Abstract
A significant advancement in understanding mitochondrial gene expression is the recent identification of two new human mitochondrial transcription factors, h-mtTFB1 and h-mtTFB2. Both proteins stimulate transcription in collaboration with the high-mobility group box transcription factor, h-mtTFA, and are homologous to rRNA methyltransferases. In fact, the dual-function nature of h-mtTFB1 was recently demonstrated by its ability to methylate a conserved rRNA substrate. Here, we demonstrate that h-mtTFB1 binds h-mtTFA both in HeLa cell mitochondrial extracts and in direct-binding assays via an interaction that requires the C-terminal tail of h-mtTFA, a region necessary for transcriptional activation. In addition, point mutations in conserved methyltransferase motifs of h-mtTFB1 revealed that it stimulates transcription in vitro independently of S-adenosylmethionine binding and rRNA methyltransferase activity. Furthermore, one mutation (G65A) eliminated the ability of h-mtTFB1 to bind DNA yet did not affect transcriptional activation. These results, coupled with the observation that h-mtTFB1 and human mitochondrial RNA (h-mtRNA) polymerase can also be coimmunoprecipitated, lead us to propose a model in which h-mtTFA demarcates mitochondrial promoter locations and where h-mtTFB proteins bridge an interaction between the C-terminal tail of h-mtTFA and mtRNA polymerase to facilitate specific initiation of transcription. Altogether, these data provide important new insight into the mechanism of transcription initiation in human mitochondria and indicate that the dual functions of h-mtTFB1 can be separated.
Expression of the human mitochondrial genome is initiated with transcription at promoters in the D-loop regulatory region of the mitochondrial DNA (mtDNA) molecule (28). Mitochondrial transcription requires a relatively small set of organelle-dedicated regulatory proteins, which are encoded by nuclear genes and imported into the mitochondrial matrix, where they orchestrate the initial stages of mitochondrial gene expression (26). The basic transcription machinery consists of a single-subunit, mitochondrial RNA (mtRNA) polymerase molecule that is homologous to the bacteriophage T7 family of RNA polymerases (18, 30), the DNA-binding transcription factor h-mtTFA (10), and two related transcription factors, h-mtTFB1 (19) and h-mtTFB2 (9). In the presence of human mtRNA (h-mtRNA) polymerase, h-mtTFB1 and h-mtTFB2 can each activate transcription from the human light-strand promoter (LSP) and heavy-strand promoter (HSP) in vitro in a manner that requires h-mtTFA (9, 19). These data indicate that transcription initiation requires critical interactions between these basic protein components and the promoter DNA; however, the mechanism underlying transcriptional initiation has yet to be determined in this system. An understanding of the mechanism of mitochondrial gene expression is required to uncover, and perhaps develop methods to counteract, the deleterious effects of loss of mitochondrial function encountered in numerous disease states and the aging process (8, 29, 32).
In a series of seminal experiments, Clayton and colleagues defined two essential components required for transcription initiation in human mitochondria, an h-mtRNA polymerase activity and a DNA-binding protein that engages the LSP and HSP directly upstream of the initiation site to stimulate transcription (10, 11, 13). The ability of this protein, now known as h-mtTFA, to activate transcription is presumably linked in some manner to its ability to bend and/or contort the DNA near the initiation site (12). The sequence and spacing of the h-mtTFA binding site relative to the transcription initiation site are also critical (6, 31), suggesting that h-mtTFA must be in proximity to the incoming mtRNA polymerase during initiation. Cloning of the cDNA encoding h-mtTFA revealed that it contains a mitochondrial matrix-localization signal at its amino terminus, two high-mobility group (HMG) box DNA-binding domains that are separated by a short linker region, and a C-terminal tail that is relatively rich in basic amino acids (21). Mutagenesis and domain-swapping experiments subsequently revealed that, while the HMG boxes are required for nonspecific DNA binding properties, the C-terminal tail is absolutely required for specific DNA binding and transcriptional activation (7), indicating that h-mtTFA may make contacts with other factors involved in initiation through this domain.
Studies of Saccharomyces cerevisiae (15, 16, 27, 33) and Xenopus laevis (1, 2) model systems revealed the requirement for a mitochondrial transcription factor (mtTFB) that is distinct from mtTFA, suggesting the existence of a second mitochondrial transcription factor in vertebrates. This prediction was recently realized with the cloning of a cDNA encoding the first human homolog of yeast mtTFB (sc-mtTFB/Mtf1p), which also revealed that this transcription factor is related in primary sequence to a large family of RNA methyltransferases and binds the requisite S-adenosylmethionine (SAM) cofactor used by this class of enzymes (19). Recently, we reported that h-mtTFB1 can indeed methylate tandem adenine residues located within a conserved stem-loop in the small rRNA subunit of ribosomes, demonstrating that it is likely a dual-function protein, acting as both an rRNA methyltransferase and a transcription factor (24). Consistent with these findings is the three-dimensional structure of sc-mtTFB, which reveals striking similarity to the bacterial ErmC′ rRNA methyltransferase (23). Isolation of a second human homolog of sc-mtTFB, h-mtTFB2, revealed that it is also related to rRNA methyltransferases and possesses marked transcriptional activation properties in vitro (9).
