Abstract
The prevailing model of polytopic membrane protein insertion is based largely on the in vitro analysis of polypeptide chains trapped during insertion by arresting translation. To test this model under conditions of active translation in vivo, we have used a kinetic assay to determine the order and timing with which transmembrane segments of bacterioopsin (BO) are inserted into the membrane of the archaeon Halobacterium salinarum. BO is the apoprotein of bacteriorhodopsin, a structurally well characterized protein containing seven transmembrane α-helices (A-G) with an N-out, C-in topology. H. salinarum strains were constructed that express mutant BO containing a C-terminal His-tag and a single cysteine in one of the four extracellular domains of the protein. Cysteine translocation during BO translation was monitored by pulse–chase radiolabeling and rapid derivatization with a membrane-impermeant, sulfhydryl-specific gel-shift reagent. The results show that the N-terminal domain, the BC loop, and the FG loop are translocated in order from the N terminus to the C terminus. Translocation of the DE loop could not be examined because cysteine mutants in this region did not yield a gel shift. The translocation order was confirmed by applying the assay to mutant proteins containing two cysteines in separate extracellular domains. Comparison of the translocation results with in vivo measurements of BO elongation indicated that the N-terminal domain and the BC loop are translocated cotranslationally, whereas the FG loop is translocated posttranslationally. Together, these results support a sequential, cotranslational model of archaeal polytopic membrane protein insertion in vivo.
A critical step in the biogenesis of polytopic membrane proteins is the insertion of transmembrane segments into the lipid bilayer. Although in vitro studies have yielded profound insights into the molecular mechanism of this process (1, 2), much remains to be learned about how it occurs in vivo. In particular, little is known about the order with which transmembrane segments are inserted or the timing of insertion with respect to translation in intact cells. Resolving these issues under cellular conditions is essential for understanding other aspects of membrane protein biogenesis, such as the pathway by which tertiary structure is formed.
In a widely accepted model of polytopic membrane protein insertion, transmembrane segments are inserted cotranslationally in the same order as in the primary sequence (1–3). The timing and order of insertion are attributable to the extrusion of nascent polypeptides from the ribosome into an aqueous pore formed by the secretory translocase, a membrane-bound translocation complex. Evidence for this model has been obtained from in vitro studies of nascent polytopic membrane proteins trapped at intermediate stages of insertion into the eukaryotic endoplasmic reticulum membrane (4, 5) and from a study of the order with which transmembrane segments insert during active translation (6). These studies should be interpreted with a degree of caution, because the methods used to assess protein insertion require extensive incubation periods, during which the topology of a nascent polypeptide chain might be rearranged. Also, the lower efficiency of translation and translocation in vitro may alter the observed order and timing of insertion. Studies of polytopic membrane protein insertion during active translation in vivo are needed to resolve these concerns.
Prokaryotes with a single cellular membrane offer an advantage for analyzing membrane protein insertion, because insertion can be monitored directly with external reagents. The insertion mechanism in prokaryotes and eukaryotes is expected to be similar, because core components of the secretory translocase are conserved (7). In Escherichia coli, polytopic membrane protein insertion seems to require the secretory translocase (8–10) as well as other proteins needed for insertion in eukaryotes, such as the signal recognition particle (10, 11). In addition, insertion of E. coli polytopic membrane proteins in vitro occurs cotranslationally (12–14). Thus, the mechanism of polytopic membrane protein insertion in E. coli and other prokaryotes may be universally relevant. However, up until now, it has not been possible to exploit the advantage of prokaryotes to test whether insertion occurs cotranslationally and sequentially in vivo.
We have studied the insertion of bacterioopsin (BO), a seven-transmembrane α-helical polypeptide. BO is the apoprotein of bacteriorhodopsin (BR), which contains a covalently bound molecule of retinal. BR functions as a light-driven proton pump in the sole membrane of the archaeon Halobacterium salinarum. Under low-oxygen conditions, it accumulates at high levels and forms the purple membrane, a two-dimensional crystal. BR is attractive for insertion studies, because its topology in the native membrane is known from high-resolution structural analysis (15, 16).
