Abstract
Heme-responsive motifs (HRMs) mediate heme regulation of diverse regulatory proteins. The heme activator protein Hap1 contains seven HRMs, but only one of them, HRM7, is essential for heme activation of Hap1. To better understand the molecular basis underlying the biological significance of HRMs, we examined the effects of various mutations of HRM7 on Hap1. We found that diverse mutations of HRM7 significantly diminished the extent of Hap1 activation by heme and moderately enhanced the interaction of Hap1 with Hsp90. Furthermore, deletions of nonregulatory sequences completely abolished heme activation of Hap1 and greatly enhanced the interaction of Hap1 with Hsp90. These results show that the biological functions of HRMs and Hsp90 are highly sensitive to structural changes. The unique role of HRM7 in heme activation stems from its specific structural environment, not its mere presence. Likewise, the role of Hsp90 in Hap1 activation is dictated by the conformational or structural state of Hap1, not by the mere strength of Hap1-Hsp90 interaction. It appears likely that HRM7 and Hsp90 act together to promote the Hap1 conformational changes that are necessary for Hap1 activation. Such fundamental mechanisms of HRM-Hsp90 cooperation may operate in diverse regulatory systems to mediate signal transduction.
Heme serves key roles in many fundamental biological processes in virtually all living organisms. It is essential for the transport and storage of oxygen, for the generation of cellular energy by respiration, for the synthesis and degradation of steroids, lipids and certain neurotransmitters, for the detoxification of xenobiotics, and for controlling oxidative damage (4). Heme not only serves as an essential prosthetic group in many proteins and enzymes that transport or use oxygen, but it also directly controls the levels and functional activities of such proteins and enzymes and their upstream regulators (28, 38, 39). For example, in reticulocytes, heme controls protein synthesis by directly regulating the activity of the heme-regulated inhibitor kinase HRI, which phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF-2) at serine 51, causing the formation of an eIF2α (phosphorylated)/eIF-2B complex and inhibition of protein synthesis (2, 7, 8). Notably, heme directly binds to and inhibits the mammalian transcriptional repressor Bach1 (27), a dimerization partner of the Maf family proteins, which controls the expression of a wide array of genes, including oxidative stress-responsive genes and globin genes (1, 18-22, 27, 41, 46). Other heme-regulated transcription factors include the CAAT box-binding factor NF-Y (25), the heme-responsive Ku protein complex (36, 37), the bacterial iron response regulator protein Irr (34), and the yeast heme activator protein Hap1 (9, 29). Remarkably, recent studies have also shown that heme is critical for the activation of the Ras-MAPK signaling pathway and the expression of many neuron-specific genes induced by nerve growth factor (53, 54). It is increasingly clear that heme controls the activity of numerous regulatory and signaling pathways by interacting directly with key pathway regulatory proteins.
How does heme control the activity of diverse regulatory proteins? Heme regulation of many proteins appears to be mediated by a short heme-binding motif, termed the heme-responsive motif (HRM) (27, 34, 40, 49). Most, if not all, heme-regulated proteins have been shown to contain HRMs, which consist of two invariable CP residues and other more variable residues, such as D and H (Fig. 1) (27, 34, 40, 49). For example, Bach1 contains four HRMs, at least one of which is critical for heme binding and regulation (27). Irr contains one HRM that is critical for heme binding and regulation (34). Hap1 contains seven HRMs, of which six (HRM1 to -6) are clustered near the DNA-binding domain, and one (HRM7) is located near the transcription activation domain (Fig. 1) (49). Other hemoproteins that contain HRMs include HRI, heme oxygenase, and heme lyases (35, 49). In vitro, all these HRMs bind heme reversibly with micromolar affinity (34, 49). However, it is unlikely that all HRMs in a protein are equally important for biological function in vivo. If HRMs possess different functional roles, how are their functional roles determined? Existing data in the literature provide little mechanistic insight into how heme binding to HRMs may alter the structure of regulatory proteins and cause changes in their activity. Thus, a more detailed analysis of the functional roles of HRMs and their modes of action is necessary.
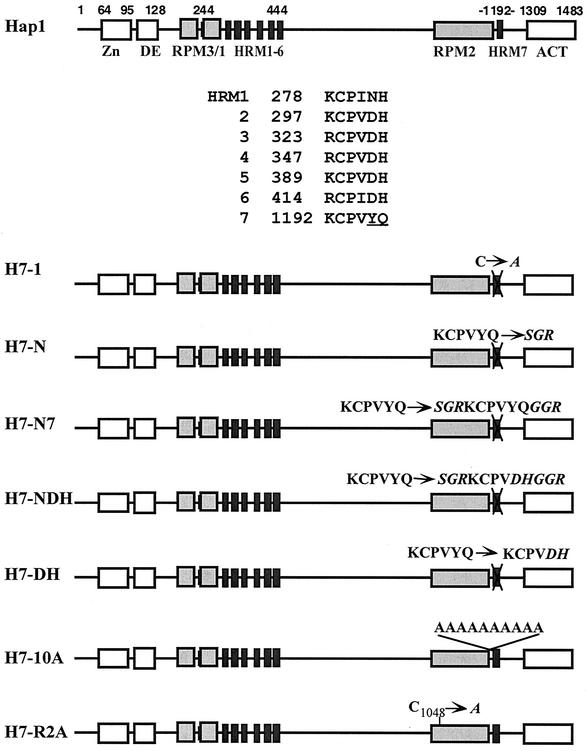
FIG. 1.
Hap1 domain structure and schematic diagrams of Hap1 mutants with mutations in or near HRM7. Shown are the C6 zinc cluster motif (Zn), the dimerization element (DE), three repression modules (RPM1 to 3), seven heme-responsive motifs (HRM1 to -6 and HRM7), and the activation domain (ACT). Also shown are the amino acid residues of HRM1 to -7. Mutant H7-1 contains a substitution of the Cys residues in HRM7 with Ala. Mutant H7-N contains a substitution of the HRM7 residues KCPVYQ with residues SGR; mutant H7-N7 contains residues SGRKCPVYQGGR in place of HRM7 residues KCPVYQ; mutant H7-NDH contains residues SGRKCPVDHGGR in place of HRM7 residues KCPVYQ; mutant H7-DH contains residues DH in place of residues YQ in HRM7; mutant H7-10A contains 10 Ala residues inserted in front of HRM7; mutant H7-R2A contains a substitution of the Cys residue at position 1048 in RPM2 with Ala.