How the RNA methyltransferase structural features and/or enzymatic activity of h-mtTFB1 impinges on the function of the protein as a transcription factor is unknown. In particular, whether SAM binding and/or RNA methyltransferase activity is associated with the transcriptional activation properties of h-mtTFB1 has not been addressed. Here we report that h-mtTFB1 interacts directly with h-mtTFA and, despite the fact that this protein is also a SAM-dependent rRNA methyltransferase (24), can activate transcription independently of SAM binding and RNA methyltransferase activity. These data support a new model for transcription initiation in human mitochondria in which h-mtTFB proteins act to bridge an interaction between h-mtRNA polymerase and a promoter-bound h-mtTFA complex.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
The plasmid pGST-CGI75 used to express wild-type and mutant forms of h-mtTFB1 in Escherichia coli is a derivative of pGEX4T (Promega, Inc.) and has been described previously (19). Site-directed mutagenesis was accomplished using the following PCR-based protocol. Pairs of overlapping oligonucleotides that contained the desired nucleotide changes were added together with pGEMT plasmid (Promega, Inc.) harboring the h-mtTFB1 open reading frame (ORF) to a PCR that amplified the entire plasmid sequence (PCR conditions were optimized for each oligonucleotide pair). After confirmation that PCR was successful, a sample of the PCR mix was digested to completion with DpnI to restrict the methylated parental plasmid DNA containing the wild-type sequence, leaving only the unmethylated amplified DNA containing the desired point mutations. The DpnI-digested sample was then used to transform E. coli, which enabled retrieval of the corresponding mutated pGEMT plasmid. The mutagenic oligonucleotides used were as follows: G65A mutation, oligonucleotide 1, 5′-GTTTACGAAGTGGCCCCTGGGCCAGGGG-3′, and oligonucleotide 2, 5′-CCCCTGGCCCAGGGGCCACTTCGTAAAC-3′; N141A mutation, oligonucleotide 1, 5′-GTACATATTATTGGAGCTCTGCCTTTTAGTGTTTCAACTCC-3′, and oligonucleotide 2, 5′-GGAGTTGAAACACTAAAAGGCAGAGCTCCAATAATATGTAC-3′; K220Α mutation, oligonucleotide 1, 5′-CAAGCTTTTGTCCCCGCGCCAGAGGTGGACGTG-3′, and oligonucleotide 2, 5′-CACGTCCACCTCTGGCGCGGGGACAAAAGCTTG-3′. Site-directed mutations were confirmed by sequencing the isolated plasmids. Once confirmed, the mutant alleles were subcloned into the plasmid pGEX-CGI75 on a BamHI-NotI restriction fragment, allowing expression of the corresponding glutathione S-transferase (GST)::h-mtTFB1 fusion proteins in E. coli.
The h-mtTFB2 ORF was isolated by PCR amplification of a human fetal brain cDNA library with the primers 5′-ATGTGGATCCCAGTGGTCGGGCTTCC-3′ and 5′-CTACCTATCTTCCAGGGTTTCATC-3′ and cloned into the plasmid pGEMT to create the plasmid pGEMT-HB2. This plasmid was subsequently used as a template for PCR with oligonucleotides 5′-TCCCCCCGGGTCATGTGGATCCCAGTGGTCGGG-3′ and 5′-ACGCGTCGACCTACCTATCTTCCAGGGTTTC-3′, which introduced a SmaI site at the 5′ end and a SalI site at the 3′ end to facilitate cloning of the h-mtTFB2 ORF into pGEX4T. The resulting plasmid (pGST-HB2) directs expression of the ORF as a fusion protein with GST in E. coli. T4 DNA ligase and restriction enzymes were obtained from either New England Biolabs or Promega.
Purification of recombinant proteins.
GST-tagged and untagged (thrombin-cleaved) h-mtTFB1 and h-mtTFB2 and the h-mtTFB1 site-directed variants were each produced in bacteria and purified by glutathione-Sepharose chromatography as described previously (19). Full-length and C-terminally truncated recombinant h-mtTFA proteins were expressed in E. coli, gel purified, and renatured as described previously (7).
Antibody production.
GST-tagged h-mtTFB1 was produced in bacteria, purified by glutathione-Sepharose chromatography, and cleaved with thrombin as previously described (19). The recombinant, untagged protein was resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis after which the gel was stained with 0.25 M KCl and the h-mtTFB1 protein was excised and eluted into dilution buffer (0.1% SDS, 50 mM Tris [pH 7.9], 0.1 mM EDTA, 150 mM NaCl, 5 mM dithiothreitol) as described (14). The eluted protein was sent to Rockland Immunochemicals for the production of rabbit polyclonal antibodies. Antibodies were affinity purified from the provided rabbit crude serum by protein A-Sepharose (Amersham Pharmacia Biotech, Inc.) chromatography. The protein A-purified antibody was then affinity purified on an Aminolink column (Pierce, Inc.) that had recombinant untagged h-mtTFB1 cross-linked to it as described by the manufacturer.
Immunodepletion of mitochondrial extracts and mitochondrial transcription assays.
Mitochondrial protein extracts were prepared as described previously (20) from ∼3 × 109 HeLa cells grown in Joklik's modified minimal essential medium (Sigma Chemical, Inc.) plus 10% fetal calf serum. Protein A-purified h-mtTFB1 antibodies or the corresponding preimmune serum was bound to protein A-Sepharose in mitochondrial lysis buffer (MLB; 25 mM HEPES-KOH [pH 7.6], 5 mM MgCl2, 10% glycerol, 0.125% Tween 20, 0.125 M KCl, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride). Unbound antibodies were removed by sequential washes with MLB after incubation. Mitochondrial extracts were diluted 1:4 with MLB to a final volume of 200 μl and incubated with 50 μl of antibody-bound beads for 1 h at 4°C with rotation. The beads were removed by centrifugation at 2,000 × g for 2 min at room temperature. The singly depleted extract was subjected to a second depletion with a new batch of beads under conditions identical to those for the first depletion. The resulting h-mtTFB1 immunodepleted extract was used in the transcription assays and for Western blot analysis (Fig. 1).
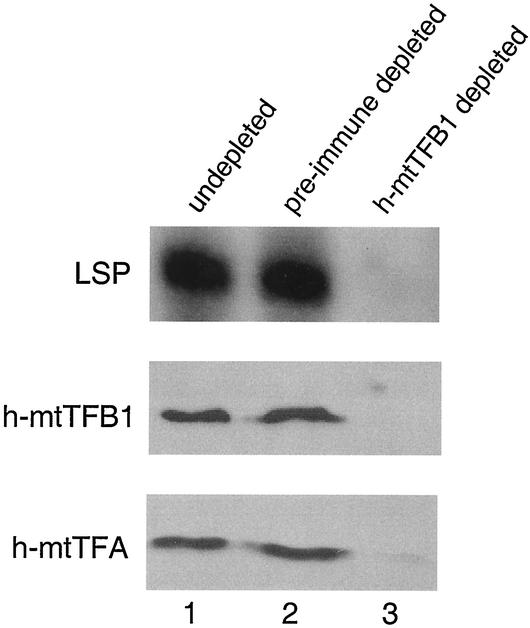
FIG. 1.
