Abstract
Loss of p53 sensitizes to antimicrotubule agents in human tumor cells, but little is known about its role during mitosis. We have identified the Polo-like kinase family member serum inducible kinase (Snk/Plk2) as a novel p53 target gene. Snk/Plk2 mutagenesis demonstrated that its kinase activity is negatively regulated by its C terminus. Small interfering RNA (siRNA)-mediated Snk/Plk2 silencing in the presence of the mitotic poisons paclitaxel (Taxol) or nocodazole significantly increased apoptosis, similar to p53 mutations, which confer paclitaxel sensitivity. Furthermore, we have demonstrated that the apoptosis due to silencing of Snk/Plk2 in the face of spindle damage occurs in mitotic cells and not in cells that have progressed to a G1-like state without dividing. Since siRNA directed against Snk/Plk2 promoted death of paclitaxel-treated cells in mitosis, we envision a mitotic checkpoint wherein p53-dependent activation of Snk/Plk2 prevents mitotic catastrophe following spindle damage. Finally, these studies suggest that disruption of Snk/Plk2 may be of therapeutic value in sensitizing paclitaxel-resistant tumors.
The p53 tumor suppressor plays a key role in the cell's response to genotoxic stress, and loss of p53-dependent checkpoints is an important step in carcinogenesis.
Mutation or loss of p53 has been observed in over 50% of all tumors and in almost every tumor type (16). Furthermore, it has been estimated that the p53 pathway is disrupted by mutation or inhibition of its function in the vast majority of tumors (48). In response to a variety of genotoxic stresses (DNA-damaging agents, UV damage, antimicrotubule agents, and hypoxia) or inappropriate proliferative signals (c-Myc, E2f-1, E1A, or Ras), p53 protein is stabilized and activated, allowing it to transactivate its target genes. Upon stabilization and activation, p53 mediates several cellular responses including growth arrest, DNA repair, apoptosis and senescence depending on the cellular context. These cellular responses allow p53 to inhibit tumorigenesis and genomic instability. Loss of p53 has been shown to lead to genomic instability in both normal and tumor cells (49). Although p53 plays an important function in maintaining genomic stability, little is known about a role in enforcing the mitotic spindle checkpoint.
Although several studies have shown a key role for p53 in mediating chemosensitivity in response to DNA-damaging agents, loss of p53 appears to sensitize cells to antimicrotubule agents, including paclitaxel (Taxol) (15, 47, 50). Previous studies had suggested that after treatment with spindle inhibitors, cells initially arrest in mitosis and then undergo adaptation and exit mitosis (51). Cells with wild-type p53 undergo a G1 arrest after adaptation leading to resistance to paclitaxel and other antimicrotubule agents. Cells which lack p53 do not arrest in G1 phase after exposure to antimicrotubule agents and endoreduplicate their DNA, leading to massive apoptosis (7, 9, 22, 32). The p53 target gene p21WAF1 has been implicated in mediating this G1 arrest, as p21 knockout mouse embryonic fibroblasts or p21 somatic knockout cells undergo a similar endoreduplication after treatment with spindle inhibitors (20, 22, 41). An additional role for p21 during mitosis has been found in breast cancer cells, as p21 appears to mediate paclitaxel resistance through inhibition of p34cdc2 kinase activity during G2/M (56).
The Polo-like kinase family is a conserved family of serine-threonine protein kinases that are required for mitotic progression. Members of the Polo-like kinase family are characterized by a conserved N-terminal catalytic domain and a C-terminal Polo box domain, which allows these kinases to localize to mitotic structures (14, 34). This kinase family is named after the founding member, polo, identified in Drosophila melanogaster as a recessive maternal-effect lethal mutation that displayed spindle defects (43). Homologs have since been identified in yeast, worms, Xenopus and mammalian systems. Most higher organisms have three family members although Plk1 appears to be the most functionally homologous to polo and the plk genes in Saccharomyces cerevisiae (Cdc5p) and Schizosaccharomyces pombe (Plo1p). Two other family members have been identified in mammals: Serum inducible kinase, also known as Polo-like kinase 2 (Snk/Plk2), and Fnk/Prk (Plk3) (6, 24, 39). Plk1 is the best characterized family member and appears to play a role in mediating the onset of mitosis, centrosome duplication and separation, regulation of the anaphase promoting complex, and mitotic exit (10). Recent studies have demonstrated that Plk3 is activated during the DNA damage checkpoint in G2 phase and inhibits entry into mitosis through phosphorylation of cdc25c (35, 54) (55). Although very little is known about the function or regulation of Snk/Plk2, both Snk/Plk2 and Plk3 have been implicated in synaptic plasticity and associate with CIB, a calmodulin-related protein (19).
Since loss of p53 can sensitize cells to antimicrotubule agents, we examined the mechanisms through which this sensitization might occur. p53 is a transcription factor that prevents neoplasia and genomic instability primarily through the activation of its target genes. Therefore, to identify novel p53 target genes which may explain p53-mediated resistance to paclitaxel and other spindle poisons, we performed an Affymetrix microarray analysis of gene expression patterns in wild-type and p53-null animals before and after irradiation (4). Our analysis of the spleen and thymus revealed that serum inducible kinase also known as Polo-like kinase 2 (Snk/Plk2), was induced after irradiation in a p53-dependent manner in vivo. We further found that Snk/Plk2 is a transcriptional target of p53 after genotoxic stress. A mutational analysis of the human Snk/Plk2 cDNA has revealed that it has kinase activity that is negatively regulated by its C-terminal domain. In order to elucidate the function of Snk/Plk2 we have silenced its expression in cell lines using small interfering RNA (siRNA) oligonucleotides. Upon silencing Snk/Plk2 in the presence of the mitotic poisons paclitaxel or nocodazole, we observed a significant increase in apoptosis, similar to what is observed when p53 is mutated. Furthermore, we found that the increase in apoptosis occurred during mitosis. Our studies suggest that p53 may play a role in a mitotic checkpoint, and this represents a novel mechanism by which p53 prevents genomic instability. Because siRNA directed against Snk/Plk2 promoted death of paclitaxel-treated cells in mitosis, we envision a mitotic checkpoint wherein p53-dependent activation of Snk/Plk2 prevents mitotic catastrophe following spindle damage. Finally, our findings suggest the intriguing possibility that disruption of Snk/Plk2 may be of therapeutic value in sensitizing paclitaxel resistant tumors.
MATERIALS AND METHODS
Plasmids.
The wild-type p53 expression vector pCEP4-p53 and pCEP4 were generously supplied by Bert Vogelstein (Johns Hopkins University). The pCMV-β-Gal plasmid and pEGFP plasmid were obtained from Clontech. The pWWP-Luc plasmid has been describe previously (12). Human Snk/Plk2 was amplified from a human fetal brain cDNA library (Clontech) using PFU Turbo (Invitrogen) using the following primers: 5′ CGC GGA TCC GCG ACC ATG GAG CTT TTG 3′ and 5′ CCG CTC GAG GTT ACA TCT TTG TAA GAG CAT 3′. This full-length Snk/Plk2 cDNA was then cloned in frame into the pcDNA3.1/V5-His (Invitrogen) as a BamHI/XhoI fragment. The C-terminally tagged V5-His plasmid was subsequently mutagenized using the QuikChange site-directed mutagenesis kit (Stratagene) as previously described (30). All constructs were sequenced to verify the authenticity of cloned cDNAs or mutants.
RNA preparation.
Twenty-one-base pair siRNA duplexes were synthesized by Dharmacon Research (Lafayette, Colo.). The targeted sequence for Snk/Plk2 siRNA duplex was 5′ AAG CGC UAC UGC CGG GGC AAA 3′. An siRNA targeting a sequence in firefly (Photinus pyralis) luciferase (accession no. X65324) mRNA (11) served as a negative control.
Cell lines, transfection, and adenoviral infections.
The human cell lines U20S (osteosarcoma), H460 (non-small cell lung carcinoma), HeLa (cervical carcinoma), SKOV3 (ovarian carcinoma), SKBR3 (breast carcinoma), SW480 (colon carcinoma), and HEK 293 (human embryonic kidney) were obtained from the American Type Culture Collection. An Epstein-Barr virus-immortalized lymphoblastoid cell line from a healthy individual (2184D) was obtained from the Human Genetic Mutant Cell Repository (Camden, N.J.). The U20S Neo and E6 cells were maintained as previously described (44). 293 cells and Saos2 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. For siRNA duplex and vector cotransfection experiments, U2OS and HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. For siRNA duplex transfection experiments, Lipofectamine 2000 (Invitrogen) was used for U20S cells and Oligofectamine (Invitrogen) was used for HeLa cells and H460 cells according to the manufacturer's specifications. Ad-LacZ and Ad-p53 were obtained from B. Vogelstein (Johns Hopkins University). All viruses were propagated, their titers were determined, and they were amplified as previously described (12). Cells were infected at a multiplicity of infection to ensure >90% infectivity. Adriamycin, etoposide, and paclitaxel were obtained from the University of Pennsylvania Cancer Center Pharmacy, Philadelphia. Nocodazole was obtained from Sigma. For colony formation assays, U2OS Neo and E6 cells were plated at 2.4 × 104 cells per well in a 12-well dish and treated with increasing doses of paclitaxel for 24 h. After 24 h of treatment, the medium was removed and replaced with fresh medium without paclitaxel. Cells were grown for 7 days and then stained with Coomassie blue as previously described (52).
Western blotting.
Total cellular protein was harvested in 1× Laemmli sample buffer and quantitated using the amido black staining method as previously described (31). A total of 80 μg of protein per lane was loaded on a sodium dodecyl sulfate (SDS)-polyacrylamide gel and electrophoresed and electroblotted as previously described (31). The following antibodies were used in these experiments: mouse monoclonal antiactin (C-2; Santa Cruz), mouse monoclonal anti-Ran (BD Pharmingen), mouse monoclonal anti-p53 (Ab-2; Oncogene Sciences), mouse monoclonal anti-p21 (Ab-1; Oncogene Sciences), mouse monoclonal anti-V5 (Invitrogen), mouse monoclonal anti-V5-HRP (Invitrogen), rabbit polyclonal anti-cyclin B1 (H-433; Santa Cruz), and mouse monoclonal anti-cdc2 p34 (no. 17; Santa Cruz).