Hap1 is a well-studied heme-regulatory protein. It contains seven HRMs (Fig. 1) (47, 51). Thus, Hap1 provides an excellent model for studying the functional role of HRMs and the molecular events underlying heme regulation. Previous studies showed that HRM7 of Hap1, located near the activation domain, plays a key role in heme activation of Hap1 in vivo, whereas HRM1 to -6, which are located near the DNA-binding domain, play only a minor, auxiliary role in heme activation (12, 15). It was further shown that the molecular chaperone Hsp90 plays a positive role in promoting heme activation of Hap1 (24).
Hsp90 plays critical roles in the proper functioning of diverse signal transducers, such as steroid receptors and protein kinases (26, 30-33). Our previous studies showed that Hsp90 plays a positive role in heme regulation of Hap1 (17, 24, 52). Hsp90 is critical for heme activation of Hap1 but does not appear to be important for Hap1 repression in the absence of heme (17, 24, 52). Deciphering the mode of Hsp90 action in heme activation of Hap1 may help to clarify Hsp90 action in the regulation of many other signal transducers. To understand the determinants that underlie the functional role of HRMs, we generated, for this report, a series of targeted mutations in HRM7 and its surrounding sequences. We analyzed the effects of these mutations on heme activation of Hap1 in vivo, on Hap1 DNA-binding in vitro, and on the interaction between Hap1 and the molecular chaperone Hsp90. Our results suggest that the structural environment of HRM7—not its mere presence—determines its functional role in heme activation. Likewise, a positive role of Hsp90 in Hap1 activation entails the presence of specific Hap1 structural states. The mere association of Hsp90 with Hap1 does not lead to the potentiation of Hap1 activation by heme. Together, our data suggest that Hsp90 interacts intimately with the HRM7 region of Hap1 to promote Hap1 conformational changes that are necessary for Hap1 activation, resulting in heme activation of Hap1-enhanced transcription of target genes.
MATERIALS AND METHODS
Yeast strains and reporters.
Saccharomyces cerevisiae strains used were MHY200 (MATaura3-52 leu2-3,112 his4-519 ade1-100 hap1::LEU2hem1-Δ100) (13) and JEL1 (MATα leu2 trp1 ura3-52 nprb1-1122 pep4-3 ΔHis3::pGAL10-GAL4) (52). The reporter used was the UAS1/CYC1-lacZ reporter described previously (42).
Construction of expression plasmids for Hap1 mutants.
We used a previously established site-directed mutagenesis method (12, 15) to construct mutants H7-N, H7-DH, H7-10A, H7-R2A, H7-dd1, H7-dd2, H7-dd3, H7-dd4, and H7-dd5. Briefly, the BglII-KpnI fragment encoding Hap1 residues 746 to 1309 from the HAP1 expression vector (SD5-HAP1) (42) was inserted into a vector derived from the Stratagene Bluescript II KS+ vector. Single-stranded uracil DNA was then generated in the dut ung bacterial strain CJ 236 (Bio-Rad). Oligonucleotides that encoded mutated amino acid residues or that lacked coding sequence for the deleted residues and that had 16 bases complementary to the region on both sides of the deleted or mutated sequence were annealed to the single-stranded uracil DNA. Double-stranded DNA was synthesized and transformed into the wild-type MV 1190 bacterial strain, and plasmid DNA was isolated from the transformants. Mutants were verified by restriction digestion and sequencing. Confirmed mutant constructs were cut with BglII and KpnI and inserted back into SD5-HAP1 (42). Mutants H7-N7 and H7-NDH were generated by inserting a double-stranded oligonucleotide that encoded the corresponding amino acid residues into the NotI site generated in mutant H7-N. Mutant smHap1 was generated by ligating a fragment that encoded residues 1000 to 1309 to the SD5-HAP1 expression vector cut with BamHI and KpnI. All mutants were confirmed by sequencing. Oligonucleotide sequences are available upon request.
β-Galactosidase assays.
To determine the β-galactosidase levels from the UAS1/CYC1-lacZ reporter gene (42) in cells expressing wild-type Hap1 and various mutants, SD5-HAP1 plasmids expressing wild-type Hap1 and mutants from the GAL1 promoter were transformed into the yeast strain MHY200 (13) bearing the UAS1/CYC1-lacZ reporter. Cells were grown in synthetic complete medium containing 2% raffinose, 1 μg of heme precursor per ml, and 5-aminolevulinic acid (mixture brought to A0.5), and the cells were then induced with 1% glucose plus 1% galactose (12) and designated concentrations of heme precursor, deuteroporphyrin IX (dpIX) (15). To achieve a low-to-intermediate level of Hap1 expression (which is comparable to the normal expression level from its own promoter), 1% glucose plus 1% galactose were used. After 7 h of induction, cells were collected and subjected to β-galactosidase assays as described previously (12, 15).
Preparation of yeast extracts and Western blotting.
Extracts were prepared according to previously established protocols (12, 15, 17, 52). Briefly, yeast JEL1 cells bearing various HAP1 expression plasmids were grown to A0.3 and induced with 2% galactose for about 6 h. Cells were harvested and resuspended in four packed cell volumes of buffer (20 mM Tris, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.3 M NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml). Cells were then permeabilized by agitation with three packed cell volumes of glass beads. Extracts were collected and centrifuged at 4°C at 13,000 rpm for 15 min in an Eppendorf 5417R centrifuge and then centrifuged at 55K in a Beckman Optima TLX centrifuge for 30 min to clear large aggregates. Protein concentrations were determined by Bradford assays (Bio-Rad).
For Western blotting, whole-cell extracts were first separated on 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF). Hap1 and Hsp90 were detected by using antibodies against GST-Hap1 (residues 1 to 171) (52) and part of Hsp90 (5, 6), respectively. The signals were revealed by using a chemiluminescence Western blotting kit (Boehringer Mannheim) as described previously (12, 15, 17, 52).
Electrophoretic mobility shift assays and DNA pull down.
DNA-binding reactions were carried out in a 20-μl volume with 5% glycerol, 4 mM Tris (pH 8), 40 mM NaCl, 4 mM MgCl2, 2 ng of heme per ml, 10 mM dithiothreitol, 3 μg of salmon sperm DNA, 10 μM ZnOAc2, and protease inhibitor cocktail (Boehringer Mannheim) as described previously (12, 15, 17, 52). Approximately 0.03 pmol of labeled DNA and 20 μg of protein extracts were used in each reaction. The reaction mixtures were incubated at 4°C for 1 h and then loaded onto 4% polyacrylamide gels in 1/3× Tris-borate-EDTA for gel electrophoresis at 4°C. The radioactive bands were visualized and quantified by using a PhosphorImager and the accompanying software from Molecular Dynamics.