Coimmunoprecipitation of h-mtTFB1 and h-mtTFA from a transcriptionally active mitochondrial lysate from HeLa cells. A soluble HeLa cell mitochondrial extract was prepared that efficiently initiated transcription from a linear DNA template containing the human LSP (top panel). The lysate (undepleted, lane 1) produced a specific radiolabeled runoff transcript (labeled LSP) that was visualized by autoradiography. Transcription activity was also assayed from lysates that were immunodepleted with affinity-purified h-mtTFB1 antibody (h-mtTFB1 depleted, lane 3) or a control affinity-purified preimmune serum (pre-immune depleted, lane 2). Western immunoblot analysis of the corresponding extracts with an antibody to h-mtTFB1 (middle panel) or h-mtTFA (bottom panel) is also shown.
All transcription reactions were performed using a linearized mtDNA template containing the human LSP as described previously (19). Each extract was first tested empirically for dependence on addition of recombinant h-mtTFA protein to achieve a minimal amount of specific transcription initiation and then subsequently for stimulation of this level by addition of recombinant wild-type h-mtTFB1. This procedure was carried out for each extract and preparation of recombinant h-mtTFA before the effects of the h-mtTFB1 mutants were assessed. In all of the transcription assays, additions of identical amounts (50 or 250 ng) of wild-type and mutant h-mtTFB1 were always used for comparison.
The conditions used for the detection of h-mtRNA polymerase by coimmunoprecipitation with h-mtTFB1 were similar to those used above, except that the soluble HeLa cell mitochondrial extracts were diluted 1:8 in MLB to a final concentration of 62.5 mM KCl and 0.06% Tween 20. The extract was then precleared by incubation with rabbit preimmune serum, which was bound to protein A-Sepharose beads (Amersham Pharmacia) at 4°C for 1 h with rotation. The extract was then centrifuged at 3,000 × g for 2 min to pellet the beads, and the supernatant was transferred to new 1.5-ml tubes. A portion of this precleared soluble mitochondrial protein extract (75 μg of total protein) was incubated with protein A-Sepharose bound to either rabbit preimmune serum or anti-h-mtTFB1 antibody at 4°C for 2 h with rotation. The preimmune serum and h-mtTFB1 antibody used at this step in the experiment were purified over a protein A-Sepharose column and then cross-linked to the protein A-Sepharose beads with dimethyl pimelimidate (Pierce) according to the manufacturer's instructions. The beads were then centrifuged at 3,000 × g for 2 min, and the supernatant was removed. The beads were then washed five times with 1 ml of MLB (with 62.5 mM KCl and 0.0625% Tween 20). Finally, an equal volume of 2× Laemmli gel loading buffer was added to the beads. The eluted proteins corresponding to the immunoprecipitate were separated by electrophoresis on an 8% polyacrylamide gel and subjected to Western analysis with an anti-h-mtRNA polymerase peptide antibody (25).
Direct protein-protein interaction assays.
Recombinant wild-type or mutant GST::h-mtTFB1 fusion proteins were isolated from soluble crude E. coli extracts and bound to glutathione-Sepharose beads as described previously (19). The protein-bound beads were then incubated with full-length protein or each of the C-terminal deletion mutants of h-mtTFA in MLB with rocking at 4°C for 2 h. The samples were then washed five times with 1 ml of MLB. After washing, an equal volume of gel loading buffer was added to the beads and the eluted proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with an antibody directed against h-mtTFA. When the h-mtTFB1 mutants were tested for h-mtTFA binding (see Fig. 5C), the amounts of each h-mtTFB1 variant bound to the beads were checked by Western blotting to ensure that similar amounts of protein were analyzed.
FIG. 5.
Effects of h-mtTFB1 RNA methyltransferase motif point mutations on DNA binding, h-mtTFA binding, and transcriptional stimulation from the human LSP. (A) Electrophoretic mobility shift assays. Recombinant wild-type GST::h-mtTFB1 (B1, lanes 1 and 2) and the indicated point mutants (G65A, lanes 3 and 4; N141A, lanes 5 and 6; and K220A, lanes 7 and 8) were tested for their ability to bind to a radiolabeled linear DNA fragment containing the human LSP in the presence (+; lanes 2, 4, 6, and 8) or absence (−; lanes 1, 3, 5, and 7) of poly(dI-dC) competitor DNA. A shift in mobility of the LSP-containing fragment is indicated by the arrowhead. The position of migration of the unbound, end-labeled probe is shown by the diamond, and its migration in the absence of added protein is shown in lane 9. A slower-migrating band (indicated by the arrow) is commonly observed with this probe under these conditions. The physical nature of this species that causes its altered mobility is unknown. (B) Transcription assays. Wild-type GST::h-mtTFB1 (B1) and the indicated mutated proteins (G65A, N141A, and K220A) were tested in a transcription factor-dependent transcription assay that measures specific initiation from the LSP. The assay results in the production of a specific radiolabeled transcript from the LSP (arrowhead). A human HeLa cell mitochondrial extract deficient in specific LSP activity was used as a source of human mtRNA polymerase (assayed in lane 1). The addition of recombinant GST::h-mtTFB1 alone and recombinant h-mtTFA alone to the extract is indicated in lanes 2 and 3, respectively. The ability of GST::h-mtTFB1 (B1, lanes 4 and 5) and the indicated mutants (lanes 6 to 11) to stimulate LSP transcription was assessed in the presence of h-mtTFA (indicated by the bracket labeled +h-mtTFA). Two amounts (50 ng, lanes 4, 6, 8, and 10, and 250 ng, lanes 5, 7, 9, and 11) of each h-mTFB1 protein were tested. (C) h-mtTFA binding assays. The indicated amount of recombinant h-mtTFA (input h-mtTFA) was incubated with equal amounts of GST::mtTFB1 (B1) or the indicated mutated proteins bound to glutathione-agarose beads. The amount of h-mtTFA bound (bound h-mtTFA) by each was assessed by Western blotting with an antibody to h-mtTFA.
Other assays.