Northern analysis and Taqman real-time quantitative RT-PCR.
RNA isolation, Northern blotting, and hybridization were performed as previously described (12). The 2,058-bp BamHI/XhoI fragment from pcDNA3.1-V5His-Snk/Plk2 was used as a probe for Snk/Plk2 expression. The full-length mouse Snk/Plk2 cDNA was amplified from a mouse kidney cDNA library (Clontech) using the following primers: 5′ CGC GGA TCC GCC ACC ATG GAG CTC CTG CGG ACT 3′ and 5′ CGC CTC GAG TCA GTT ACA TCT CTG TAA GAG 3′. This full-length mouse Snk/Plk2 cDNA was then cloned into pcDNA3 (Invitrogen) as a BamHI/XhoI fragment and sequenced. A 2,050-bp BamHI/XhoI fragment from pcDNA3-mSnk/Plk2 was used as a probe for Snk/Plk2 expression. The TaqMan reverse transcription (RT)-PCR assay was conducted as previously described (4). In brief, oligonucleotides (probes) for TaqMan RT-PCR were labeled with FAM (6-carboxyfluorescin) (mouse Snk/Plk2 and Snk/Plk2) or VIC (mouse GAPDH and GAPDH) and 3′ prime quencher, TAMRA (6-carboxyl-N,N,N′,N′-tetra-methyl-rhodamine). The following primer and probe sequences were used for each gene, respectively: mouse Snk/Plk2 primers, 5′ AGC AGC GAA TGC CTT GAA G 3′ and 5′ TCC TCG AAG GAC TCT TGC CA 3′; mSnk/Plk2 probe, 6FAM CAG CAC CAT GGG AAG TGT GGC AGA C-TAMRA; Snk/Plk2 primers, 5′ TCG GAT GAT AGT CAG AGG GAC TC 3′ and 5′ TCT TTG GGA ATG CAA TCA GCT T 3′; Snk/Plk2 probe 6FAM CTT CAA GGC ATT CAC TGC TGC TGC TAC A-TAMRA. GAPDH primer and VIC-labeled probe were obtained from PE Applied Biosystems. All primers and probes were designed with the use of Primer Express (version 1.0; PE Applied Biosystems). Total RNA was isolated from individual tissues as previously described (12), and 1 μg was used for reverse transcription and amplification using TaqMan Reverse Transcription Reagents according to the manufacturer's protocol (PE Applied Biosystems). A master mix of TaqMan reagents was prepared and 10 ng of each RT sample was used in the Taqman PCR. Each tube contained both a gene probe and primers and a GAPDH control probe and primer. Each sample was analyzed in quadruplicate. Reactions in which reverse transcriptase was not added to the RT reaction where used to control for genomic contamination. The increase in fluorescence (Δ Rn) was proportional to the concentration of template in the PCR. The PCR cycle number at the threshold represents the threshold cycle. The standard curve method was used to quantitate amounts of each gene relative to the GAPDH amount in each reaction according to the manufacturer's protocol (PE Applied Biosystems). Reactions were carried out in 96-well plates using the ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems).
Animals and treatments.
Healthy 6- to 7-week-old female wild-type (+/+) and p53-null (−/−) animals were obtained from Jackson Laboratories (Bar Harbor, Maine). Total body irradiation was performed using a 137cesium γ-source at a dose rate of 1.532 Gy/min. At 0, 6, and 24 h, the mice were euthanized using an approved Institutional Animal Care and Use Committee Protocol, which followed recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. Tissues were either snap frozen in liquid nitrogen or paraffin embedded (after an overnight fixation in 4% paraformaldehyde at 4°C).
In situ hybridization.
The mSnk/Plk2 expression vector pcDNA3-mSnk/Plk2 was digested and used to make either antisense (SP6) or sense (T7) RNA probes with a Dig RNA labeling kit (SP6/T7) (Roche). In situ hybridization was performed as described previously (13, 18).
Flow cytometry.
Cells were harvested after the indicated time periods and prepared for detection of active caspase 3 by flow cytometry or stained with propidium iodide and analyzed by flow cytometry for sub-G1 content as previously described (36). Cyclin B1 staining was performed using the fluorescein isothiocyanate-labeled anti-human cyclin B1/isotype control reagent set according to the manufacturer's protocol (BD Pharmingen). In addition cells were stained with propidium iodide to measure DNA content. Cell sorting was performed on a Beckman Coulter Epics Elite flow cytometer.
Luciferase assays.
A 2,540-bp genomic fragment containing three candidate p53 binding sites (BS1, BS2, and BS3) was amplified from Fadu genomic DNA and cloned into the pGL3-Basic vector (Promega) to obtain pGL3-Snk/Plk2 promoter using the following primers: 5′ CGG GGT ACC AGG TTT GCA CAA ATC TGG GA 3′ and 5′ CTA GCT AGC GCC GAC TAG CAC CCA ACA 3′. Similarly genomic fragments lacking BS1 (pGL3-Snk/Plk2 promoter-Δ1); BS1 and BS2 (pGL3-Snk/Plk2 promoter-Δ1,2); and BS1, BS2, and BS3 (pGL3-Snk/Plk2 promoter-Δ1,2,3) were also constructed, using the following primer sets, respectively: for Δ1, 5′ CGG GGT ACC CTT AGG CCA CAA ATG AAT ACA 3′ and 5′ CTA GCT AGC GCC GAC TAG CAC CCA ACA 3′; for Δ1,2, 5′ CGG GGT ACC TAA ATT TGA CAG TGG ACT 3′ and 5′ CTA GCT AGC GCC GAC TAG CAC CCA ACA 3′; for Δ1,2,3, 5′ CGG GGT ACC ATC TCC CAT GTA TAT ATG AGA C 3′ and 5′ CTA GCT AGC GCC GAC TAG CAC CCA ACA 3′. All constructs were sequenced to verify the authenticity of the cloned fragments. These reporters (2.88 μg) were cotransfected with pCEP4 or pCEP4-p53 (0.72 μg) and pCMV-β-Gal (0.4 μg) into Saos2 cells using Lipofectamine 2000 (Invitrogen). Luciferase activity and β-galactosidase activity were determined 24 h later. All samples were normalized to β-galactosidase activity. Empty vector (pGL3-Basic) and the p21WAF1 promoter (pWWP-Luc) were used as negative and positive controls, respectively.
ChIP assays.
Chromatin immunoprecipitation (ChIP) was carried out as described previously (44). Briefly, U2OS cells were untreated or treated with adriamycin (200 ng/ml) for 24 h and fixed in 1% formaldehyde for 10 min at room temperature. Formaldehyde was neutralized with 0.125 M glycine for 5 min at room temperature, and cells were washed with ice-cold phosphate-buffered saline. Cells were then scraped in 1 ml of RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0], 5 mM EDTA) supplemented with Complete protease inhibitor (Roche Diagnostics, Indianapolis, Ind.), 5 μM trichostatin A, 50 mM sodium fluoride, and 200 μM sodium orthovanadate and incubated on ice for 10 min. DNA was sonicated to a range of 200- to 1-kb fragments, and lysates were cleared by centrifugation for 15 min at 4°C. After preclearing with protein G-agarose beads for 1 h at 4°C, lysates were centrifuged and divided evenly between two tubes. One set of tubes received anti-p53 antibodies (10 μg each of Ab-1 and Ab-2; Oncogene Research Products) and protein G-agarose beads, while the other set of tubes served as the bead-only negative controls. After overnight precipitation at 4°C, the immune complexes were collected by centrifugation at 4°C and washed twice with RIPA buffer, four times with IP wash buffer (100 mM Tris [pH 8.5], 500 mM lithium chloride, 1% NP-40, and 1% deoxycholate), and two more times with RIPA buffer. All washes were for 5 min at 4°C. Cross-linking was reversed by boiling the immune complexes for 30 min in reversal buffer (125 mM Tris [pH 6.8], 10% β-mercaptoethanol, 4% SDS). DNA was extracted with phenol-chloroform, and this was followed by ethanol precipitation using glycogen as a carrier. DNA was resuspended in 50 μl of distilled water and amplified by PCR. The BS1 site was amplified using BS1ChipF (5′ CTT AGG CCA CAA ATG AAT ACA 3′) as a forward primer and BS1ChipR (5′-TCCCAGATTTGTGCAAACCT-3′) as the reverse primer. The BS2 site was amplified using BS2ChipF (5′-CACAAATCTGGGAAGCAAAGGTC-3′) as the forward primer and BS2ChipR (5′-CATTAGAGAGGAGAAAGGGAAAGGC-3′) as the reverse primer.
Immunoprecipitations and kinase assays.
Equal amounts of 293 cell lysate (2.5 × 106 cell equivalents) and U2OS cell lysate (1.0 × 106 cells) were harvested and lysed on ice for 30 min in a solution containing 50 mM Tris, 5 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.1% Triton X-100, 0.1 mM Na3VO4, and 0.5 mM phenylmethylsulfonyl fluoride supplemented with Complete protease inhibitor (Roche Diagnostics). Lysates were clarified by centrifugation for 10 min. For Snk/Plk2 kinase assays, lysates were divided into two samples and then incubated with mouse monoclonal anti-polyhistidine agarose (Sigma) for 3 h at 4°C. Beads were then washed three times in lysis buffer and twice in kinase buffer (20 mM HEPES [pH 7.4], 10 mM magnesium acetate) and then incubated in 30 μl of reaction buffer (50 mM HEPES [pH 7.4], 25 mM magnesium acetate, 10 μM ATP, 2.5 mM dithiothreitol, 5 μCi of [γ-32P]ATP, 20 μg of casein [Sigma]) at 30°C for 30 min. Thirty microliters of 2× Laemmli sample buffer was added to stop the reaction, and the sample was separated on an SDS-15% polyacrylamide gel and electrophoresed. For cyclin B1/cdc2 kinase assays, lysates were divided into two samples and precleared with protein G-agarose beads (Invitrogen) for 1 h at 4°C. Lysates were then incubated with 6 μg of a mouse monoclonal cyclin B1 antibody (GNS1; Santa Cruz) for 3 h at 4°C. Beads were then washed three times in lysis buffer and twice in kinase buffer (20 mM HEPES [pH 7.4], 10 mM magnesium acetate) and then incubated in 30 μl of reaction buffer (50 mM HEPES [pH 7.4], 25 mM magnesium acetate, 50 μM ATP, 2.5 mM dithiothreitol, 5 μCi of [γ-32P] ATP, 1 μg of histone H1 [Sigma]) at 30°C for 20 min as previously described (28). Thirty microliters of 2× Laemmli sample buffer was added to stop the reaction, and the sample was separated on an SDS-15% polyacrylamide gel and electrophoresed.