For DNA pull down (17, 24), extracts prepared from cells expressing wild-type or mutant Hap1 were incubated with streptavidin-conjugated magnetic beads (DYNAL) prebound with the biotinylated DNA containing the high-affinity (CGGACTTATCGG) or mutated (CGGACTCATCCG) Hap1-binding site (16) under the conditions described above for electrophoretic mobility shift assays. The reaction mixture contained 0.5% NP-40. The beads were extensively washed and boiled in SDS gel loading buffer to release the bound proteins (36). Proteins were then analyzed by SDS-polyacrylamide gel electrophoresis, which was followed by Western blotting analysis. The intensity of proteins bands was measured by using NIH Image software.
RESULTS
Different mutations of HRM7 diminish heme activation of Hap1 to the same extent in vivo.
Previous data showed that HRM7 is critical for Hap1 activation, whereas HRM1 to -6 play only an auxiliary role in Hap1 activation (12, 15). Which characteristic of HRM7 accounts for the critical role of HRM7? HRM7 is unique, perhaps because of its physical location or because of its specific sequence (Fig. 1). The last two residues of HRM7 are Y and Q—instead of the consensus D and H— of HRM2 to -6 (Fig. 1). Previous in vitro studies that used a synthetic peptide showed that mutation of the Cys residue completely abolishes heme binding by an HRM, while mutation of other residues has a much less severe effect on its heme-binding affinity (49). The mutation of the Cys residue also greatly reduces heme activation of Hap1 in vivo (see H7-1, Fig. 1 and 2) (12, 15). This result is consistent with the idea that the Cys residue directly chelates iron in heme (49). However, in vivo, in the context of the whole protein and its interacting partners, certain residues may play a much greater role than that which can be inferred from heme-synthetic peptide interactions.
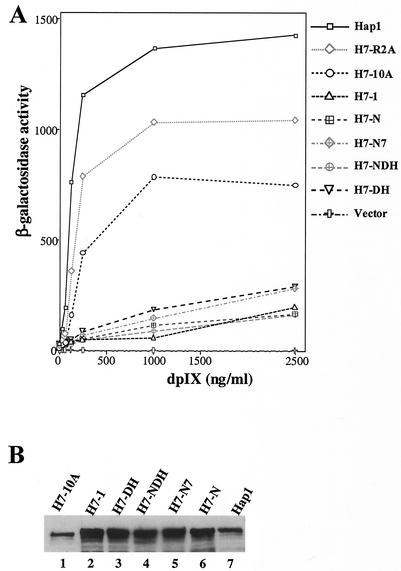
FIG. 2.
(A) The activities of wild-type Hap1 and various mutants at different concentrations of the heme analogue, deuteroporphyrin IX. Yeast cells bearing the UAS1/CYC1-lacZ reporter plasmid and the expression plasmid for wild-type Hap1 and mutants (Fig. 1) were grown to A0.5 and then induced with the indicated concentrations of dpIX for 7 h. The cells were subsequently collected, and β-galactosidase assays were performed. The data plotted here are averages of data from three experiments. The standard deviations are not shown, for clarity, but they were within 30%. (B) Western blot showing the protein levels of wild-type Hap1 and mutants in yeast cell extracts. Extracts were prepared from yeast cells expressing wild-type Hap1 (lane 7), H7-10A (lane 1), H7-1 (lane 2), H7-DH (lane 3), H7-NDH (lane 4), H7-N7 (lane 5), or H7-N (lane 6). Approximately 30 μg of total proteins from each extract was loaded onto 7.5% SDS-polyacrylamide gels, and the proteins were electrophoresed, transferred to PVDF membranes, and probed with an anti-Hap1 antibody (22). Hap1 proteins were visualized by using a chemiluminescence Western blotting kit (Boehringer Mannheim).
To ascertain the relative importance of residues of HRM7 in heme activation of Hap1 and to pinpoint the key factor that determines the critical role of HRM7, we generated a series of targeted mutations in and around HRM7 (Fig. 1). Initially, to make it easy for extensive mutagenesis of HRM7, we substituted a NotI restriction site for the HRM7 coding sequence in Hap1. As a result, we generated a mutant (H7-N, Fig. 1) in which the HRM7 residue KCPVYQ is replaced by residues SGR. As expected, this mutant, like H7-1, was severely defective in heme activation (Fig. 2A). The protein level of H7-N in extracts prepared from cells expressing H7-N was as high as that of H7-1 and wild-type Hap1 (Fig. 2B), thus showing that the defect in heme activation of H7-N was not caused by a reduced protein level. Next, we inserted the HRM7 residues KCPVYQ into the NotI site, thereby generating mutant H7-N7 containing HRM7 but with three additional surrounding residues on each side (Fig. 1). H7-N7 was still defective in heme activation (Fig. 2A) and was stably expressed (Fig. 2B), although it was slightly more activated than H7-N and H7-1 (Fig. 2A). These results show that insertion of HRM7 back into H7-N did not rescue the function of HRM7. We reasoned that this loss of HRM7 function may be attributable to changes in the structural environment of HRM7 that were caused by the introduction of the extra amino acid residues of the NotI site (Fig. 1).
To further ascertain the role of the specific residues YQ in HRM7 function (Fig. 1), we generated two additional mutants, H7-NDH and H7-DH, with the YQ residues replaced by the DH residues found in HRM2 to -6 at these positions (Fig. 1). H7-DH contains changes of YQ to DH residues, while H7-NDH also contains three additional residues at each side of the HRM (Fig. 1). Both mutants were defective in heme activation (Fig. 2A), even though both were stably expressed (Fig. 2B). In contrast, insertion of 10 Ala residues between RPM2 and HRM7 in mutant H7-10A and Ala substitution in mutant H7-R2A (Fig. 1) did not greatly reduce the extent of Hap1 activation by heme. H7-10A appeared to be as stable as wild-type Hap1 (Fig. 2B), while H7-R2A was more labile in extracts, as detected by Western blotting (data not shown). These results show that the biological function of HRM7 is selectively hypersensitive to certain kinds of—but not all—structural alterations.
To better understand the molecular basis underlying the defect of HRM7 mutants in heme activation, we examined the effects of increasing heme concentrations on the formation of Hap1-DNA complexes by using electrophoretic mobility shift assays (10, 12, 16, 17, 48, 52). In the absence of heme, Hap1 is bound by proteins, including Hsp90, Hsp70, and Ydj1, thereby forming a high-molecular-weight complex (HMC) that was detected by electrophoretic mobility shift assays and by chromatography (10, 16, 17, 48, 52). Heme disrupts the HMC and allows Hap1 to form a faster-migrating complex, designated the dimeric complex (DC) (Fig. 3) (10, 12, 16, 17, 48, 52). In response to increasing heme concentrations, the HMC gradually transforms into DC (Fig. 3) (10, 12, 16, 17, 48, 52). The HMC is associated with low DNA-binding and transcriptional activities, while DC is associated with high DNA-binding and transcriptional activities (16, 48).