The solid-phase SAM-binding assay was performed as described previously (19). Electrophoretic mobility shift assays were performed essentially as described previously (19), except that a final concentration of 10 nM end-labeled LSP-containing DNA fragment was incubated with 250 ng of recombinant wild-type or mutant GST::h-mtTFB1. Where indicated 45 nM poly(dI-dC) (Amersham Pharmacia Biotech, Inc.) was also added to the reaction mixture.
RESULTS
Antibodies to h-mtTFB1 coimmunoprecipitate h-mtTFB1 and h-mtTFA from a transcription-competent soluble mitochondrial extract.
A rabbit polyclonal antibody was generated against recombinant h-mtTFB1 and affinity purified (see Materials and Methods). This preparation was used to immunodeplete a soluble mitochondrial extract derived from HeLa cells that was capable of supporting robust transcription from the human mitochondrial LSP in vitro (Fig. 1, lane 1). Depletion with affinity-purified preimmune serum did not result in any loss of transcription activity compared to an untreated control extract (Fig. 1, lane 2), while treatment with the h-mtTFB1 antibody resulted in complete inhibition of transcription (Fig. 1, lane 3). Western analysis revealed that h-mtTFB1 was in fact largely, if not completely, depleted from the extract in this experiment (Fig. 1, lane 3). These results are entirely consistent with our previously published data showing that h-mtTFB1 functions in mitochondrial transcription (19). However, repeated attempts to reconstitute transcription activity by adding recombinant h-mtTFB1 back to the depleted extract were unsuccessful (data not shown), suggesting that other required factors were being coimmunoprecipitated along with h-mtTFB1 in this experiment.
We reported previously that h-mtTFB1 activates transcription only in collaboration with h-mtTFA (19), suggesting to us that loss of transcription in the h-mtTFB1 immunodepletion experiment described above could be explained by coimmunoprecipitation of h-mtTFA from the extract. Indeed, Western analysis revealed that treatment with h-mtTFB1 antibody also resulted in severe depletion of h-mtTFA from the extract that was not observed in the preimmune control depletion (Fig. 1, compare lane 3 to lane 2). Thus, h-mtTFB1 and h-mtTFA appear to be present in a complex in this transcriptionally active mitochondrial extract.
h-mtTFB1 and h-mtTFB2 interact with the C-terminal activation region of h-mtTFA.
Given that h-mtTFB1 is capable of forming an immunoprecipitable complex containing h-mtTFA, we next determined whether the two proteins interact directly in an in vitro protein-binding assay. Recombinant GST::h-mtTFB1 fusion protein was produced in E. coli and bound to glutathione-Sepharose beads (see Materials and Methods). When gel-purified, renatured recombinant h-mtTFA protein was incubated with the h-mtTFB1-bound beads, a significant fraction remained bound to the beads after several stringency washes (Fig. 2A).
FIG. 2.
Direct interaction between h-mtTFB1 or h-mtTFB2 and h-mtTFA requires the C-terminal activation domain of h-mtTFA. The results of solid-phase protein-protein interaction assays are shown. (A) The amount of recombinant gel-purified h-mtTFA that remained stably associated with a GST::h-mtTFB1 fusion protein that was bound to glutathione-Sepharose beads was visualized by Western immunoblotting with an antibody to h-mtTFA (lanes labeled as “bound h-mtTFA”). Full-length h-mtTFA (indicated as such) as well as the following three C-terminal deletion mutants was analyzed: 1-199 (missing C-terminal 5 amino acids), 1-194 (missing C-terminal 10 amino acids), and 1-179 (missing entire C-terminal 25-amino-acid tail). Twenty-five percent of the total amount of protein initially incubated with the beads in each assay is also shown (indicated as “input h-mtTFA”). (B) The results of a direct protein-binding assay between h-mtTFB2 and h-mtTFA C-terminal deletion mutants are shown in the same manner as described for panel A above.
The ability of h-mtTFA to activate transcription in vitro has been attributed to the presence of a 25-amino-acid C-terminal tail domain (7). In that study, it was demonstrated that incremental 5-amino-acid deletions in the tail correspondingly decrease the ability of h-mtTFA to activate transcription from the LSP, while complete removal of the tail (deleting the C-terminal 25 amino acids) results in a protein that binds DNA only nonspecifically and is incapable of activating transcription in vitro. To test the hypothesis that the C-terminal tail of h-mtTFA is required for transcriptional activation because it is an interaction point for h-mtTFB1, we performed direct-binding assays with h-mtTFA C-terminal deletion mutants described by Dairaghi et al. (7). As was the case for full-length h-mtTFA (1-204), a protein missing the C-terminal 5 amino acids (1-199) was still capable of interacting with h-mtTFA (Fig. 2A), while those missing the C-terminal 10 amino acids (1-194) or the entire C-terminal tail (1-179) resulted in severe, if not complete, loss of the interaction with h-mtTFB1 in vitro (Fig. 2A). Virtually identical results were obtained with h-mtTFB2 (Fig. 2B). As reported by Dairaghi et al. (7), all of the h-mtTFA mutant proteins were still capable of binding DNA in an electrophoretic mobility shift assay (data not shown), indicating that the inability of the 1-194 and 1-179 proteins to bind h-mtTFB1 is not a consequence of improper folding or instability of the renatured protein. In addition, the lack of interaction observed with the 1-194 and 1-179 proteins demonstrates that the observed interaction with the wild-type and 1-199 proteins is not due to nonspecific binding of h-mtTFA to the beads or the GST peptide in the fusion protein. Altogether, these binding data mirror the transcriptional activation properties of the h-mtTFA mutants (see Discussion) and demonstrate that the C-terminal tail of h-mtTFA is an important physical and likely functional interaction point for h-mtTFB1 and h-mtTFB2. In addition, these data strongly suggest that h-mtTFB1 and h-mtTFB2 interact directly with h-mtTFA and that this interaction is responsible for the coimmunoprecipitation of these two proteins from mitochondrial extracts with h-mtTFB1 antibodies (Fig. 1).
Antibodies to h-mtTFB1 also coimmunoprecipitate h-mtRNA polymerase.