RESULTS
Snk/Plk2 is a p53 target gene.
To identify novel p53 target genes in vivo, we performed an Affymetrix microarray analysis using a variety of tissues from wild-type and p53-null animals after irradiation (4). Our analysis of the spleen and thymus revealed that Snk/Plk2 was induced after irradiation in a p53-dependent manner in vivo (Fig. 1 and data not shown). Although very little is known about the function and regulation of Snk/Plk2, it belongs to a conserved family of kinases, the Polo-like kinases, which have been demonstrated to mediate mitotic progression. Members of the Polo-like kinase family are characterized by a conserved N-terminal catalytic domain and a C-terminal Polo box domain, which allows these kinases to localize to mitotic structures (14, 34).
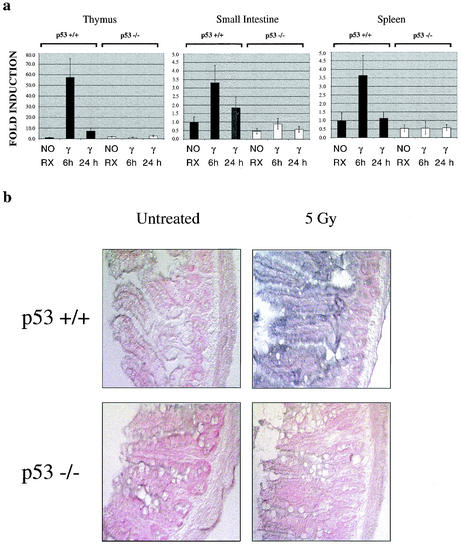
FIG. 1.
Snk/Plk2 is a p53 target gene in vivo. (a) mSnk/Plk2 mRNA expression in the thymus, small intestine, and spleen of wild-type (+/+) and p53 null (−/−) animals after irradiation (5 Gy). mRNA levels were determined by a TaqMan real-time quantitative RT-PCR assay. All expression levels were normalized to GAPDH in each well and are defined as the increases for samples relative to the level for p53+/+ untreated animals. NO RX, untreated animals. (b) In situ hybridization of irradiated ileum in wild-type (+/+) and p53 null (−/−) mice. The signal appears as blue or dark purple with pink or red counterstaining. Light microscopy images were taken using Nomarski optics (magnification, ×180).
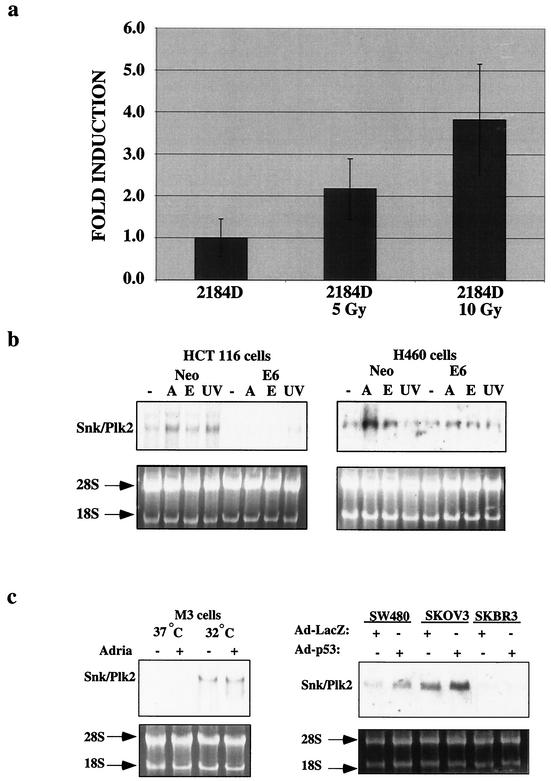
To confirm the observation from our microarray analysis that Snk/Plk2 is a p53 target gene in vivo, we examined Snk/Plk2 mRNA levels after irradiation of the thymus, small intestine, and spleen from wild-type and p53-null animals. Similar to the microarray data, we observed a p53-dependent induction of Snk/Plk2 after irradiation in vivo (Fig. 1a). Furthermore, through in situ hybridization we observed a p53-dependent induction of Snk/Plk2 mRNA in the epithelial layer of the ileum after irradiation (Fig. 1b). In addition, we found that Snk/Plk2 was induced after ionizing radiation in a normal human diploid lymphoblastoid cell line, 2184D after ionizing radiation (Fig. 2a). Previous studies demonstrated that p53 is stabilized after ionizing radiation in this cell line and induces its transcriptional target genes (5, 53). To demonstrate that Snk/Plk2 could be induced in a p53-dependent manner in tumor cell lines, we treated the colon carcinoma cell line, HCT116; the non-small cell lung carcinoma cell line, H460; and their isogenic clones expressing HPV E6 with several DNA-damaging agents. After treatment with adriamycin, Snk/Plk2 mRNA was induced in a p53-dependent manner in both tumor cell lines. In addition, a p53-dependent induction of Snk/Plk2 mRNA by etoposide and UV irradiation was noted in HCT116 cells (Fig. 2b). We also observed p53-dependent regulation of Snk/Plk2 in a murine lymphoma cell line stably expressing a temperature-sensitive p53 mutant, Val 135. At the permissive temperature, Snk/Plk2 was induced in a p53-dependent manner even in the absence of DNA damage (Fig. 2c). Furthermore, we observed Snk/Plk2 mRNA induction after overexpression of p53 from an adenovirus (Ad-p53) but not with a control adenovirus (Ad-LacZ) in two mutant p53-expressing cell lines (Fig. 2b). Interestingly we did not observe induction or basal expression of Snk/Plk2 mRNA in the breast carcinoma cell line SKBR3.
FIG. 2.
Snk/Plk2 is a p53 target gene in response to DNA-damaging agents. (a) hSnk/Plk2 mRNA expression after ionizing radiation in the normal human diploid lymphoblastoid cell line 2184D. 2184D cells were untreated or exposed to irradiation at 5 or 10 Gy, and total RNA was harvested 10 h after treatment. mRNA levels were determined by a TaqMan real-time quantitative RT-PCR assay. All expression levels were normalized to GAPDH in each well and are defined as the increases for samples relative to the level for untreated 2184D cells. (b) HCT116 Neo and E6 cells and H460 Neo and E6 cells were untreated or treated with adriamycin (0.2 μg/ml) (A), 0.2 μM etoposide (E), or 50 J/m2 (UV), and total RNA was harvested 10 h after treatment. Northern blotting was performed for Snk/Plk2. An ethidium stain of the RNA confirms equivalent loading. (c) (Left panel) Mouse M3 lymphoma cells expressing mutant p53 (37°C) or wild-type p53 (32°C) were untreated or treated with adriamycin (0.2 μg/ml) and harvested after 10 h. Northern blotting was performed for mSnk/Plk2. (Right panel) Three mutant p53-expressing colon (SW480), ovarian (SKOV3), and breast cancer cell lines (SKBR3) were infected with either Ad-LacZ or Ad-p53, and total RNA was harvested 10 h after infection. Northern blotting was performed for Snk/Plk2. An ethidium stain of the RNA confirms equivalent loading.
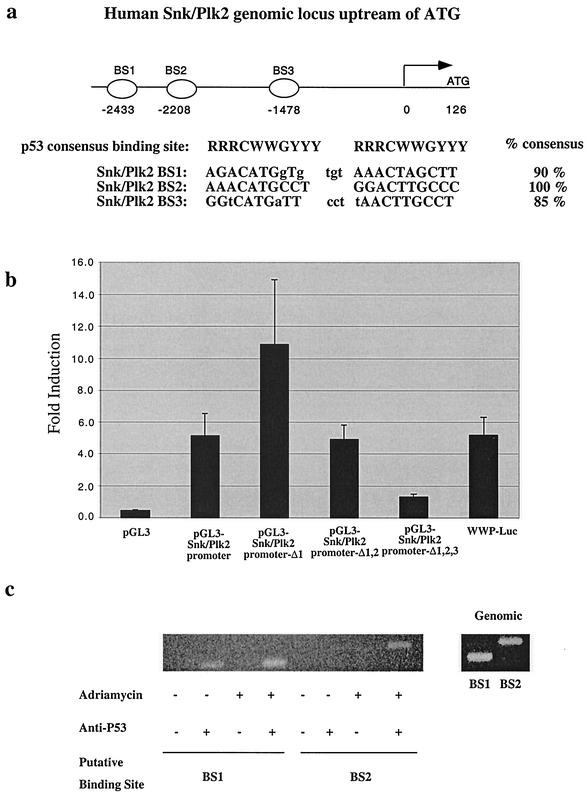
Since Snk/Plk2 mRNA could be induced by DNA damage in a p53-dependent manner or following exogenous p53 overexpression, we determined whether Snk/Plk2 is a direct p53 transcriptional target. We searched the NCBI database for potential p53 DNA-binding sites in the Snk/Plk2 genomic locus. We identified three candidate p53 binding sites in the promoter region of the Snk/Plk2 locus, hereafter referred to as BS1, BS2, and BS3. These sites were all located within 2,433 bp of the start of transcription and were 90, 100, and 85% homologous to the p53 consensus binding-site sequence (5′-RRRCWWGYYY[N0-13]RRRCWWGYYY-3′) (Fig. 3a). Similar sites were also found within the promoter region of the mouse Snk/Plk2 locus (data not shown). To determine whether p53 could transactivate the Snk/Plk2 promoter, we cloned the promoter region into a luciferase reporter construct. Using the p53-null cell line Saos2, we observed induction of the Snk/Plk2 promoter region by p53 overexpression compared to vector transfection. This induction was similar in magnitude to the induction observed with the p21WAF1 promoter construct pWWP-Luc (Fig. 3b). To determine which candidate p53 DNA binding sites were responsible for the p53 transactivation, we constructed a series of deletions within the Snk/Plk2 promoter region which removed either one (Δ1), two (Δ1,2), or all three (Δ1,2,3) p53-binding sites. Interestingly, deletion of the first p53-binding site actually increased the induction by p53, suggesting that the first p53 binding site (BS1) or other elements in that region may negatively regulate p53-dependent transactivation. Deletion of the first (BS1) and second (BS2) sites resulted in reduced induction by p53 that was completely abolished by deletion of the third site (BS3) (Fig. 3b). Similar results were observed in two other cell lines (data not shown). Therefore, it appears that all three sites may contribute to the p53-dependent regulation of Snk/Plk2.