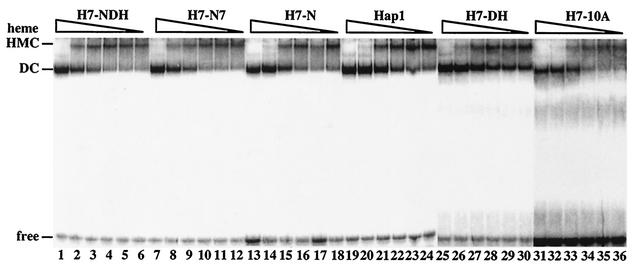
FIG. 3.
The effect of increasing heme concentrations on the transformation of mutant HMCs to DCs. Approximately 20 μg of protein extracted from cells expressing wild-type Hap1 (lanes 19 to 24), H7-NDH (lanes 1 to 6), H7-N7 (lanes 7 to 12), H7-N (lanes 13 to 18), H7-DH (lanes 25 to 30), or H7-10A (lanes 31 to 36) was incubated with radiolabeled DNA in the presence of 0 (lanes 6, 12, 18, 24, 30, and 36), 0.1 (lanes 5, 11, 17, 23, 29, and 35), 0.2 (lanes 4, 10, 16, 22, 28, and 34), 0.5 (lanes 3, 9, 15, 21, 27, and 33), 1 (lanes 2, 8, 14, 20, 26, and 32), or 2 (lanes 1, 7, 13, 19, 25, and 31) μg of heme per ml. The reaction mixtures were loaded onto 3.5% nondenaturing polyacrylamide gels and electrophoresed, and the bands were visualized by using a PhosphorImager. These experiments were repeated at least twice.
We therefore examined the effects of increasing heme concentrations on the transformation of various mutant HMCs to DCs (Fig. 3). Figure 3 shows that the levels of heme required to completely transform mutant HMCs to DCs were the same or slightly higher than that required to transform wild-type HMC to DC. The HMC formed by H7-NDH appeared to be most resistant to heme. The heme level (2 μg/ml) required for the transformation of mutant H7-NDH HMC (see lane 1, Fig. 3) is twofold higher than that required for the transformation of wild-type HMC (see lane 20, Fig. 3). These results suggest that HRM7 mutants did not cause drastic structural changes that broadly alter the nature of protein-protein interactions in the HMC. However, it is conceivable that HRM7 mutants may cause localized changes in Hap1 conformation and in Hap1-Hsp90 interaction.
Deletions of nonregulatory sequences of Hap1 render Hap1 heme inactivable.
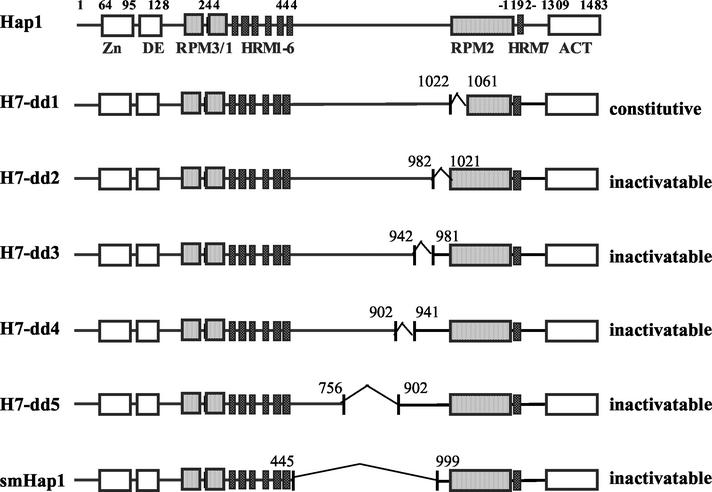
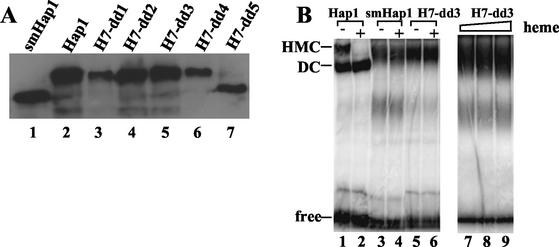
Previous studies identified two classes of Hap1 elements critical for heme regulation—repression modules (RPMs) (Fig. 1 and 4) and HRMs (Fig. 1 and 4) (12, 15). To better understand how these elements work together to promote heme regulation and how HRM7 promotes heme activation, we examined the effects of deletions of nonregulatory sequences between HRM1 to -6 and RPM2 on Hap1 activation. Deletion of residues 1022 to 1061 in mutant H7-dd1 (Fig. 4), like deletion of other residues in RPM2 (12), caused Hap1 to be constitutively active, even in heme-deficient cells. The activity of mutant H7-dd1, measured as β-galactosidase activity expressed from the UAS1/CYC1-lacZ reporter gene (42), was 399 ± 34 (Miller units) in heme-deficient cells in the absence of the heme analogue deuteroporphyrin IX (dpIX) and was 330 ± 20 (Miller units) in heme-sufficient cells in the presence of 2,500 ng of dpIX per ml. In contrast, wild-type Hap1 exhibited an activity of 34 ± 3 (Miller units) in heme-deficient cells and an activity of 1,217 ± 190 (Miller units) in heme-sufficient cells. The data suggest that residues 1022 to 1061 are part of RPM2. Strikingly, deletions of residues 982 to 1021 (mutant H7-dd2), 942 to 981 (mutant H7-dd3), 902 to 941 (mutant H7-dd4), 756 to 902 (mutant H7-dd5), and 445 to 999 (smHap1) all caused Hap1 to be inactivable. The UAS1/CYC1-lacZ reporter activity in cells expressing one of these mutants was as low as that in cells bearing the empty expression vector and remained unchanged by the addition of increasing concentrations of dpIX. Notably, all these mutant proteins were expressed at levels comparable to that of wild-type Hap1, as shown by Western blotting (Fig. 5A).
FIG. 4.
Hap1 domain structure and schematic diagrams of Hap1 mutants with deletions between HRM1 to -6 and RPM2. Shown are the C6 zinc cluster motif (Zn), the dimerization element (DE), three repression modules (RPM1 to -3), seven heme-responsive motifs (i.e., HRM1 to -6 and HRM7), and the activation domain (ACT). The deleted regions in mutants are marked. Residues 1022 to 1061, 982 to 1021, 942 to 981, 902 to 941, 756 to 902, and 445 to 999 are deleted in mutants H7-dd1, H7-dd2, H7-dd3, H7-dd4, H7-dd5, and smHap1, respectively. Mutant H7-dd1 was constitutively active—its activity in the presence of the heme analogue dpIX was as high as that in the absence of dpIX. All other mutants are inactivable—their activity was as low as the basal level activity and remained unchanged in cells grown in increasing concentrations of dpIX. Hap1 activity was measured as β-galactosidase activity expressed from the UAS1/CYC1-lacZ reporter gene.