Given that h-mtTFB1 and h-mtTFA were coimmunoprecipitated from our transcriptionally active mitochondrial extract (Fig. 1), we attempted to reconstitute transcription activity in the depleted extract by adding back both recombinant h-mtTFB1 and h-mtTFA. However, we were still unable to reconstitute transcription activity in the extract in this manner (data not shown). This suggested to us that, in addition to h-mtTFA, h-mtRNA polymerase may also be coimmunoprecipitated from the extract with h-mtTFB1 antisera. Unfortunately, we were unable to confidently detect the h-mtRNA polymerase polypeptide in the immunoprecipitate from this experiment (data not shown). Therefore, to address this possibility, we prepared a new mitochondrial extract that contained amounts of h-mtRNA polymerase detectable by Western analysis (Fig. 3, lane 1) and repeated the immunoprecipitation with h-mtTFB1 antibodies under conditions better optimized for detecting h-mtRNA polymerase in the immunoprecipitate (see Materials and Methods). Under these conditions, we were able to demonstrate that h-mtRNA polymerase can be coimmunoprecipitated with h-mtTFB1 (Fig. 3, lane 2). This interaction was specific for h-mtTFB1 in that no coimmunoprecipitation of h-mtRNA polymerase was observed with the corresponding control preimmune serum (Fig. 3, lane 3). These data demonstrate that h-mtTFB1 and h-mtRNA polymerase can exist as a part of a complex in the soluble mitochondrial extract and also likely explain our inability to reconstitute transcription activity by adding back recombinant h-mtTFB1 and h-mtTFA to the original h-mtTFB1 immunodepleted extract.
FIG. 3.
Coimmunoprecipitation of human mtRNA polymerase with h-mtTFB1. A soluble HeLa cell mitochondrial extract was prepared as described in Materials and Methods. This extract was probed for the presence of h-mtRNA polymerase by Western analysis (lane 1). The same extract was immunodepleted with a polyclonal h-mtTFB1 antibody (lane 2) or the control preimmune serum (lane 3) and probed for the presence of the h-mtRNA polymerase polypeptide (∼139 kDa) in the corresponding immunoprecipitates (IP) by Western analysis. Positions of migration of molecular weight standards (in kilodaltons) are shown to the right of the figure.
Mutational analysis of h-mtTFB1 reveals that transcriptional activation in vitro is independent of SAM and DNA binding.
Our previous analysis of h-mtTFB1 revealed that it is homologous to a family of rRNA methyltransferases, binds the requisite SAM cofactor used by this class of enzymes (19), and methylates a conserved rRNA stem-loop substrate in vivo (24). Sequence comparisons and structural analysis of methyltransferases in general have revealed the presence of eight conserved motifs, designated I to VIII (17, 22) (Fig. 4). In the rRNA methyltransferases, motifs I to III comprise a major portion of the SAM-binding site, with specific residues within these motifs apparently providing direct bonding interactions with this cofactor (3). Motif IV may have a role in SAM binding as well, but it also contains conserved residues that are believed to be important for catalysis for other reasons (17). Many of these essential residues in motifs I to VIII are conserved in h-mtTFB1 (19); however, the role of the rRNA methyltransferase motifs in transcription factor function has not been determined.
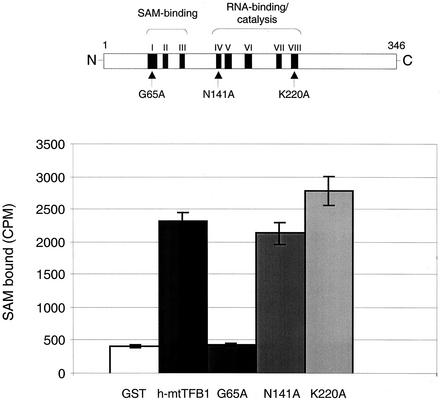
FIG. 4.
Effects of h-mtTFB1 RNA methyltransferase motif point mutations on SAM binding. A schematic diagram of the 346-amino-acid h-mtTFB1 protein is shown at the top of the figure with its RNA methyltransferase motifs I to VIII (17, 19) indicated by the black boxes. The positions of the three point mutations characterized in this study (G65A, motif I; N141A, motif IV; and K220A, motif VIII) are indicated by the arrows. The results of a solid-phase SAM-binding assay that involves the binding of radiolabeled ligand to immobilized GST::h-mtTFB1 proteins are shown in the bar graph. The ordinate represents the amount of labeled SAM (in counts per minute) bound to beads containing GST peptide alone (GST), GST::h-mtTFB1 (h-mtTFB1), and each mutated GST::h-mtTFB1 fusion protein (G65A, N141A, and K220A). That similar amounts of control and h-mtTFB1 proteins were loaded onto the beads was confirmed by Western analysis prior to each assay (data not shown).
In order to address this issue, we analyzed the effects of point mutations in three specific rRNA methyltransferase motifs in h-mtTFB1 on SAM binding, DNA binding, and transcriptional activation functions of h-mtTFB1 in vitro. The first mutation changes glycine 65 to an alanine (G65A) in motif I, which we showed previously abolishes rRNA methyltransferase activity in vivo (24). The second mutation changes the asparagine residue at position 141 to alanine (N141A) in motif IV, which is highly conserved in rRNA methyltransferases. In the crystal structure of the ErmC′ rRNA methyltransferase, this amino acid appears to form hydrogen bonds with SAM (3, 22). It is also conserved in all of the mtTFB homologs identified thus far (9, 19, 23). The third mutation changes the lysine residue at 220 to alanine (K220A) in motif VIII. Based on the crystal structure of sc-mtTFB, this amino acid is predicted to be located in a region of the protein that forms a highly charged groove that has been hypothesized to bind RNA (23). We have shown that the K220A mutation markedly decreases the RNA methyltransferase activity of h-mtTFB1 (24).
By the use of a solid-phase SAM-binding assay described previously (19), the ability of the three mutated h-mtTFB1 proteins to bind the cofactor was assessed. In accordance with previous results, wild-type GST::h-mtTFB1 fusion protein bound approximately fivefold-more SAM than did a GST peptide used as a negative control (Fig. 4). Similar results were obtained with the N141A and K220A mutant proteins (Fig. 4). In contrast, the G65A mutant in motif I resulted in no detectable SAM-binding activity in this assay. The inability of this mutant to bind SAM provides a logical explanation for the previously reported loss of RNA methyltransferase activity as a result of this substitution (24). Lastly, h-mtTFB2 was found to bind SAM in a similar manner as h-mtTFB1 in this assay (data not shown).