Previous studies are consistent with a model in which BO is inserted into the membrane by the secretory translocase. BO is synthesized with a 13 amino acid presequence at its N terminus (17), which may act as a signal sequence for membrane targeting. The presequence may be recognized by the signal recognition particle, which in other systems initiates cotranslational insertion by the secretory translocase (18). In accord with this hypothesis, the RNA component of the signal recognition particle is localized to the membrane on induction of BO synthesis (19). The model also is supported by our previous work, in which we used an in vivo kinetic assay to show that the N terminus of BO is translocated cotranslationally, indicating that the first transmembrane segment is inserted cotranslationally (20). In this study, we examine the translocation order of other extracellular domains of the protein and the timing of translocation with respect to translation. These findings are discussed in light of current models for polytopic membrane protein insertion.
Materials and Methods
Materials.
Oligonucleotides were obtained from Operon Technologies (Alameda, CA), Taq polymerase and ligase from Promega, and restriction endonucleases from New England Biolabs. Ni2+-nitrilotriacetic acid Superflow was obtained from Qiagen (Chatsworth, CA). Redivue 35S-methionine (Met) [>1,000 Ci/mmol (1 Ci = 37 GBq)] was obtained from Amersham Pharmacia and Tris(2-carboxyethyl)phosphine was purchased from Sigma. 4-Acetamido-4′maleimidylstilbene-2,2′-disulfonic acid disodium salt (AMS) and fluorescein-5-maleimide (FM) were obtained from Molecular Probes.
Plasmid and Strain Construction.
H. salinarum strains expressing BR:H6, I4C:H6, T5C:H6 (H6 refers to a C-terminal His-tag), and A103C were described previously (20, 21). H. salinarum strains expressing the BR variants M68C:H6, E74C:H6, Q75C:H6, K129C:H6, V13°C:H6, Y131C:H6, A196C:H6, I198C:H6, V199C:H6, T5C:A196C:H6, E74C:A196C:H6, and G116C were created as follows. PCR was used to introduce mutations into the bop gene in E. coli plasmids. Cysteine substitution at position 68 of mature BO was achieved with the primers 5′-GCGCGGATCCGACGTGAAGA-3′ and 5′-CCACCGAACTGTACACATGTGAGGCC-3′ (underlined bases correspond to codon changes); at positions 74 or 75 with 5′-TACAAGACCGAGTGGGGGACT-3′ and 5′-CCTCACAATGGTACCGTTCGGTGGGTGCCAGAACC-3′ or 5′-CCTCACAATGGTACCGTTCGGTGGGGAGTGCAACCCC-3′, respectively; and at positions 129, 130, or 131 with 5′-TGTTGAGCGACGCTGGAAAG-3′ and 5′-GCCGACGGCATCATGATCGGGACCGGCCTGGTCGGCGCACTGACGTGCGTCTACTCGTACCG-3′, 5′-GCCGACGGCCATCATGATCGGGACCGGCCTGGTCGGCGCACTGACGAAGTGCTACTC-3′, or 5′-GCCGACGGCATCATGATCGGGACCGGCCTGGTCGGCGCACTGACGAAGGTCTGCTCGTACCG-3′, respectively. Two-step PCR was used to substitute cysteine at positions 196, 198, or 199. Megaprimers were created by using the primers 5′-TGTTCTTCGGGTTCACCTCG-3′ and 5′-GATTCCGCAACCTTCGCTG-3′, 5′-CGGCACGCATCCCGCACCTTC-3′, or 5′-CGGGCAGATTCCCGCACCTTCG-3′, respectively. The megaprimers were combined with 5′-TACAAGACCGAGTGGGGGACT-3′ in the second PCR step. PCR products were digested with restriction enzymes, combined with the appropriate fragment encoding a C-terminal His-tag (20), and cloned in pMPK85, an H. salinarum integrating vector (22). Cysteine was substituted at position 116 by mutagenesis with the primer 5′-GCGCGGATCCGACGTGAAGA-3′ and the degenerate primer 5′-GGCGGTTCAGATCATGATVVNGTCGGCACCGACGAGCGCAA-3′ (V = A, C, or G). The PCR product was digested with restriction enzymes and cloned in pMPK62 (23). Double mutants with cysteines at positions 5 and 74 or 74 and 196, as well as with the C-terminal His-tag, were constructed from the single mutants. H. salinarum strains expressing the single cysteine mutants were created by targeted gene replacement as described (23). The H. salinarum strain MPK406, which contains a single copy of ura3 at the bop locus, was used to construct the double cysteine mutants on 5-fluororotic acid as described (24). Southern analysis confirmed a single copy of bop in the recombinant strains. Fluorescent terminator cycle sequencing (Prism; Applied Biosystems) confirmed the bop sequences of the recombinant strains and revealed the presence of a silent mutation at position 201 in I198C:H6 and V199C:H6.