FIG. 3.
The Snk/Plk2 promoter contains three p53 binding sites and is regulated by p53. (a) Schematic representation of the human Snk/Plk2 promoter region. Three p53 DNA binding sites are located at bp −2433 (BS1), bp −2208 (BS2), and bp −1478 (BS3) upstream from the start of transcription (indicated by arrow). An alignment of the p53 consensus binding element with the p53 binding sites present in the Snk/Plk2 promoter is shown. (b) pGL3-Snk/Plk2 promoter-Δ1, pGL3-Snk/Plk2 promoter-Δ1,2, or pGL3. Snk/Plk2 promoters were cotransfected (2.88 μg) with pCEP4 or pCEP4-p53 (0.72 μg) and pCMV-β-Gal (0.4 μg) into Saos2 cells. Luciferase activity was determined 24 h later. Empty vector (pGL3-Basic) and the p210WAF1 promoter (pWWP-Luc) were used as a negative and a positive control, respectively. Induction is indicated as levels for reporters with pCEP4-p53 divided by levels for reporters with pCEP4 (empty vector). (c) Adriamycin treatment enhances p53 binding to the Snk/Plk2 promoter in vivo. Untreated or adriamycin-treated U20S cells were harvested for chromatin immunoprecipitation as described in Materials and Methods. The left panel shows DNA that coprecipitated with the immunocomplexes and was amplified by PCR. The right panel shows amplification of genomic DNA using BS1 and BS2 primers.
To demonstrate that p53 binds these p53-binding elements in the Snk/Plk2 promoter region in vivo, we performed a ChIP assay. U2OS cells containing wild-type p53 were treated with adriamycin and then harvested for ChIP analysis after immunoprecipitation with an antibody against p53. Both BS1 and BS2 were specifically amplified by PCR after immunoprecipitation of p53 following adriamycin treatment (Fig. 3c). For BS1 we also detected a weak binding by p53 in the absence of DNA damage, probably due to the basal levels of p53 present in this wild-type p53 expressing cell line. These results suggest that Snk/Plk2 is a direct transcriptional target of p53 both in tumor cell lines and in vivo.
Snk/Plk2 has kinase activity that is negatively regulated by its carboxy-terminal region.
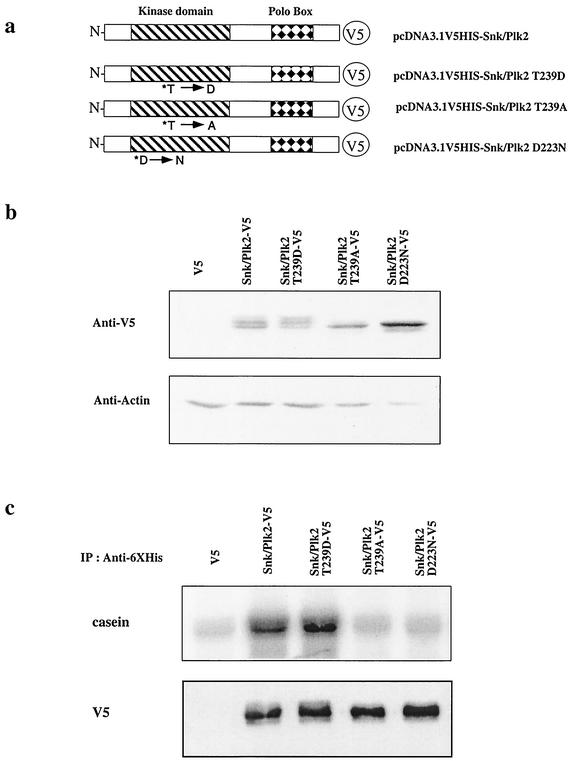
Members of the Polo-like kinase family are characterized by a conserved N-terminal catalytic domain and a C-terminal Polo-box domain. Previous studies with another Polo-like kinase family member, Plk1, have demonstrated that phosphorylation of a conserved Thr (homologous to residue 239 in Snk/Plk2) within kinase subdomain VIII (T-loop region) was necessary for full kinase activity (23). Further studies have also demonstrated that the carboxy-terminal sequences in Plk1 can inhibit kinase activity and that phosphorylation of this conserved Thr in the catalytic domain can overcome the inhibitory effects of the carboxy-terminal region (17, 33). To investigate whether Snk/Plk2 has kinase activity and more importantly whether its kinase activity is regulated similar to other Polo-like kinase members, we constructed a series of V5 epitope-tagged wild-type and mutant Snk/Plk2 constructs and expressed them in 293 cells (Fig. 4a). To test whether the conserved Thr in Snk/Plk2 was critical for kinase activity, we substituted Asp for Thr at residue 239 (T239D), which would mimic phosphorylation at this site and would be predicted to be a constitutively active mutant. Thr 239 was also mutated to an Ala (T239A), which is predicted to prevent phosphorylation at this site. Finally we constructed a predicted kinase-dead mutant by substituting the conserved Asp in the DFG sequence of kinase subdomain VII to Asn (D223N) (23). Although all constructs expressed similar amounts of protein only the wild-type and Thr 239-to-Asp constructs displayed three bands, suggesting that Snk/Plk2 may undergo autophosphorylation upon activation (Fig. 4b). In vitro kinase assays revealed that phosphorylation at Thr 239 is required for Snk/Plk2 kinase activity, as the kinase activity of the T239D mutant was significantly elevated compared to wild type. Conversely, the kinase activity of the T239A mutant was similar to that of the kinase-dead mutant, D223N, despite approximately equal amounts of Snk/Plk2 protein immunoprecipitated (Fig. 4c). Similar elevation of kinase activity was also observed when the carboxyl terminus was deleted (data not shown), supporting previous findings that phosphorylation of this conserved Thr in other Polo-like kinase family members could prevent the autoinhibitory effects of the carboxyl terminus (17, 33).
FIG. 4.
Snk/Plk2 has kinase activity that is regulated by phosphorylation in its kinase domain. (a) Schematic of Snk/Plk2 mutants. Snk/Plk2 contains an N-terminal kinase domain and a C-terminal Polo box. Single-amino-acid residue substitutions in the kinase domain of conserved residues are indicated with an asterisk. Constructs contain a C-terminal V5-six-histidine tag. (b) 293 cells were transfected with the indicated Snk/Plk2 constructs or empty vector. At 24 h, the cells were harvested for protein and analyzed for expression with mouse anti-V5 antibody. Actin was used to confirm that an equivalent amount of protein was loaded in each lane. (c) 293 cells were transfected with the indicated Snk/Plk2 constructs or empty vector for 48 h. Equal amounts of 293 cell lysates (2.5 × 106 cell equivalent) were harvested and immunoprecipitated with mouse monoclonal anti-polyhistidine agarose and subjected to Western blot analysis (lower panel) or an in vitro protein kinase assay (upper panel) using casein as a substrate. Lysates were immunoblotted for exogenously immunoprecipitated Snk/Plk2 using mouse monoclonal anti-V5-HRP.
Silencing of Snk/Plk2 sensitizes tumor cells to antimicrotubule agents through induction of apoptosis.
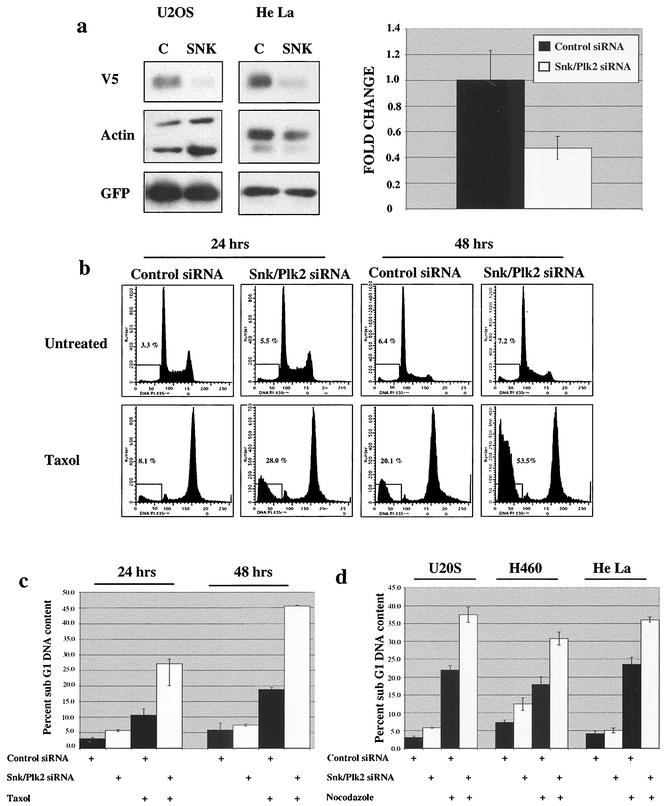
Our data demonstrated that Snk/Plk2 is a p53 target gene and that the kinase activity of Snk/Plk2 is regulated in a manner similar to other Polo-like kinase family members. Two Polo-like kinase members, Plk1 and Prk/Plk3, have previously been shown to positively and negatively regulate progression through G2 phase of the cell cycle, respectively (1, 21, 35, 38, 46, 54). Interestingly, Plk1 is required for progression through multiple phases of mitosis (10). We therefore explored the possibility that Snk/Plk2 may negatively regulate progression through mitosis under conditions of mitotic stress. We hypothesized that the p53-dependent regulation of Snk/Plk2 may explain the observed resistance to antimicrotubule agents in wild-type p53-containing cell lines. To elucidate the function of Snk/Plk2 in mitosis we designed an siRNA duplex that targeted the 5′ end of the coding sequence of Snk/Plk2. Transfection of U20S and HeLa cells with the siRNA duplex specifically silenced expression of exogenously expressed Snk/Plk2 protein compared to a control duplex designed against firefly luciferase (11) (Fig. 5a). Furthermore, the Snk/Plk2 siRNA duplex also efficiently silenced endogenous Snk/Plk2 mRNA in U20S, H460, and HeLa cells (Fig. 5a and data not shown). As expected the amount of silencing observed for endogenous Snk/Plk2 mRNA correlated with the observed transfection efficiency (data not shown).