Using an electrophoretic mobility shift assay, we showed previously that wild-type h-mtTFB1 binds to an LSP-containing DNA fragment in an apparently nucleotide sequence-independent manner (19). Using this same assay, we next determined the ability of the three mutated h-mtTFB1 proteins to bind DNA. The results of this analysis were similar to that of the SAM-binding experiment. That is, when added at equal concentrations, the N141A and K220A mutants bound DNA in a similar manner as the wild-type GST::h-mtTFB1 fusion protein (Fig. 5A), while the G65A protein was deficient in DNA binding (Fig. 5A, lane 3). Notably, the G65A protein was unable to significantly bind the probe even when substantially higher protein concentrations were tested (data not shown). These data indicate that, in addition to eliminating SAM binding, the G65A mutation in h-mtTFB1 also eliminated its double-stranded DNA-binding activity.
Next, we tested the ability of mutant h-mtTFB1 proteins to activate transcription from the mitochondrial LSP in a transcription factor-dependent transcription assay. In this assay, recombinant h-mtTFA and h-mtTFB1 were added to a soluble mitochondrial extract from HeLa cells that served as a source of human mtRNA polymerase (see Materials and Methods). As described previously (19), addition of recombinant GST::h-mtTFB1 protein alone to the extract did not result in stimulation of transcription initiation (Fig. 5B, compare lane 2 with lane 1), while addition of recombinant h-mtTFA alone resulted in a minimal, yet significant, amount of specific transcription initiation (Fig. 5B, compare lane 3 with lane 1). When wild-type protein or any one of the mutant GST::h-mtTFB1 fusion proteins was added in addition to recombinant h-mtTFA, transcription initiation was significantly increased above that seen with h-mtTFA alone (Fig. 5B, lanes 4 to 11). In addition, stimulation by all the mutant h-mtTFB1 proteins was dose dependent, similar to that observed with the wild-type protein (Fig. 5B, lanes 4 to 11). Finally, consistent with their ability to activate transcription, all of the mutant h-mtTFB1 proteins retained the ability to bind h-mtTFA in vitro in a manner similar to the wild-type protein (Fig. 5C). Altogether, these data indicate that mutations in three conserved RNA methyltransferase motifs do not affect the transcriptional activation properties of h-mtTFB1, despite the facts that a mutation in motif I (G65A) resulted in loss of SAM binding and the ability to interact with DNA and that the G65A and K220A mutations both resulted in lack of significant RNA methyltransferase activity (24).
DISCUSSION
The recent identification of two human homologs of yeast mitochondrial transcription factor B, h-mtTFB1 (19) and h-mtTFB2 (9), has provided the opportunity to gain a more complete understanding of the mechanism of transcriptional regulation in human mitochondria and its influence on human disease and aging. In our initial characterization of h-mtTFB1 we noted its remarkable primary sequence similarity to a class of rRNA methyltransferase enzymes and demonstrated that it binds the requisite SAM cofactor used by this class of enzymes (19). We went on to show that this protein actually possesses rRNA methyltransferase activity and is capable of methylating two adjacent adenine residues in a stem-loop structure that is conserved in bacterial 16S and human mitochondrial 12S rRNA molecules (24). Thus, h-mtTFB1 is apparently a dual-function protein capable of activating transcription and modifying rRNA. The goal of this study was to determine the extent to which these two activities are linked: that is, to determine whether the methyltransferase structural features and/or activity is required for the transcription factor function of h-mtTFB1. In addition, our initial characterizations also revealed a weak and apparently sequence-nonspecific DNA-binding activity of h-mtTFB1 (19). The relevance of this activity to the ability of h-mtTFB1 to activate transcription was also addressed in this study.
The first main conclusion from this work is that the ability of h-mtTFB1 to activate transcription in collaboration with h-mtTFA is largely, if not completely, independent of its activity as an rRNA methyltransferase. The data supporting this conclusion are as follows. First, point mutations in three sequence motifs that are conserved between rRNA methyltransferases and h-mtTFB1 had no effect on the ability of h-mtTFB1 to activate transcription from the mitochondrial LSP in vitro (Fig. 5B). Two of these, K220A and G65A, were shown previously to greatly reduce and eliminate RNA methyltransferase activity, respectively (24). The G65A mutation is in conserved motif I, which is implicated in binding SAM directly (3, 17), and resulted in the predicted defect in cofactor binding (Fig. 4), thus providing a logical explanation for why this mutation results in loss of RNA methyltransferase activity. We conclude from these data that the ability of h-mtTFB1 to activate transcription does not require RNA methyltransferase activity, SAM binding, or an intact SAM-binding pocket.
The second main conclusion from this work is that the C-terminal domain of h-mtTFA is a physical and likely functional interaction point for h-mtTFB1 and h-mtTFB2. The data supporting this conclusion are as follows. First, immunodepletion of h-mtTFB1 from a transcription-competent HeLa cell mitochondrial extract not only immunoprecipitates h-mtTFB1 but also coimmunoprecipitates most of the h-mtTFA as well (Fig. 1). The resulting depleted extract is incapable of transcription initiation from the human LSP (Fig. 1). The coimmunoprecipitation of these two factors was confirmed by our demonstration that they also interact directly in vitro (Fig. 2). Of particular significance is the fact that at least one important interaction point for h-mtTFB1 is the C-terminal tail of h-mtTFA that has been implicated previously in the transcriptional activation function of this HMG box, mtDNA-binding protein (7). When we tested a series of h-mtTFA C-terminal tail deletion mutations for their interaction with h-mtTFB1 in vitro, the results were in remarkable correspondence with the ability of these mutant proteins to activate transcription in vitro reported by Dairaghi et al. (7). That is, a deletion of 5 amino acids (h-mtTFA 1-199) was still capable of interacting with h-mtTFB1 (Fig. 2) and maintained wild-type transcriptional activation function (7). Deletion of 10 (h-mtTFA 1-194) or 25 (h-mtTFA 1-179) C-terminal residues resulted in a severe reduction, if not complete loss, of interaction with h-mtTFB1 in vitro (Fig. 2) that correlates with the reported loss of transcriptional activation function (7). These data strongly suggest that the direct interaction between h-mtTFA and h-mtTFB1 reported here is an important determinant of the ability of these two proteins to cooperate during transcription initiation in human mitochondria.