Expression, Characterization, and Purification of Mutant BO.
BR synthesis was induced and quantified as described (20). For derivatives of the Δura3 strain MPK406, the growth medium was supplemented with 50 μg of uracil per ml. Full-length His-tagged BO was Ni2+-affinity-purified as previously described (20).
Translocation and Elongation Assays.
BO translocation and elongation were measured as described (20) with slight modifications to improve the assay. Briefly, cells expressing BO cysteine mutants were incubated in AMS, then radiolabeled with a pulse of 35S-Met, followed with a nonradioactive Met chase. At various times after the pulse, aliquots were removed and combined with DTT to quench the AMS. The cells then were incubated to allow completion of radiolabeled polypeptides. Cells were lysed, and His-tagged protein was purified from lysates by Ni2+-affinity chromatography, electrophoresed, and examined by autoradiography and PhosphorImager analysis (Molecular Dynamics). To improve reproducibility, cells were grown to an OD660 = 0.40 ± 0.02, and temperature was maintained at 37 ± 0.1°C. To improve the extent of derivatization, AMS was used at a final concentration of 2.5 mM. Elongation was analyzed as described (20), except that the purified proteins were electrophoresed on a 5% SDS/PAGE gel and excised after Coomassie staining. Gel slices were dissolved in 30% H2O2 at 50°C for >4 h, and radioactivity was counted with a liquid scintillation analyzer. BO recovery was normalized by including 4 μg of FM-derivatized T5C:H6 per ml in the cell lysis solution (20). T5C:H6 was derivatized with 2 mM FM at 37°C for >2 h in 100 mM Tris⋅Cl, pH 7.0/0.1% SDS after pretreatment with 2.5 mM Tris(2-carboxyethyl)phosphine in the same buffer for 5 h at 37°C. Fluorescence was quantified with a Hitachi FMBIO MultiView Fluoroimager before staining.
Data Analysis.
Gel lanes were analyzed with imagequant (Molecular Dynamics), and the distribution of radiolabel in derivatized and underivatized protein was fit with two Gaussian peaks by using igor pro Version 3.3 (WaveMetrics, Lake Oswego, OR). Elongation data were fit with excel (Microsoft) with an algorithm that accounts for the spacing of the nine Met residues in mature BO (details available on request). In the algorithm, changes in cellular 35S-Met levels during the pulse and chase are modeled as monoexponential processes reaching >98% completion in 10 sec as measured previously (20).
Results
Cysteine Mutants.