FIG. 5.
Loss of Snk/Plk2 sensitizes tumor cells to spindle inhibitors through apoptosis.
(a) (Left panel) U2OS cells and HeLa cells were transfected with Snk/Plk2 V5-six-histidine tag (4.5 μg), pEGFP (0.5 μg), and control siRNA duplex or Snk/Plk2 siRNA duplex (200 nmol). After 48 h, cells were harvested for protein and analyzed for expression with anti-V5 antibody. Green fluorescent protein was used to control for possible differences in transfection efficiencies. Actin was used as to confirm that an equivalent amount of protein was loaded in each lane. (Right panel) U2OS cells were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). After 48 h, cells were harvested for total RNA,and Snk/Plk2 mRNA levels were determined by a TaqMan real-time quantitative RT-PCR assay. All expression levels are normalized to GAPDH in each well, and changes are defined as levels of Snk/Plk2 mRNA with Snk/Plk2 siRNA duplex divided by levels of Snk/Plk2 mRNA with control siRNA duplex. (b) U2OS cells were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). Twenty-four hours after transfection, cells were treated with 1 μM paclitaxel. After 24 or 48 h of paclitaxel treatment, cells were harvested and collected for fluorescence-activated cell sorting analysis. Cell death was scored as the percentage of cells with a DNA content of less than 2N. (c) Quantitation of experiment in panel b. Depicted in the chart is one of several experiments each done in triplicate. (d) The osteosarcoma cell line U2OS, the non-small cell lung cancer cell line H460, and the cervical carcinoma cell line HeLa, were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). Twenty-four hours after transfection, cells were treated with nocodazole (1 μg/ml). After 24 h of treatment, cells were harvested and collected for fluorescence-activated cell sorting analysis. Cell death was scored as the percentage of cells with a DNA content of less than 2N. Depicted in the chart is one of several experiments, each done in triplicate.
To examine whether loss of Snk/Plk2 may negatively regulate progression through mitosis under conditions of mitotic stress, U2OS cells were transfected with Snk/Plk2 siRNA duplexes, and after 24 h of transfection the cells were treated with paclitaxel for either 24 or 48 h. In the absence of paclitaxel, we observed no significant difference in the percentage of cells with a sub-G1 DNA content, nor were there any differences in the cell cycle profiles compared to the control duplex (Fig. 5b and c). This suggested that Snk/Plk2 was not required for normal progression through the cell cycle and mitosis. However, in the presence of paclitaxel we observed a significant increase in apoptosis in the cells in which Snk/Plk2 had been silenced (Fig. 5b and c). This sensitization to paclitaxel was even more dramatic after 48 h of treatment and could be observed as early as 16 h after treatment (Fig. 5b and c and data not shown). A similar sensitization was observed at lower doses of paclitaxel (data not shown). To investigate whether this sensitization was specific to paclitaxel, we treated three tumor cell lines (osteosarcoma, non-small cell lung cancer, and cervical carcinoma) with nocodazole after transfection of Snk/Plk2 siRNA. Silencing of Snk/Plk2 also sensitized these tumor cell lines to nocodazole by significantly increasing apoptosis (Fig. 5d). Although paclitaxel and nocodazole have opposing effects on microtubule dynamics, both agents lead to disruption of the mitotic spindle and activate the mitotic spindle checkpoint.
Loss of p53 sensitizes tumor cells to antimicrotubule agents.
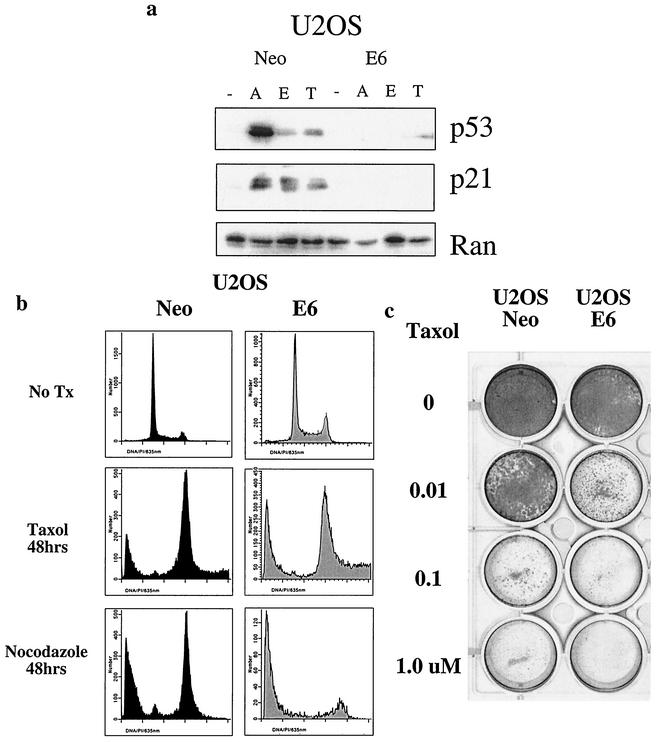
Several studies have shown that cell lines with mutant or deleted p53 are more sensitive to antimicrotubule agents, including paclitaxel (15, 47, 50). Since loss of the p53 target gene Snk/Plk2 could sensitize cells to antimicrotubule agents, we further investigated whether loss of p53 might also sensitize cells to these agents in U2OS cells. We first determined whether p53 could be stabilized and transactivate its target genes after treatment with antimicrotubule agents. In response to adriamycin, etoposide, and paclitaxel, p53 is stabilized and induces expression of its target gene p21 in the osteosarcoma cell line, U2OS (Fig. 6a). However, in the presence of human papillomavirus E6, p53 is degraded and cannot be stabilized by DNA-damaging agents or antimicrotubule agents (Fig. 6a). Since p53 was stabilized in response to paclitaxel, the effect of loss of p53 on sensitivity to antimicrotubule agents was evaluated. Upon treatment with paclitaxel, U20S cells expressing human papillomavirus E6 demonstrated increased apoptosis compared to the isogenic clone that contains wild-type p53. The effect did not appear to be agent specific, as a similar sensitization was observed with nocodazole, which activates the mitotic spindle checkpoint through destabilizing microtubules (Fig. 6b). Therefore, as expected, loss of p53 appears to sensitize cells to antimicrotubule agents in short term-assays. To measure whether loss of p53 could influence long-term outcome after treatment with paclitaxel, we examined the ability of U2OS Neo and U2OS E6 to form colonies after treatment with increasing doses of paclitaxel. Loss of p53 significantly increased sensitivity of this tumor cell line to paclitaxel as measured by the colony formation assay (Fig. 6c).
FIG. 6.
Loss of p53 sensitizes tumor cells to spindle inhibitors. (a) U2OS Neo and E6 cells were untreated (−) or treated for 24 h with adriamycin (0.2 μg/ml) (A), 0.2 μM etoposide (E), or 1 μM paclitaxel (T). Western blot analysis of lysates was performed for p53 and p21. Ran was used as to confirm that an equivalent amount of protein was loaded in each lane. (b) U2OS Neo and E6 were treated with 1 μM paclitaxel or nocodazole (1 μg/ml). After 48 h cells were harvested and collected for fluorescence-activated cell sorting analysis. Cell death was scored as the percentage of cells with a DNA content of less than 2N. (c) U2OS Neo and E6 cells were plated at 2.4 × 104 cells per well in a 12-well dish and treated with the indicated doses of paclitaxel for 24 h. After 24 h of treatment, the medium was removed and replaced with fresh medium without paclitaxel. Cells were grown for 7 days and then stained with Coomassie blue.
Silencing of Snk/Plk2 leads to apoptosis during mitosis after treatment with antimicrotubule agents.