FIG. 5.
(A) Western blot showing the protein levels of wild-type Hap1 and Hap1 mutants in yeast cell extracts. Extracts were prepared from yeast cells expressing wild-type Hap1 (lane 2), smHap1 (lane 1), H7-dd1 (lane 3), H7-dd2 (lane 4), H7-dd3 (lane 5), H7-dd4 (lane 6), or H7-dd5 (lane 7). Approximately 30 μg of total proteins from each extract was loaded onto 7.5% SDS-polyacrylamide gels, and the proteins were electrophoresed, transferred to PVDF membranes, and probed with an anti-Hap1 antibody (22). Hap1 proteins were visualized by using a chemiluminescence Western blotting kit (Boehringer Mannheim). (B) DNA-bound complexes formed by wild-type Hap1 and mutants. Approximately 20 μg of protein extracted from cells expressing wild-type Hap1 (lanes 1 and 2), smHap1 (lanes 3 and 4), or H7-dd3 (lanes 5 and 6) was incubated with radiolabeled DNA in the absence (lanes 1, 3, and 5) or presence of 2 (lanes 2, 4, 6, and 7), 5 (lane 8), or 10 (lane 9) μg of heme per ml. The reaction mixtures were loaded onto 3.5% nondenaturing polyacrylamide gels and electrophoresed, and the bands were visualized by using a PhosphorImager. These experiments were repeated at least twice.
We also examined the effect of heme on the DNA-Hap1 complexes formed by these mutants, although these mutant complexes are quite labile in cell extracts. Figure 5B shows complexes formed by two representative mutants—smHap1 and H7-dd3. In the absence of heme, these mutants formed a complex that is similar to the HMC (Fig. 5B, compare lanes 3 and 5 with lane 1). However, unlike wild-type HMC, this complex formed by smHap1 (Fig. 5B, lanes 3 and 4) and H7-dd3 (Fig. 5B, lanes 5 to 9) was not transformed to a faster-migrating DC-like complex, even at much higher levels of heme than that used to disrupt wild-type HMC (Fig. 3 also shows wild-type HMC; the lower band in lanes 3, 4, and 7 to 9 is due to smaller Hap1 fragments from degradation). Quantification of the bands (Fig. 5B, lanes 5, 6, and 7 to 9) showed that high levels of heme did not alter the intensity of the Hap1-DNA complexes; the variations in the intensity of the band were less than 10%. These results show that these mutants cannot be activated by heme in vitro or in vivo, although their HRMs, including the critical HRM7, are intact, suggesting that the biological function of HRMs is highly sensitive to their structural environment.
Hap1 mutants defective in heme activation enhance Hap1-Hsp90 association.
Previously, we showed that Hsp90 plays an indispensable role in heme activation of Hap1 (24, 52). To better understand how Hsp90 may act together with HRM7 to promote heme activation, we examined the interactions of the aforementioned Hap1 mutants defective in heme activation. To this end, we used a previously established DNA pull-down technique (17, 24). Previous studies on Hsp90 mutants have shown that DNA pull down is a more sensitive technique for detecting changes in Hap1-Hsp90 interaction than electrophoretic mobility shift assays (24). Furthermore, this technique is much more reliable and free of background than similar techniques, such as His6 pull down or antibody pull down, when used to examine Hap1-Hsp90 interaction (17, 24). Briefly, extracts prepared from cells expressing wild-type or mutant Hap1 were incubated with streptavidin-conjugated magnetic beads bound by biotinylated DNA containing an optimal, high-affinity Hap1-binding sequence (16, 17, 50) under conditions that allow formation of the HMC and maintenance of Hap1-Hsp90 interaction. Hap1 and associated Hsp90 were subsequently detected by Western blotting with antibodies against Hap1 and Hsp90, respectively. As a control, streptavidin-conjugated magnetic beads bound with a mutated Hap1-binding site, to which Hap1 binds with 20-fold lower affinity (16, 17, 50), were incubated with the extracts, and bound proteins were analyzed in parallel.
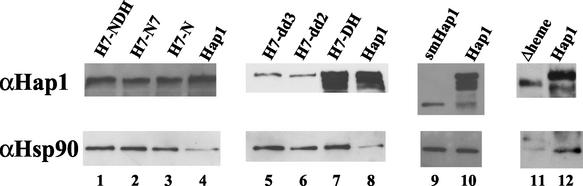
Figure 6 shows the results of DNA pull down when a high-affinity Hap1-binding site was used. When a mutated Hap1-binding site was used, Hap1 and Hsp90 signals were not detected under conditions in which strong signals were detected when the high-affinity Hap1-binding sequence was used. When the chemiluminescence signals were greatly enhanced, Hap1 and Hsp90 signals from the mutated Hap1-binding sequence were at least 20-fold weaker than those obtained from the optimal, high-affinity Hap1-binding sequence, as would be predicted from the Hap1-binding affinity of the mutated site (16, 17, 50). These results show that the Hsp90 signals detected were due to its association with Hap1.
FIG. 6.
Analysis of the interactions of Hap1 mutants with Hsp90. Extracts prepared from cells expressing wild-type or the indicated Hap1 mutant were incubated with streptavidin-conjugated magnetic beads (DYNAL) prebound with the biotinylated wild-type Hap1-binding site. The beads were extensively washed and boiled in SDS gel loading buffer to release the bound proteins (36). Hap1 and Hsp90 protein levels were detected by Western blotting. The membranes were first probed with an anti-Hap1 antibody and then stripped and probed with an anti-Hsp90 antibody. When a mutated Hap1-binding site was used in the pull-down assay, the amounts of bound Hap1 and Hsp90 proteins were undetectable or negligible (data not shown). These experiments were repeated at least twice.
The data show that similar levels of mutant H7-NDH (lane 1), H7-N7 (lane 2), H7-N (lane 3), and H7-DH (lane 7) pulled down at least threefold more Hsp90 than that pulled down by wild-type Hap1 (lanes 4 and 8). This suggests that H7-NDH, H7-N7, H7-N, and H7-DH had enhanced association with Hsp90. For mutants H7-dd2 (lane 6) and H7-dd3 (lane 5), although the levels of mutant proteins pulled down were at least eightfold less than that of wild-type Hap1 (lane 8), the levels of Hsp90 pulled down were fourfold more than that pulled down by wild-type Hap1. These results suggest that mutants H7-dd2 and H7-dd3 enhanced their association with Hsp90 by 30-fold. Similarly, about the same levels of Hsp90 were pulled down by 10-fold less mutant smHap1 (lane 9) compared to wild-type Hap1 (lane 10), suggesting that the association between smHap1 and Hsp90 was about 10-fold greater than the association between wild-type Hap1 and Hsp90. For comparison, we show that the previously described (52) constitutively active mutant Δheme (lane 11) greatly reduced its interaction with Hsp90 compared to wild-type Hap1 (lane 12). These results together demonstrate that mutants defective in heme activation, unlike Δheme, enhance their interaction with Hsp90.