Finally, we found that the G65A mutation also resulted in dramatic loss of DNA-binding activity in an electrophoretic mobility shift assay (Fig. 5A), leading us to conclude that the transcription factor function of h-mtTFB1 is independent of not only RNA methyltransferase activity but also its double-stranded DNA-binding ability. This suggests that the formation of a closed transcription complex at the mitochondrial LSP can occur without a DNA binding contribution from h-mtTFB1. However, we do acknowledge that we have not eliminated the possibility that h-mtTFB1 may contribute to activation of initiation through binding of single-stranded DNA during initiation (e.g., during open complex formation). A potential function involving single-stranded DNA binding is perhaps more likely for a transcription factor that binds RNA, a nucleic acid substrate with at least some single-stranded character. If such an interaction is occurring, it is possible that the G65A mutation has differentially affected single-stranded and double-stranded DNA binding of h-mtTFB1, perhaps indicating the existence of two separate nucleic acid binding sites on the molecule. In the absence of this speculation, however, the simplest explanation of our results is that transcriptional activation by h-mtTFB1 is independent of its only documented DNA-binding activity.
Based on the data presented in this report and the present state of knowledge, we propose the following model describing the fundamental interactions required for transcription initiation at a human mitochondrial promoter (Fig. 6). The basic premise of this model is that h-mtTFB1 (and by analogy h-mtTFB2) functions to bridge an interaction between h-mtTFA and h-mtRNA polymerase at the promoter to facilitate transcription initiation. We propose that the interaction between h-mtTFB1 and h-mtTFA is mediated in large part by the C-terminal tail of h-mtTFA based on the data presented herein and on the documented strict requirement for this domain in transcriptional activation (7). The proposed direct interaction between h-mtTFB1 and mtRNA polymerase is supported by our data showing that antibodies to h-mtTFB1 coimmunoprecipitate h-mtRNA polmerase (Fig. 3) and on the indirect, yet compelling, evidence reported by others that a one-to-one complex can form between h-mtTFB1 or h-mtTFB2 and h-mtRNA polymerase (9). Such a complex is also consistent with the fact that the S. cerevisiae homologs of these proteins, sc-mtTFB (Mtf1p) and mtRNA polymerase (Rpo41p), have been shown to interact (4, 5). A final aspect of this model is based on our observation that activation of transcription by h-mtTFB1 is independent of its normal double-stranded DNA-binding activity. This suggests that promoter recognition by h-mtRNA polymerase (i.e., closed complex formation) per se is facilitated not by h-mtTFB1 DNA-binding activity but rather by the sequence-specific DNA binding of h-mtTFA at the promoter and/or its generation of a specific protein-DNA conformation that is accomplished at that site through its ability to bend and wrap DNA (12). According to this model, h-mtTFB1 has an “adapter” function that facilitates delivery of mtRNA polymerase to the promoter, which is demarcated by a specific configuration of the C-terminal domain of h-mtTFA bound at the promoter. The ability of h-mtTFB1 to activate transcription independently of its RNA methyltransferase activity is consistent with the proposed adapter function in that it suggests that this function involves the regions of the protein that are unique to this class of transcription factors and absent in the related RNA methyltransferase proteins (23). While this model directly implicates h-mtTFA as an important player in the promoter recognition process, our data do not discount the possibility that h-mtRNA polymerase and/or h-mtTFB1 and h-mtTFB2 also contribute to promoter specificity in some manner. Additional experiments are needed to determine precisely how these four factors cooperate to achieve promoter-specific transcription initiation from the LSP and HSP.
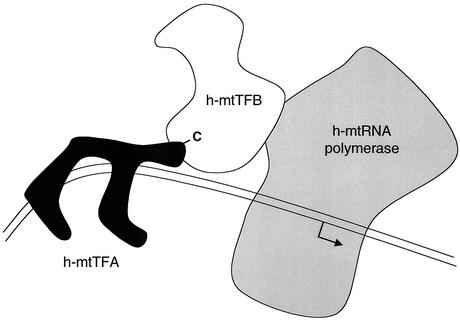
FIG. 6.
Proposed model of the interactions between human mitochondrial transcription proteins during initiation of transcription at the human LSP. Human mtTFA (black) is shown bound to the LSP upstream of the site of transcription initiation (bent arrow). It is shown bending the DNA (parallel lines) at the promoter as part of its ability to activate transcription initiation as proposed by Fisher et al. (12). The C-terminal activation region of h-mtTFA (-C) is shown binding to h-mtTFB1 (or h-mtTFB2) based on the results of this study. In this model, h-mtTFB1 serves an adapter function to bridge an interaction between h-mtTFA and h-mtRNA polymerase at the promoter, which is demarcated by a specific h-mtTFA/DNA complex. The 1:1 complex shown between h-mtTFB1 (or h-mtTFB2) and h-mtRNA polymerase is drawn based on our ability to coimmunoprecipitate these proteins (Fig. 3), the report of Falkenberg et al. (9) that these proteins interact in vitro during copurification, and on the reports of Cliften et al. that sc-mtTFB physically interacts with mtRNA polymerase in S. cerevisiae (4, 5).