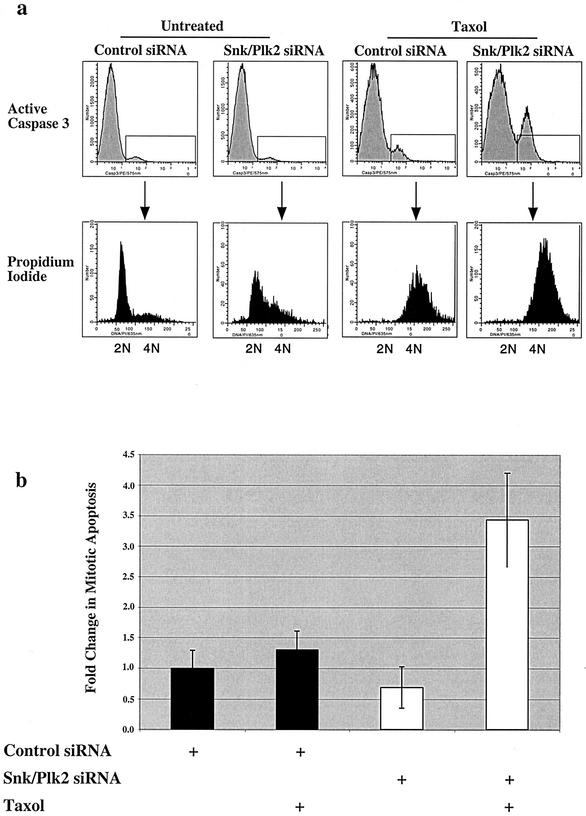
Previous studies have suggested that p53 confers resistance to paclitaxel and other antimicrotubule agents by mediating a G1 arrest after treatment of cells with antimicrotubule agents. After treatment with spindle inhibitors, cells initially arrest in mitosis and then undergo adaptation and exit mitosis into a G1-like phase (51). Cells which lack p53 do not arrest in G1 after exposure to antimicrotubule agents and endoreduplicate their DNA, leading to polyploidy and massive apoptosis (9, 22, 32). Our data suggest that p53 may contribute to a checkpoint during mitosis after exposure to antimicrotubule agents through its target gene, Snk/Plk2. We therefore investigated whether loss of Snk/Plk2 sensitizes cells to antimicrotubule agents through induction of apoptosis in mitosis or after progression to the subsequent G1-like phase. To confirm that the increase in sub-G1 DNA content was due to apoptosis, cells that had been transfected with Snk/Plk2 siRNA duplex followed by paclitaxel treatment were stained with an antibody directed against active caspase 3, a marker of apoptosis. As expected, silencing of Snk/Plk2 dramatically increased the fraction of cells that were positive for active caspase 3 (Fig. 7a). Propidium iodide staining of active caspase 3-positive cells revealed that cells treated with paclitaxel or nocodazole underwent apoptosis with a 4N DNA content (Fig. 7a and data not shown). However, after treatment with antimicrotubule agents, cells with a 4N DNA content could either be arrested in mitosis or in a G1-like phase of the cell cycle if they had progressed without dividing.
FIG. 7.
Loss of Snk/Plk2 leads to apoptosis during mitosis after treatment with antimicrotubule agents. (a) U2OS cells were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). Twenty-four hours after transfection, cells were treated with 1 μM paclitaxel. After a 24-h treatment, cells were harvested and stained for active caspase 3. Cells were also stained with propidium iodide to measure DNA content. The upper panel depicts active caspase 3 staining, while in the lower panel, DNA content of active caspase 3-positive cells was determined. (b) U2OS cells were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). Twenty-four hours after transfection, cells were treated with 1 μM paclitaxel. After 20 h of treatment, cells were harvested and stained for active caspase 3 and cyclin B. Cells were also stained with propidium iodide to measure DNA content. Depicted in the chart is the change in mitotic apoptosis (active caspase 3 positive, cyclin B positive, 4N DNA content) after paclitaxel treatment and siRNA directed against luciferase or Snk/Plk2. Changes are relative to untreated control siRNA. Depicted in the chart is one of several experiments, each performed in triplicate.
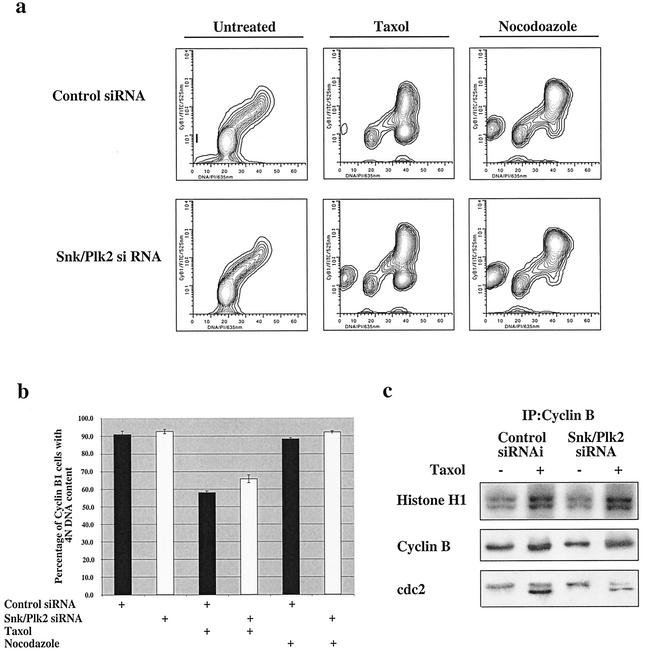
To determine whether these cells are undergoing apoptosis in mitosis or during a G1-like phase, U20S cells were transfected with Snk/Plk2 siRNA and treated with paclitaxel or nocodazole and stained for active caspase 3 and cyclin B1. The cells were also stained with propidium iodide to measure DNA content (Fig. 7b). Previous studies have demonstrated that cells arrested in mitosis maintain high levels of cyclin B1, whereas cyclin B1 is rapidly degraded in cells that have adapted and progressed to the G1 phase after treatment (32, 41). Therefore, cells that have a 4N DNA content and are cyclin B1 positive are in mitosis, while cells with a 4N DNA content that are cyclin B1 negative have progressed to a G1-like phase without dividing. After transfection with control or Snk/Plk2 siRNA and treatment with paclitaxel for 20 h, the active caspase 3-positive cells with a 4N DNA content were examined for cyclin B1 expression. More than 80% of the active caspase 3-positive cells with a 4N DNA content were positive for cyclin B1 and therefore represented cells that were undergoing apoptosis during mitosis (data not shown). Furthermore, cells in which Snk/Plk2 expression was silenced displayed a highly significant increase in “mitotic apoptosis” compared to control siRNA (Fig. 7b). Therefore, it appears that silencing of the direct p53 target gene, Snk/Plk2 lead to apoptosis during mitosis after exposure to antimicrotubule agents. These results were further supported by the observation that loss of Snk/Plk2 did not increase the number of cells that adapted to spindle damage and progressed to G1 phase after exposure to paclitaxel or nocodazole as measured by cyclin B1 staining (Fig. 8a and b). After transfection with Snk/Plk2 siRNA or control siRNA and treatment with either paclitaxel or nocodazole, the majority of the cells remain cyclin B1 positive and have a 4N DNA content even in the absence of Snk/Plk2 (Fig. 8a). After paclitaxel treatment, there is a small but significant cyclin B1-negative population with a 4N DNA content, which probably represents cells that have progressed to the G1 phase (Fig. 8b). However, almost 90% of nocodazole-treated cells are cyclin B1 positive and therefore are still in mitosis. Finally, loss of Snk/Plk2 did not lead to decreased levels of cdc2 kinase activity, further corroborating that its loss did not lead to progression into the G1 phase (Fig. 8c). Taken together these results suggest that p53, through its target gene, Snk/Plk2, may mediate a mitotic checkpoint after treatment with antimicrotubule agents.
FIG. 8.
Loss of Snk/Plk2 does not lead to progression into anaphase or G1 phase after spindle inhibitors. (a) U2OS cells were transfected with control siRNA duplex or Snk/Plk2. siRNA duplex (300 nmol). Twenty-four hours after transfection, cells were treated with 1 μM paclitaxel or nocodazole (1.0 μg/ml). After a 24-h treatment, cells were harvested and stained for cyclin B1 expression. Cells were also stained with propidium iodide to measure DNA content. (b) Depicted in the chart is the percentage of cells with 4N DNA content that are cyclin B1 positive. Results are from one of several experiments each done in triplicate. (c) U2OS cells were transfected with control siRNA duplex or Snk/Plk2 siRNA duplex (300 nmol). Thirty-six hours after transfection, cells were treated with 1 μM paclitaxel. After 12 h, equal amounts of U2OS cell lysate were harvested and immunoprecipitated with mouse monoclonal anti-cyclin B1 antibody and subjected to Western blot analysis or an in vitro protein kinase assay using histone H1 as a substrate. Lysates were immunoblotted for cyclin B1 and cdc2.
DISCUSSION
An indirect role for p53 in the response to spindle damage through induction of a G1 arrest has been previously suggested (9, 22, 32). We now propose that p53 may play a direct role in the response to spindle damage during mitosis through the regulation of a polo-like kinase. We have demonstrated that Snk/Plk2 is a p53 target gene and that its induction in response to genotoxic stress occurs in vivo. Furthermore, we have established that Snk/Plk2 has kinase activity and this activity is regulated similarly to other family members. Importantly we have demonstrated that loss of Snk/Plk2 phenocopies loss of p53 by dramatically increasing sensitivity to paclitaxel and other antimicrotubule agents. Interestingly, we demonstrated that this sensitization occurs during mitosis before the cells have adapted and progressed to the G1 phase. We believe this represents a novel mechanism through which p53 maintains genomic integrity. Since siRNA directed against Snk/Plk2 promoted death of paclitaxel-treated cells in mitosis, we envision a mitotic checkpoint wherein p53-dependent activation of Snk/Plk2 prevents mitotic catastrophe following spindle damage. Finally, we believe inhibition of Snk/Plk2 may be of therapeutic value when using taxanes or other classes of antimicrotubule agents in tumors with wild-type p53 or to enhance killing in tumors with p53 mutations.
Initial studies with p53-null mouse embryonic fibroblasts suggested a role for p53 in the mitotic spindle checkpoint (7). However, several subsequent studies using normal mouse and human fibroblast cell lines or a mouse prolymphocytic cell line have suggested that p53 did not have a direct role in mitosis but rather induced a G1 arrest after treatment with antimicrotubule agents (9, 22, 32). Our data suggest that p53 may in addition directly mediate a checkpoint in mitosis through its regulation of Snk/Plk2. In the human tumor cell lines used in these studies, we have observed an extended arrest in mitosis at least for the first 24 h after treatment (Fig. 8). Furthermore, the sensitization to both paclitaxel and nocodazole was observed prior to adaptation, when the p53 target gene Snk/Plk2 was silenced (Fig. 7 and 8). Previous studies with the colon cancer cell line HCT116 have also shown arrest in mitosis before progression to the G1 phase to be longer than what had previously been observed in fibroblasts (41). This suggests that there may be a difference in the length of the mitotic spindle checkpoint in normal versus tumor cells. Interestingly, phosphorylation of p53 in response to microtubule inhibitors appears to be differentially regulated in normal fibroblasts compared to epithelial tumor cell lines (42). It is possible that this differential phosphorylation of p53 may explain why this p53-dependent checkpoint was not observed in normal cells. This is the first study directly linking p53 to a mitotic checkpoint after treatment with antimicrotubule agents and suggests a novel mechanism by which p53 maintains genomic stability.