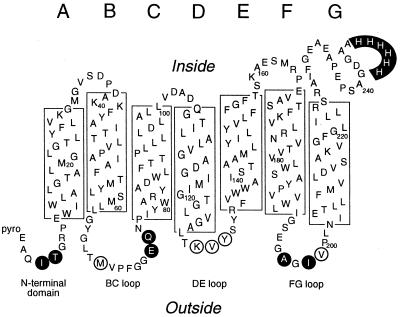
In the translocation assay, insertion of transmembrane segments of BO into the H. salinarum cytoplasmic membrane is monitored by rapid derivatization of unique cysteines in extracellular domains of the protein with AMS, a membrane-impermeant, sulfhydryl-specific gel-shift reagent. The assay was developed with I4C:H6 and T5C:H6 (20), which contain a cysteine at positions 4 or 5 in the N-terminal domain of mature BO (Fig. 1). To apply the assay to other extracellular domains, cysteines were introduced in the BC, DE, and FG loops (Fig. 1). Several cysteine mutants were examined in each loop, because it was unknown which proteins would be expressed or would exhibit a gel shift with AMS. M68C:H6, E74C:H6, and Q75C:H6 in the BC loop; K129C:H6, V130C:H6, and Y131C:H6 in the DE loop; and A196C:H6, I198C:H6, and V199C:H6 in the FG loop (Fig. 1) yielded purple colonies, indicating that the mutant proteins are expressed at relatively high levels.
Figure 1.
Location of cysteine substitutions. The secondary structure of mature BR is shown, with transmembrane α-helices A-G (16) indicated by boxes. Loops are designated by the transmembrane helices they connect. Extracellular residues substituted with cysteine are circled, and the C-terminal His tag is highlighted. A subset of the mutant proteins (closed circles) was examined with the in vivo kinetic assay of translocation.
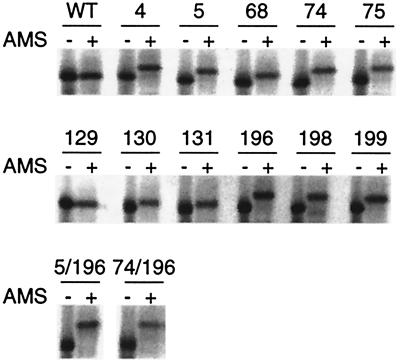
To test for a gel shift, the mutant proteins were purified, solubilized in SDS, treated with AMS, and electrophoresed. Relative to the untreated samples, a large shift was observed for cysteine mutants in the N-terminal domain (Fig. 2, samples 4 and 5) as reported (20). Shifts also were observed for cysteine mutants in the BC loop (Fig. 2, samples 68, 74, and 75) and the FG loop (Fig. 2, samples 196, 198, and 199). In the DE loop, no shift was detected for K129C:H6 and only slight shifts were observed for V130C:H6 and Y131C:H6 (Fig. 2, samples 129–131). All of the DE-loop mutants reacted with FM to yield a fluorescent product (data not shown). These results suggest that all of the cysteines in the mutant proteins are derivatizable, but that the magnitude of the gel shift depends on the cysteine location. Similar variation has been observed with other proteins derivatized with reagents related to AMS (25).
Figure 2.
AMS derivatization of mutant BR. Purified mutant proteins were incubated in 100 mM Tris⋅Cl (pH 8.0), 0.4% SDS, and 2 mM Tris(2-carboxyethyl)phosphine overnight at 37°C, then treated with 20 mM AMS (+) or mock-treated with DMSO (−) for 5 h at 37°C. After quenching with 40 mM DTT, the proteins were electrophoresed and Coomassie-stained. Numbers indicate the position of the cysteine substitution in each mutant protein.
Two cysteine mutants from each loop showing large gel shifts were selected for further analysis (Fig. 1, filled circles). The expression levels and spectral properties of these proteins were comparable to those of BR:H6, suggesting that they bind retinal and have a topology and structure similar to that of the wild-type protein (Table 1). Thus, the mutant proteins are suitable for insertion studies.
Table 1.