DISCUSSION
HRMs play an essential role for heme regulation of many important proteins, including the mammalian transcriptional regulator Bach1 (27), the bacterial iron response regulator protein Irr (34), and the mammalian 5-aminolevulinic acid synthase (23). HRMs are short sequence motifs containing a critical Cys residue (Fig. 1) (49). The mode of heme binding by HRMs is simple and is well understood (34, 49). However, how heme binding to this motif causes drastic changes in the functions of various proteins is a much more complex process and remains unknown. Our studies, presented in this report, may provide important insights into how heme binding to HRMs causes changes in the activity of Hap1, how Hsp90 promotes heme activation of Hap1 and, analogously, how HRMs and Hsp90 may work to promote the functions of other signal transducers.
First, various mutations in HRM7, including the complete removal of HRM7 (mutant H7-N), insertion of certain residues surrounding HRM7 (mutant H7-N7), and the replacement of YQ residues with DH (mutants H7-DH and H7-NDH), all greatly reduced the extent of Hap1 activation by heme in vivo (Fig. 1 and 2). These mutants did not significantly affect the formation of DNA-Hap1 complexes and the transformation of the HMC to the DC (Fig. 3). However, DNA pull down, a more sensitive technique for detecting changes in Hap1-Hsp90 interaction (24), revealed that these mutants moderately enhanced Hap1-Hsp90 interaction (Fig. 6). These results suggest that the biological functions of HRM7 and Hsp90 are highly sensitive to structural or conformational states of Hap1. The mere presence of HRM7 or the mere association of Hap1 with Hsp90 does not necessarily lead to the proper functioning of Hsp90 and HRM7 and to the activation of Hap1 by heme. These results also suggest that HRM1 to -6, in the absence of HRM7, are able to partially substitute the role in heme binding and activation. Even when the HRM7 sequence is completely removed (H7-N), the mutant is able to bind heme and allow the transformation of the HMC to the DC in vitro (Fig. 3) and partial activation by heme in vivo (Fig. 2A). These results are consistent with the previous finding that HRM1 to -6 play an auxiliary role in heme binding and heme activation (12). In the presence of HRM7, HRM1 to -6 may play a role in maintaining Hap1 overall structure and in maintaining a low level of Hap1 activity, as suggested previously (15).
Second, deletions of sequences outside HRMs and heme regulatory regions abolished heme activation of Hap1 (Fig. 4). These deletion mutations had a much greater effect (Fig. 4) on Hap1 structure and function than those mutations in HRM7 (Fig. 1). These deletions likely affect the tertiary structure of Hap1. Evidently, the biological function of HRMs is greatly affected by changes in the tertiary structure of Hap1. Even in vitro, heme did not cause changes in the DNA-protein complexes formed by these deletion mutants (Fig. 5B). Notably, these deletion mutants also completely lost their transcriptional activity, even though they can still bind to DNA (Fig. 5B). Perhaps the altered, strong association of Hsp90 (and possibly other proteins) with Hap1 sterically blocked the activation domain, thus causing the complete loss of transcriptional activity.
The effects of Hap1 mutations on Hap1-Hsp90 interaction and on the formation of Hap1-DNA complexes are linked to the severity of their defect in heme activation. Mutations in or around HRM7 (Fig. 1 and 2) did not significantly alter the transformation of the HMC to the DC by heme in vitro (Fig. 3), moderately enhanced their interaction with Hsp90 (Fig. 6), and are partially, not completely, defective in heme activation (Fig. 2A). In contrast, internal deletions shown in Fig. 4 completely abolished the ability of heme to disrupt HMC-like Hap1-DNA complexes to smaller complexes (Fig. 5), greatly enhanced Hap1-Hsp90 interaction (Fig. 6), and rendered Hap1 completely inactivable. That HRM7 mutants (Fig. 1) cause localized structural changes in Hap1 that lead to moderately enhanced Hap1-Hsp90 interaction and partially defective heme activation is very likely, while deletion mutations (Fig. 4) cause broad structural changes in Hap1 that lead to greatly enhanced Hap1-Hsp90 interaction (Fig. 6) and complete abolition of heme activation (Fig. 5).
Notably, the strong association of Hap1 mutants with Hsp90 does not cause Hsp90 to act positively to promote Hap1 activation. Rather, preexisting structural changes caused by mutations in Hap1 mutants that enhance their association with Hsp90 likely inhibit or prevent further conformational changes that are necessary for Hap1 activation to occur. These results support the idea that Hap1 activation and the biological functions of HRM7 and Hsp90 entail a series of dynamic structural or conformational changes prior to and/or following heme binding. Such changes evidently occur at the levels of secondary, tertiary, and quaternary structure (association with Hsp90).
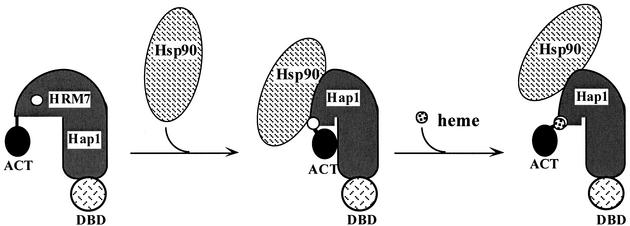
Based on these and previous results (17, 24, 52), we propose a model for how HRM7 and Hsp90 act to promote heme activation of Hap1 (Fig. 7). We postulate that in the absence of heme, Hsp90 binds to Hap1, thereby permitting Hap1 to adopt a conformation that can bind readily to heme and is activable. Upon heme binding to HRM7, Hsp90 and Hap1 likely act together to promote further conformational changes in Hap1, thus leading to the full activation of Hap1. Under such a model, defective heme activation by Hap1 mutants may be attributable to an inability to bind to heme or an inability to cause further structural changes following heme binding. In the case of HRM7 mutants (Fig. 1 and 2), it appears that these mutants largely retain their ability to bind heme, as shown by the transformation of their HMCs to DCs by heme (Fig. 3). This suggests that the defect of these mutants in heme activation is attributable to an inability to adopt a fully active Hap1 conformation following heme binding. This reasoning is also consistent with our previous studies suggesting that Hsp90 promotes heme activation of Hap1 by facilitating conformational changes following heme binding—not by facilitating heme binding (24). In the case of deletion mutants (Fig. 4), it appears that the deletions result in broad structural changes in Hap1 that may have caused it to completely lose its ability to bind and respond to heme. Evidently, these mutants are unable to adopt an active conformation, even if they are able to bind to heme.