While this report provides important new information regarding the mechanism of transcription in human mitochondria, many questions remain unanswered. For example, our data indicate that, in terms of SAM binding and its interaction with h-mtTFA (Fig. 2B), h-mtTFB2 behaves in a manner indistinguishable from h-mtTFB1 in the assays used. Thus, we have not provided an explanation for the observation that h-mtTFB2 activates transcription more efficiently in vitro with a fully recombinant transcription system (9). Nor have we yet uncovered why the system has evolved a requirement for two h-mtTFB homologs. We would argue that both h-mtTFB1 and h-mtTFB2 are involved directly in transcription and are present to provide a yet-to-be-elucidated mechanism for differential regulation of mitochondrial gene expression. However, it remains a formal possibility that h-mtTFB1 is primarily an rRNA methyltransferase in vivo (24) and that h-mtTFB2 is primarily a transcription factor in vivo, or vice versa. More detailed studies are needed in the future to decipher the individual or dual roles of these two important factors in mitochondrial gene expression. Clearly, much remains to be learned regarding the regulation of human mitochondrial transcription and its impact on human aging and disease.
Acknowledgments
We thank Bonnie Seidel-Rogol for critical comments on the manuscript and other important contributions to the project.
This work was supported by a grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health (HL-59655) awarded to G.S.S. and an NRSA (HL-68459) awarded to V.M.
REFERENCES
- 1.Antoshechkin, I., and D. F. Bogenhagen. 1995. Distinct roles for two purified factors in transcription of Xenopus mitochondrial DNA. Mol. Cell. Biol. 15:7032-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. [PubMed] [Google Scholar]
- 3.Bussiere, D. E., S. W. Muchmore, C. G. Dealwis, G. Schluckebier, V. L. Nienaber, R. P. Edalji, K. A. Walter, U. S. Ladror, T. F. Holzman, and C. Abad-Zapatero. 1998. Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37:7103-7112. [DOI] [PubMed] [Google Scholar]
- 4.Cliften, P. F., J. Y. Park, B. P. Davis, S. H. Jang, and J. A. Jaehning. 1997. Identification of three regions essential for interaction between a sigma-like factor and core RNA polymerase. Genes Dev. 11:2897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cliften, P. F., S. H. Jang, and J. A. Jaehning. 2000. Identifying a core RNA polymerase surface critical for interactions with a sigma-like specificity factor. Mol. Cell. Biol. 20:7013-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dairaghi, D. J., G. S. Shadel, and D. A. Clayton. 1995. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta 1271:127-134. [DOI] [PubMed] [Google Scholar]
- 7.Dairaghi, D. J., G. S. Shadel, and D. A. Clayton. 1995. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 249:11-28. [DOI] [PubMed] [Google Scholar]
- 8.DiMauro, S., K. Tanji, E. Bonilla, F. Pallotti, and E. A. Schon. 2002. Mitochondrial abnormalities in muscle and other aging cells: classification, causes, and effects. Muscle Nerve 26:597-607. [DOI] [PubMed] [Google Scholar]
- 9.Falkenberg, M., M. Gaspari, A. Rantanen, A. Trifunovic, N. G. Larsson, and C. M. Gustafsson. 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31:289-294. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, R. P., and D. A. Clayton. 1985. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem. 260:11330-11338. [PubMed] [Google Scholar]
- 11.Fisher, R. P., and D. A. Clayton. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, R. P., T. Lisowsky, M. A. Parisi, and D. A. Clayton. 1992. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267:3358-3367. [PubMed] [Google Scholar]
- 13.Fisher, R. P., J. N. Topper, and D. A. Clayton. 1987. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50:247-258. [DOI] [PubMed] [Google Scholar]
- 14.Hager, D. A., and R. R. Burgess. 1980. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal. Biochem. 109:76-86. [DOI] [PubMed] [Google Scholar]
- 15.Jang, S. H., and J. A. Jaehning. 1991. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 266:22671-22677. [PubMed] [Google Scholar]
- 16.Lisowsky, T., and G. Michaelis. 1988. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol. Gen. Genet. 219:125-128. [DOI] [PubMed] [Google Scholar]
- 17.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 18.Masters, B. S., L. L. Stohl, and D. A. Clayton. 1987. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51:89-99. [DOI] [PubMed] [Google Scholar]
- 19.McCulloch, V., B. L. Seidel-Rogol, and G. S. Shadel. 2002. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 22:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micol, V., P. Fernandez-Silva, and G. Attardi. 1996. Isolation and assay of mitochondrial transcription termination factor from human cells. Methods Enzymol. 264:158-173. [DOI] [PubMed] [Google Scholar]
- 21.Parisi, M. A., and D. A. Clayton. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252:965-969. [DOI] [PubMed] [Google Scholar]
- 22.Schluckebier, G., P. Zhong, K. D. Stewart, T. J. Kavanaugh, and C. Abad-Zapatero. 1999. The 2.2 Å structure of the rRNA methyltransferase ErmC′ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 289:277-291. [DOI] [PubMed] [Google Scholar]
- 23.Schubot, F. D., C. J. Chen, J. P. Rose, T. A. Dailey, H. A. Dailey, and B. C. Wang. 2001. Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 10:1980-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidel-Rogol, B. L., V. McCulloch, and G. S. Shadel. 2003. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33:23-24. [DOI] [PubMed] [Google Scholar]
- 25.Seidel-Rogol, B. L., and G. S. Shadel. 2002. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 30:1929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shadel, G. S., and D. A. Clayton. 1993. Mitochondrial transcription initiation. Variation and conservation. J. Biol. Chem. 268:16083-16086. [PubMed] [Google Scholar]
- 27.Shadel, G. S., and D. A. Clayton. 1995. A Saccharomyces cerevisiae mitochondrial transcription factor, sc-mtTFB, shares features with sigma factors but is functionally distinct. Mol. Cell. Biol. 15:2101-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 29.Shoubridge, E. A. 2001. Nuclear genetic defects of oxidative phosphorylation. Hum. Mol. Genet. 10:2277-2284. [DOI] [PubMed] [Google Scholar]
- 30.Tiranti, V., A. Savoia, F. Forti, M. F. D'Apolito, M. Centra, M. Rocchi, and M. Zeviani. 1997. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 6:615-625. [DOI] [PubMed] [Google Scholar]
- 31.Topper, J. N., and D. A. Clayton. 1989. Identification of transcriptional regulatory elements in human mitochondrial DNA by linker substitution analysis. Mol. Cell. Biol. 9:1200-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 33.Xu, B., and D. A. Clayton. 1992. Assignment of a yeast protein necessary for mitochondrial transcription initiation. Nucleic Acids Res. 20:1053-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]