This study and several others have demonstrated that loss of p53 can sensitize cells to several clinically relevant antimicrotubule agents, including paclitaxel (15, 47, 50). It is clear that in the presence of antimicrotubule agents, p53 can mediate a G1 arrest after cells have adapted and exited mitosis. This G1 arrest appears to be dependent on the p53 target gene p21WAF1 in both normal and tumor cells as loss of p21 leads to endoreduplication and massive apoptosis (Fig. 7) (20, 22, 41). We have now demonstrated that p53, through its regulation of Snk/Plk2, may mediate a mitotic checkpoint and prevent apoptosis during mitosis after exposure to spindle inhibitors (Fig. 9). In both situations the function of p53 is to prevent genomic instability by preventing progression out of mitosis or G1 phase. However, since paclitaxel exerts its cytotoxic effects through the induction of a p53-independent apoptosis (50), the p53-mediated arrest in either mitosis or G1 leads to chemoresistance. The findings presented here demonstrating that silencing of Snk/Plk2 can sensitize cells to paclitaxel and previous findings that silencing of p21 can also sensitize cells to paclitaxel (56) suggest that inhibiting these p53 target genes in the context of antimicrotubule agents may represent a novel therapeutic strategy.
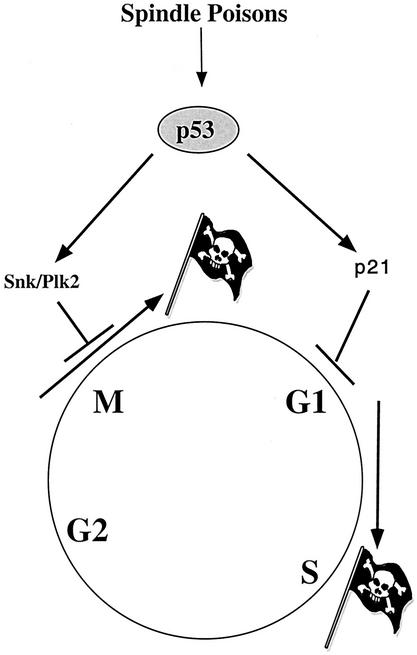
FIG. 9.
Model of p53-dependent regulation of mitotic and G1 checkpoint after cellular exposure to antimicrotubule agents.
Inhibition of Snk/Plk2 through the RNA interference strategy appears to dramatically sensitize tumor cells to antimicrotubule agents but does not appear to affect cell cycle progression in cells not exposed to microtubule poisons (Fig. 5). Interestingly, inhibition of Plk1 through RNA interference strategies resulted in arrest of tumor cells in G2/M with multiple centrosomes leading to spindle defects, arrest at anaphase, and cytokinesis defects leading to apoptosis (26, 40). These effects were observed in the absence of any genotoxic or spindle stress, unlike what was observed here with silencing of Snk/Plk2. These findings suggest that Snk/Plk2 has distinct functions from Plk1, as Snk/Plk2 does not appear to be essential for normal cell cycle progression. Rather, our data suggest that Snk/Plk2 plays a role in the mitotic spindle checkpoint, either directly or indirectly during mitosis, and loss of Snk/Plk2 leads to apoptosis in cells with damaged microtubules. Interestingly a similar sensitization to antimicrotubule agents has been observed when Mad2, a regulator of the mitotic spindle checkpoint, has been silenced (27). Previous studies have identified the IAP family member, survivin, as a key determinant of paclitaxel resistance in tumor cell lines. However, since survivin antiapoptotic activity is dependent on intact microtubules, regulation of survivin cannot explain how loss of Snk/Plk2 leads to increased apoptosis after treatment with nocodazole (Fig. 5) (25).
Prior to this study little was known about Snk/Plk2. It was originally discovered like Plk3, as an immediate-early gene, suggesting it may have a role outside of mitosis (39); however, no function for Snk/Plk2 has been suggested previously. While it is possible that Snk/Plk2, like other Plks, may have functions outside of mitosis; it is clearly an important mediator of the cellular response to antimicrotubule agents. Snk/Plk2, like other polo-like kinase family members, contains a polo box domain that has been demonstrated in other family members to localize them to the centrosomes, kinetochores, the central spindle midzone, and the midbody (8, 34). Similar to other family members, the carboxyl terminus region encompassing the polo box appears to inhibit Snk/Plk2 kinase activity (Fig. 4). These findings suggest an alternative mechanism to inhibit Snk/Plk2 function. A small peptide containing a partial sequence from the polo box of Plk1 fused to an Antennapedia peptide was shown to be efficiently internalized and inhibit Plk1 function (57). Our in vitro kinase data suggest that fusion of the Snk/Plk2 polo box to an Antennapedia peptide may be able to inhibit Snk/Plk2 function. Identification of Snk/Plk2 substrates in mitosis may also elucidate its mechanism of action after exposure to antimicrotubule agents.
Since Snk/Plk2 appears to be involved in the mitotic spindle checkpoint, it remains unclear whether long-term silencing of Snk/Plk2 may lead to increased genomic instability in tumors. In the breast cancer cell line SKBR3, Snk/Plk2 mRNA levels were undetectable and could not be induced by overexpression of p53 (Fig. 2c). Interestingly, treatment with trichostatin A, a histone deacetylase inhibitor, increased expression of Snk/Plk2, suggesting that Snk/Plk2 may be silenced in this cell line (unpublished data). Furthermore, Snk/Plk2 is located cytogenetically on chromosome 5q12.1-q13.2, a region in which translocations have been observed in acute lymphocytic leukemia, chronic myeloid leukemia, and several salivary and thyroid adenomas (2, 3, 29, 37, 45). Interestingly, comparative genomic hybridization (CGH) studies using the CGH database at the Institute of Pathology, Humboldt University of Berlin, have found that this chromosomal region is lost at a high frequency in a variety of tumor types (http://amba.charite.de/cgh/). It would be interesting to determine whether levels of Snk/Plk2 in tumor cell lines could be correlated with the level of genomic instability and/or sensitivity to antimicrotubule agents.
In summary we have demonstrated a possible role for p53 in regulating a mitotic checkpoint through its direct transcriptional regulation of Snk/Plk2, a polo-like kinase gene. This represents a novel mechanism by which p53 may maintain genomic stability. Furthermore, we have suggested a novel therapeutic intervention through silencing Snk/Plk2 that can significantly sensitize tumor cells to taxanes and other antimicrotubule agents. We believe that inhibiting p53 target genes that maintain genomic integrity in the presence of antimicrotubule agents may represent a novel therapeutic strategy.
Acknowledgments
We thank Eric Bernhard for assistance with mouse irradiation and thank the Morphology Core in the Center for Molecular Studies of Digestive and Liver Diseases for their expert assistance.
This work was supported by NIH grant T32 CA09677 (T.F.B.), NIH grant PO1 CA75138 (W.S.E.-D.), and funds from the Howard Hughes Medical Institute (W.S.E.-D.). W.S.E.-D. is an Assistant Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abrieu, A., T. Brassac, S. Galas, D. Fisher, J. C. Labbe, and M. Doree. 1998. The Polo-like kinase Plx1 is a component of the MPF amplification loop at the G2/M-phase transition of the cell cycle in Xenopus eggs. J. Cell Sci. 111:1751-1757. [DOI] [PubMed] [Google Scholar]
- 2.Ankathil, R., N. Geetha, P. Remani, V. P. Gangadharan, G. R. Pillai, and M. K. Nair. 1996. Clinical implications of cytogenetic classification in adult acute lymphoblastic leukaemia patients. J. Cancer Res. Clin. Oncol. 122:370-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belge, G., L. Roque, J. Soares, S. Bruckmann, B. Thode, E. Fonseca, A. Clode, S. Bartnitzke, S. Castedo, and J. Bullerdiek. 1998. Cytogenetic investigations of 340 thyroid hyperplasias and adenomas revealing correlations between cytogenetic findings and histology. Cancer Genet. Cytogenet. 101:42-48. [DOI] [PubMed] [Google Scholar]
- 4.Burns, T. F., E. J. Bernhard, and W. S. El-Deiry. 2001. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20:4601-4612. [DOI] [PubMed] [Google Scholar]
- 4a.Burns, T. F., and W. S. El-Deiry. Microarray analysis of p53 target gene expression patterns in the spleen and thymus in response to ionizing radiation. Cancer Biol. Ther., in press. [DOI] [PubMed]
- 5.Canman, C. E., A. C. Wolff, C. Y. Chen, A. J. Fornace, Jr., and M. B. Kastan. 1994. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 54:5054-5058. [PubMed] [Google Scholar]
- 6.Chase, D., Y. Feng, B. Hanshew, J. A. Winkles, D. L. Longo, and D. K. Ferris. 1998. Expression and phosphorylation of fibroblast-growth-factor-inducible kinase (Fnk) during cell-cycle progression. Biochem. J. 333:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross, S. M., C. A. Sanchez, C. A. Morgan, M. K. Schimke, S. Ramel, R. L. Idzerda, W. H. Raskind, and B. J. Reid. 1995. A p53-dependent mouse spindle checkpoint. Science 267:1353-1356. [DOI] [PubMed] [Google Scholar]
- 8.Dai, W., Q. Wang, and F. Traganos. 2002. Polo-like kinases and centrosome regulation. Oncogene 21:6195-6200. [DOI] [PubMed] [Google Scholar]
- 9.Di Leonardo, A., S. H. Khan, S. P. Linke, V. Greco, G. Seidita, and G. M. Wahl. 1997. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 57:1013-1019. [PubMed] [Google Scholar]
- 10.Donaldson, M. M., A. A. Tavares, I. M. Hagan, E. A. Nigg, and D. M. Glover. 2001. The mitotic roles of Polo-like kinase. J. Cell Sci. 114:2357-2358. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 12.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 13.Fei, P., E. J. Bernhard, and W. S. El-Deiry. 2002. Tissue-specific induction of p53 targets in vivo. Cancer Res. 62:7316-7327. [PubMed] [Google Scholar]