FIG. 7.
Model illustrating the action of HRMs and Hsp90 in heme activation of Hap1. Hsp90 associates with Hap1 in the absence of heme. Based on previous studies (24) and data presented in this report, we postulate that the interaction of Hsp90 with Hap1 causes Hap1 to adopt a conformation that can readily bind to heme and is activable. Upon heme binding, Hsp90 acts with Hap1 to initiate further conformational or structural changes, thereby leading to full Hap1 activation. Mutations in Hap1 may cause Hap1 to lose its ability to adopt an activable conformation so that it cannot bind to heme or cannot induce further conformational changes necessary for activation following heme binding. For simplicity, other proteins, including Ssa, Ydj1, and Sro9, are omitted from this diagram.
Our results reveal two noteworthy, putative characteristics of Hsp90 as a modulator of diverse signal transducers, including steroid receptors, kinases, and hemoproteins. First, Hsp90 action in heme regulation of Hap1, and likely in the regulation of other signal transducers (26, 30-33), entails much more than simple association. The analyses of the mutants shown in Fig. 6 suggest that even a strong association of a substrate protein with Hsp90 does not indicate an active role of Hsp90 in the regulation of the substrate protein. Whether or not Hsp90 plays an active role in the regulation of a substrate appears to be dependent on the structural states of the substrate, not on the strength of the substrate-Hsp90 interaction. This idea provides an explanation for the previous finding that the proper functioning of retinoid receptors are dependent on Hsp90, although retinoid receptors have not been shown to interact with Hsp90 directly in vitro (14). Retinoid receptors likely interact loosely with Hsp90 in vivo.
Second, Hsp90 may readily switch between its role as a positive modulator and a negative modulator of signal transducers. In the case of Hap1, Hsp90 appears to actively promote heme activation of wild-type Hap1, while Hsp90 may actually repress mutant proteins, particularly the deletion mutants (Fig. 4). Such alterations in the nature of Hsp90 function may also occur when Hsp90 acts to promote the function of diverse signal transducers (26, 30-33). This idea is consistent with previous studies suggesting that Hsp90 plays a dual role in repressing steroid receptors in the absence of ligand and in promoting ligand binding and activation of the receptors (26, 30-33), while Hsp90 plays only a positive role in Hap1 activation by heme (24). Hsp90 has been shown to be critical for the actions of other heme-responsive or hemoproteins, including the translational regulator HRI and the endothelial and neuronal nitric oxide synthases (3, 11, 43-45). Similar modes of Hsp90 action in heme regulation of Hap1 may also operate to promote the functions of HRI, NOSs, and other signal transducers.
Acknowledgments
We are grateful to S. L. Lindquist for providing antibody against Hsp90.
This work was supported by funds from NIH (GM62246 to L.Z.). L.Z. is a Monique Weill-Caulier Scholar. We thank W. Jelinek for critical reading of the manuscript.
REFERENCES
- 1.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, B. N., M. Rafie-Kolpin, L. Lu, A. Han, and J. J. Chen. 2001. Multiple autophosphorylation is essential for the formation of the active and stable homodimer of heme-regulated eIF2α kinase. Biochemistry 40:11543-11551. [DOI] [PubMed] [Google Scholar]
- 3.Bender, A. T., A. M. Silverstein, D. R. Demady, K. C. Kanelakis, S. Noguchi, W. B. Pratt, and Y. Osawa. 1999. Neuronal nitric-oxide synthase is regulated by the Hsp90-based chaperone system in vivo. J. Biol. Chem. 274:1472-1478. [DOI] [PubMed] [Google Scholar]
- 4.Bock, K. W., F. De Matteis, and W. N. Aldridge. 1978. Heme and hemoproteins. Springer-Verlag, New York, N.Y.
- 5.Chang, H. C., and S. Lindquist. 1994. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269:24983-24988. [PubMed] [Google Scholar]
- 6.Chang, H. C., D. F. Nathan, and S. Lindquist. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. J., J. S. Crosby, and I. M. London. 1994. Regulation of heme-regulated eIF-2α kinase and its expression in erythroid cells. Biochimie (Paris) 76:761-769. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. J., and I. M. London. 1995. Regulation of protein synthesis by heme-regulated eIF-2α kinase. Trends Biochem. Sci. 20:105-108. [DOI] [PubMed] [Google Scholar]
- 9.Creusot, F., J. Verdiere, M. Gaisne, and P. P. Slonimski. 1988. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol. 204:263-276. [DOI] [PubMed] [Google Scholar]
- 10.Fytlovich, S., M. Gervais, C. Agrimonti, and B. Guiard. 1993. Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 12:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Cardeña, G., R. Fan, V. Shah, R. Sorrentino, G. Cirino, A. Papapetropoulos, and W. C. Sessa. 1998. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature 392:821-824. [DOI] [PubMed] [Google Scholar]
- 12.Hach, A., T. Hon, and L. Zhang. 1999. A new class of repression modules is critical for heme regulation of the yeast transcriptional activator HapI. Mol. Cell. Biol. 19:4324-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haldi, M. L., and L. Guarente. 1995. Multiple domains mediate heme control of the yeast activator HAP1. Mol. Gen. Genet. 248:229-235. [DOI] [PubMed] [Google Scholar]
- 14.Holley, S. J., and K. R. Yamamoto. 1995. A role for Hsp90 in retinoid receptor signal transduction. Mol. Biol. Cell 6:1833-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hon, T., A. Hach, H. C. Lee, T. Chen, and L. Zhang. 2000. Functional analysis of heme regulatory elements of the transcriptional activator Hap1. Biochem. Biophys. Res. Commun. 273:584-591. [DOI] [PubMed] [Google Scholar]
- 16.Hon, T., A. Hach, D. Tamalis, Y. Zhu, and L. Zhang. 1999. The yeast heme-responsive transcriptional activator Hap1 is a preexisting dimer in the absence of heme. J. Biol. Chem. 274:22770-22774. [DOI] [PubMed] [Google Scholar]
- 17.Hon, T., H. C. Lee, A. Hach, J. L. Johnson, E. A. Craig, H. Erdjument-Bromage, P. Tempst, and L. Zhang. 2001. The Hsp70-Ydj1 molecular chaperone represses the activity of the transcriptional activator Hap1 in the absence of heme. Mol. Cell. Biol. 21:7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igarashi, K., H. Hoshino, A. Muto, N. Suwabe, S. Nishikawa, H. Nakauchi, and M. Yamamoto. 1998. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 273:11783-11790. [DOI] [PubMed] [Google Scholar]
