pathological inclusions seen in mitochondrial myopathies. Proc. Natl. Acad. Sci. USA 91:5089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, I., K. T. Seow, L. Lim, and T. Leung. 2001. Intermolecular and intramolecular interactions regulate catalytic activity of myotonic dystrophy kinase-related Cdc42-binding kinase α. Mol. Cell. Biol. 21:2767-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timchenko, L., N. W., T. Schneider, B. Patel, F. Hofmann, and C. T. Caskey. 1995. Full-length myotonin protein kinase (72 kDa) displays serine kinase activity. Proc. Natl. Acad. Sci. USA 92:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiscornia, G., and M. S. Mahadevan. 2000. Myotonic dystrophy: the role of the CUG triplet repeats in splicing of a novel DMPK exon and altered cytoplasmic DMPK mRNA isoform ratios. Mol. Cell 5:959-967. [DOI] [PubMed] [Google Scholar]
- 51.Turner, M. S., L. Fen Fen, J. W. Trauger, J. Stephens, and P. LoGrasso. 2002. Characterization and purification of truncated human Rho-kinase II expressed in Sf-21 cells. Arch. Biochem. Biophys. 405:13-20. [DOI] [PubMed] [Google Scholar]
- 52.Ueda, H., S. Ohno, and T. Kobayashi. 2000. Myotonic dystrophy and myotonic dystrophy protein kinase. Progr. Histochem. Cytochem. 35:187-251. [DOI] [PubMed] [Google Scholar]
- 53.Wansink, D. G., and B. Wieringa. Transgenic mouse models for myotonic dystrophy type 1 (DM1). Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 54.Waring, J. D., R. Haq, K. Tamai, L. A. Sabourin, J.-E. Ikeda, and R. G. Korneluk. 1996. Investigation of myotonic dystrophy kinase isoform translocation and membrane association. J. Biol. Chem. 271:15187-15193. [DOI] [PubMed] [Google Scholar]
- 55.Xu, T., W. Wang, S. Zhang, R. A. Stewart, and W. Yu. 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121:1053-1063. [DOI] [PubMed] [Google Scholar]
- 56.Yang, J., P. Cron, V. Thompson, V. Good, D. Hess, B. Hemmings, and D. Barford. 2002. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell 9:1227-1240. [DOI] [PubMed] [Google Scholar]