- 14.Glover, D. M., I. M. Hagan, and A. A. Tavares. 1998. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 12:3777-3787. [DOI] [PubMed] [Google Scholar]
- 15.Hawkins, D. S., G. W. Demers, and D. A. Galloway. 1996. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 56:892-898. [PubMed] [Google Scholar]
- 16.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 17.Jang, Y. J., C. Y. Lin, S. Ma, and R. L. Erikson. 2002. Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc. Natl. Acad. Sci. USA 99:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadkol, S., J. Juang, and T. C. Wu. 2002. In situ hybridization in cancer and normal tissue. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 19.Kauselmann, G., M. Weiler, P. Wulff, S. Jessberger, U. Konietzko, J. Scafidi, U. Staubli, J. Bereiter-Hahn, K. Strebhardt, and D. Kuhl. 1999. The polo-like protein kinases Fnk and Snk associate with a Ca2+- and integrin-binding protein and are regulated dynamically with synaptic plasticity. EMBO J. 18:5528-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan, S. H., and G. M. Wahl. 1998. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 58:396-401. [PubMed] [Google Scholar]
- 21.Kumagai, A., and W. G. Dunphy. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273:1377-1380. [DOI] [PubMed] [Google Scholar]
- 22.Lanni, J. S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18:1055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, K. S., and R. L. Erikson. 1997. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol. Cell. Biol. 17:3408-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, B., B. Ouyang, H. Pan, P. T. Reissmann, D. J. Slamon, R. Arceci, L. Lu, and W. Dai. 1996. Prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J. Biol. Chem. 271:19402-19408. [DOI] [PubMed] [Google Scholar]
- 25.Li, F., G. Ambrosini, E. Y. Chu, J. Plescia, S. Tognin, P. C. Marchisio, and D. C. Altieri. 1998. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396:580-584. [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., and R. L. Erikson. 2002. Activation of Cdc2/cyclin B and inhibition of centrosome amplification in cells depleted of Plk1 by siRNA. Proc. Natl. Acad. Sci. USA 99:8672-8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, X., Z. Tang, J. Rizo, and H. Yu. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell 9:59-71. [DOI] [PubMed] [Google Scholar]
- 28.MacLachlan, T. K., K. Somasundaram, M. Sgagias, Y. Shifman, R. J. Muschel, K. H. Cowan, and W. S. El-Deiry. 2000. BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J. Biol. Chem. 275:2777-2785. [DOI] [PubMed] [Google Scholar]
- 29.Mark, J., R. Dahlenfors, and B. Wedell. 1997. Impact of the in vitro technique used on the cytogenetic patterns in pleomorphic adenomas. Cancer Genet. Cytogenet. 95:9-15. [DOI] [PubMed] [Google Scholar]
- 30.McDonald, E. R. I., P. C. Chui, P. F. Martelli, D. T. Dicker, and W. S. El-Deiry. 2001. Death domain mutagenesis of KILLER/DR5 reveals residues critical for apoptotic signaling. J. Biol. Chem. 276:14939-14945. [DOI] [PubMed] [Google Scholar]
- 31.Meng, R. D., H. Shih, N. S. Prabhu, D. L. George, and W. S. El-Deiry. 1998. Bypass of abnormal MDM2 inhibition of p53-dependent growth suppression. Clin. Cancer Res. 4:251-259. [PubMed] [Google Scholar]
- 32.Minn, A. J., L. H. Boise, and C. B. Thompson. 1996. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10:2621-2631. [DOI] [PubMed] [Google Scholar]
- 33.Mundt, K. E., R. M. Golsteyn, H. A. Lane, and E. A. Nigg. 1997. On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun. 239:377-385. [DOI] [PubMed] [Google Scholar]
- 34.Nigg, E. A. 1998. Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol. 10:776-783. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang, B., W. Li, H. Pan, J. Meadows, I. Hoffmann, and W. Dai. 1999. The physical association and phosphorylation of Cdc25C protein phosphatase by Prk. Oncogene 18:6029-6036. [DOI] [PubMed] [Google Scholar]
- 36.Ozoren, N., M. J. Fisher, K. Kim, C. X. Liu, A. Genin, Y. Shifman, D. T. Dicker, N. B. Spinner, N. A. Lisitsyn, and W. S. El-Deiry. 2000. Homozygous deletion of the death receptor DR4 gene in a nasopharyngeal cancer cell line is associated with TRAIL resistance. Int. J. Oncol. 16:917-925. [DOI] [PubMed] [Google Scholar]
- 37.Potter, A. M., A. E. Watmore, P. Cooke, J. S. Lilleyman, and R. J. Sokol. 1981. Significance of non-standard Philadelphia chromosomes in chronic granulocytic leukaemia. Br. J. Cancer 44:51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian, Y. W., E. Erikson, C. Li, and J. L. Maller. 1998. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol. Cell. Biol. 18:4262-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simmons, D. L., B. G. Neel, R. Stevens, G. Evett, and R. L. Erikson. 1992. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol. Cell. Biol. 12:4164-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spankuch-Schmitt, B., G. Wolf, C. Solbach, S. Loibl, R. Knecht, M. Stegmuller, G. von Minckwitz, M. Kaufmann, and K. Strebhardt. 2002. Downregulation of human polo-like kinase activity by antisense oligonucleotides induces growth inhibition in cancer cells. Oncogene 21:3162-3171. [DOI] [PubMed] [Google Scholar]
- 41.Stewart, Z. A., D. Mays, and J. A. Pietenpol. 1999. Defective G1-S cell cycle checkpoint function sensitizes cells to microtubule inhibitor-induced apoptosis. Cancer Res. 59:3831-3837. [PubMed] [Google Scholar]
- 42.Stewart, Z. A., L. J. Tang, and J. A. Pietenpol. 2001. Increased p53 phosphorylation after microtubule disruption is mediated in a microtubule inhibitor- and cell-specific manner. Oncogene 20:113-124. [DOI] [PubMed] [Google Scholar]
- 43.Sunkel, C. E., and D. M. Glover. 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89:25-38. [DOI] [PubMed] [Google Scholar]
- 44.Takimoto, R., T. K. MacLachlan, D. T. Dicker, Y. Niitsu, T. Mori, and W. S. El-Deiry. 2002. BRCA1 transcriptionally regulates damaged DNA binding protein (DDB2) in the DNA repair response following UV-irradiation. Cancer Biol. Ther. 1:177-186. [DOI] [PubMed] [Google Scholar]
- 45.Teyssier, J. R., F. Liautaud-Roger, D. Ferre, M. Patey, and J. Dufer. 1990. Chromosomal changes in thyroid tumors. Relation with DNA content, karyotypic features, and clinical data. Cancer Genet. Cytogenet. 50:249-263. [DOI] [PubMed] [Google Scholar]
- 46.Toyoshima-Morimoto, F., E. Taniguchi, N. Shinya, A. Iwamatsu, and E. Nishida. 2001. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature 410:215-220. [DOI] [PubMed] [Google Scholar]
- 47.Vikhanskaya, F., S. Vignati, P. Beccaglia, C. Ottoboni, P. Russo, M. D'Incalci, and M. Broggini. 1998. Inactivation of p53 in a human ovarian cancer cell line increases the sensitivity to paclitaxel by inducing G2/M arrest and apoptosis. Exp. Cell Res. 241:96-101. [DOI] [PubMed] [Google Scholar]
- 48.Vogelstein, B., and K. W. Kinzler. 1992. p53 function and dysfunction. Cell 70:523-526. [DOI] [PubMed] [Google Scholar]
- 49.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 50.Wahl, A. F., K. L. Donaldson, C. Fairchild, F. Y. Lee, S. A. Foster, G. W. Demers, and D. A. Galloway. 1996. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat. Med. 2:72-79. [DOI] [PubMed] [Google Scholar]
- 51.Wassmann, K., and R. Benezra. 2001. Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev. 11:83-90. [DOI] [PubMed] [Google Scholar]
- 52.Wu, G. S., T. F. Burns, E. R. McDonald III, W. Jiang, R. Meng, I. D. Krantz, G. Kao, D. D. Gan, J. Y. Zhou, R. Muschel, S. R. Hamilton, N. B. Spinner, S. Markowitz, G. Wu, and W. S. El-Deiry. 1997. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17:141-143. [DOI] [PubMed] [Google Scholar]
- 53.Wu, G. S., T. F. Burns, E. R. McDonald III, R. D. Meng, G. Kao, R. Muschel, T. Yen, and W. S. El-Deiry. 1999. Induction of the TRAIL receptor KILLER/DR5 in p53-dependent apoptosis but not growth arrest. Oncogene 18:6411-6418. [DOI] [PubMed] [Google Scholar]
- 54.Xie, S., H. Wu, Q. Wang, J. P. Cogswell, I. Husain, C. Conn, P. Stambrook, M. Jhanwar-Uniyal, and W. Dai. 2001. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J. Biol. Chem. 276:43305-43312. [DOI] [PubMed] [Google Scholar]
- 55.Xie, S., H. Wu, Q. Wang, J. Kunicki, R. O. Thomas, R. E. Hollingsworth, J. Cogswell, and W. Dai. 2002. Genotoxic stress-induced activation of Plk3 is partly mediated by Chk2. Cell Cycle 1:424-429. [DOI] [PubMed] [Google Scholar]
- 56.Yu, D., T. Jing, B. Liu, J. Yao, M. Tan, T. J. McDonnell, and M. C. Hung. 1998. Overexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol. Cell 2:581-591. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, J., A. Kramer, F. Eckerdt, M. Kaufmann, and K. Strebhardt. 2002. Efficient internalization of the polo-box of polo-like kinase 1 fused to an Antennapedia peptide results in inhibition of cancer cell proliferation. Cancer Res. 62:4186-4190. [PubMed] [Google Scholar]