- 19.Inamdar, N. M., Y. I. Ahn, and J. Alam. 1996. The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem. Biophys. Res. Commun. 221:570-576. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. [DOI] [PubMed] [Google Scholar]
- 21.Kotkow, K. J., and S. H. Orkin. 1995. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol. 15:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer, M. F., P. Gunaratne, and G. C. Ferreira. 2000. Transcriptional regulation of the murine erythroid-specific 5-aminolevulinate synthase gene. Gene 247:153-166. [DOI] [PubMed] [Google Scholar]
- 23.Lathrop, J. T., and M. P. Timko. 1993. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259:522-525. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H. C., T. Hon, and L. Zhang. 2002. The Hsp90 molecular chaperone mediates heme activation of the yeast transcriptional activator Hap1. J. Biol. Chem. 277:7430-7437. [DOI] [PubMed] [Google Scholar]
- 25.Marziali, G., E. Perrotti, R. Ilari, U. Testa, E. M. Coccia, and A. Battistini. 1997. Transcriptional regulation of the ferritin heavy-chain gene: the activity of the CCAAT binding factor NF-Y is modulated in heme-treated Friend leukemia cells and during monocyte-to-macrophage differentiation. Mol. Cell. Biol. 17:1387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer, M. P., and B. Bukau. 1999. Molecular chaperones: the busy life of Hsp90. Curr. Biol. 9:R322-R325. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa, K., J. Sun, S. Taketani, O. Nakajima, C. Nishitani, S. Sassa, N. Hayashi, M. Yamamoto, S. Shibahara, H. Fujita, and K. Igarashi. 2001. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padmanaban, G., V. Venkateswar, and P. N. Rangarajan. 1989. Haem as a multifunctional regulator. Trends Biochem. Sci. 14:492-496. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer, K., K. S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56:291-301. [DOI] [PubMed] [Google Scholar]
- 30.Picard, D. 1998. The role of heat-shock protein in the regulation of steroid receptor function, p. 1-18. In L. P. Freedman (ed.), Molecular biology of steroid and nuclear hormone receptors. Birkhauser, Boston, Mass.
- 31.Pratt, W. 1997. The role of the hsp90-based chaperone system in signal transduction by nuclear receptors and receptors signaling via MAP kinase. Annu. Rev. Pharmacol. Toxicol. 37:297-326. [DOI] [PubMed] [Google Scholar]
- 32.Pratt, W., and D. Toft. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18:306-360. [DOI] [PubMed] [Google Scholar]
- 33.Pratt, W. B. 1998. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc. Soc. Exp. Biol. Med. 217:420-434. [DOI] [PubMed] [Google Scholar]
- 34.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 96:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rafie-Kolpin, M., P. J. Chefalo, Z. Hussain, J. Hahn, S. Uma, R. L. Matts, and J. J. Chen. 2000. Two heme-binding domains of heme-regulated eukaryotic initiation factor-2α kinase. N terminus and kinase insertion. J. Biol. Chem. 275:5171-5178. [DOI] [PubMed] [Google Scholar]
- 36.Reddy, S. V., O. Alcantara, and D. H. Boldt. 1998. Analysis of DNA binding proteins associated with hemin-induced transcriptional inhibition. The hemin response element binding protein is a heterogeneous complex that includes the Ku protein. Blood 91:1793-1801. [PubMed] [Google Scholar]
- 37.Reddy, S. V., O. Alcantara, G. D. Roodman, and D. H. Boldt. 1996. Inhibition of tartrate-resistant acid phosphatase gene expression by hemin and protoporphyrin IX: identification of a hemin-responsive inhibitor of transcription. Blood 88:2288-2297. [PubMed] [Google Scholar]
- 38.Sassa, S. 1996. Novel effects of heme and heme-related compounds in biological systems. Curr. Med. Chem. 3:273-290. [Google Scholar]
- 39.Sassa, S., and T. Nagai. 1996. The role of heme in gene expression. Int. J. Hematol. 63:167-178. [DOI] [PubMed] [Google Scholar]
- 40.Steiner, H., G. Kispal, A. Zollner, A. Haid, W. Neupert, and R. Lill. 1996. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J. Biol. Chem. 271:32605-32611. [DOI] [PubMed] [Google Scholar]
- 41.Talbot, D., and F. Grosveld. 1991. The 5′ HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 10:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turcotte, B., and L. Guarente. 1992. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 6:2001-2009. [DOI] [PubMed] [Google Scholar]
- 43.Uma, S., D. J. Barret, and R. L. Matts. 1998. Changes in the expression of the heme-regulated eIF-2α kinase and heat shock proteins in rabbit reticulocytes maturing during recovery from anemia. Exp. Cell Res. 238:273-282. [DOI] [PubMed] [Google Scholar]
- 44.Uma, S., S. D. Hartson, J. J. Chen, and R. L. Matts. 1997. Hsp90 is obligatory for the heme-regulated eIF-2α kinase to acquire and maintain an activable conformation. J. Biol. Chem. 272:11648-11656. [DOI] [PubMed] [Google Scholar]
- 45.Uma, S., V. Thulasiraman, and R. L. Matts. 1999. Dual role for Hsc70 in the biogenesis and regulation of the heme-regulated kinase of the α subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 19:5861-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, C., F. Tokumasu, K. I. Hohmura, J. Bungert, N. Hayashi, T. Nagasawa, J. D. Engel, M. Yamamoto, K. Takeyasu, and K. Igarashi. 1999. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4:643-655. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., and L. Guarente. 1996. The C6 zinc cluster dictates asymmetric binding by HAP1. EMBO J. 15:4676-4681. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L., and L. Guarente. 1994. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J. Biol. Chem. 269:14643-14647. [PubMed] [Google Scholar]
- 49.Zhang, L., and L. Guarente. 1995. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, L., and L. Guarente. 1994. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 8:2110-2119. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., and A. Hach. 1999. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol. Life Sci. 56:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, L., A. Hach, and C. Wang. 1998. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol. 18:3819-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, Y., T. Hon, W. Z. Ye, and L. Zhang. 2002. Heme deficiency selectively interferes with the Ras-MAPK signaling pathway and the expression of a subset of neuronal genes. Cell Growth Differ. 13:431-439. [PubMed] [Google Scholar]
- 54.Zhu, Y., H. C. Lee, and L. Zhang. 2002. An examination of heme action in gene expression: heme and heme deficiency affect the expression of diverse genes in erythroid K562 and neuronal PC12 cells. DNA Cell Biol. 21:333-346. [DOI] [PubMed] [Google Scholar]