Expression levels and spectral characteristics
| Protein | Expression levels*, %BR:H6 |
Absorption maximum, nm†
|
|
|---|---|---|---|
| Dark-adapted | Light-adapted | ||
| BR:H6 | 558‡ | 569‡ | |
| I4C:H6 | 53‡ | 558‡ | 568‡ |
| T5C:H6 | 79‡ | 558‡ | 568‡ |
| E74C:H6 | 86 ± 9 | 557 | 567 |
| Q75C:H6 | 113 ± 14 | 557 | 568 |
| A196C:H6 | 91 ± 21 | 558 | 568 |
| I198C:H6 | 83 ± 7 | 558 | 569 |
| T5C:A196C:H6 | 34 ± 1 | 556 | 568 |
| E74C:A196C:H6 | 41 ± 7 | 556 | 568 |
BR expression levels from 2-day cultures (OD660 ≈ 0.50) were determined by light/dark spectroscopy of crude cell lysates as described (20). Values reported are the average of two to four experiments with one SD.
Absorption maxima of the purified lattice form of the proteins were obtained in 30 mM sodium phosphate, pH 6.9. Dark-adapted, incubated for >8 h at room temperature in the dark; light-adapted, illuminated for 5 min with >520 nm of light.
Values reported previously (20).
Translocation Time Course.
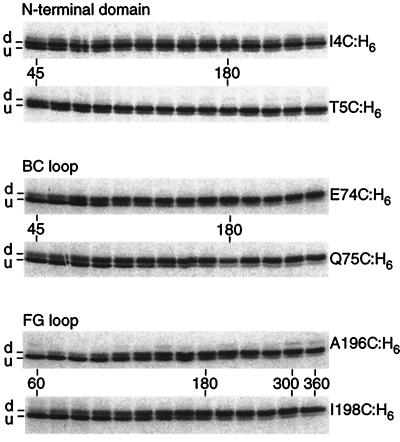
To examine translocation, cells expressing the selected cysteine mutants were pulse–chase radiolabeled with 35S-Met in the presence of AMS. At various times, aliquots were removed, treated with DTT to quench the AMS reaction, and incubated to allow completion of the nascent radiolabeled chains. Full-length protein was recovered from each aliquot by Ni2+-affinity chromatography and then electrophoresed. Two major radiolabeled bands were observed that changed in intensity with time (Fig. 3). Samples radiolabeled in the absence of AMS yielded a single band that comigrated with the lower band (data not shown), indicating that it corresponds to underivatized protein and that the presequence is efficiently processed. [An exception was I4C:H6, which showed increased levels of unprocessed protein on Coomassie-stained gels (data not shown) and an additional upper band in the translocation time course (Fig. 3) that is likely to be derivatized, unprocessed protein.] Thus, the upper band corresponds to derivatized protein.
Figure 3.
Autoradiograms of typical translocation time courses for I4C:H6, T5C:H6, E74C:H6, Q75C:H6, A196C:H6, and I198C:H6. Cells expressing the mutant proteins were preincubated with AMS for 1 min, pulse-radiolabeled with 35S-Met for 20 sec, then chased with nonradioactive Met. The final concentration of AMS was 2.5 mM. Aliquots were removed every 15 sec starting 45 or 60 sec after pulse addition, and every 30 sec starting 180 sec after pulse addition. Each aliquot was combined with an equal volume of 20 mM DTT in medium salts to quench the AMS reaction. The cells were further incubated for 10 min and lysed. Full-length protein was Ni2+-affinity-purified and electrophoresed. d, derivatized; u, underivatized.
At the earliest times, cysteine mutants in the N-terminal domain showed the greatest extent of derivatization, followed by mutants in the BC and FG loops (Fig. 3, 45 and 60 sec). At the latest times, derivatization of all samples was nearly complete (Fig. 3, 300 and 360 sec). In Fig. 3, the final extent of derivatization was 93–95%; other experiments yielded similarly high values (80–97%).
To ensure that translocated cysteines only were derivatized, cells expressing BO mutants containing a cytoplasmic- or membrane-localized cysteine were radiolabeled in the presence of AMS. Neither A103C nor G116C exhibited a gel shift, except after membranes from cells expressing these proteins were solubilized in SDS (data not shown). Thus, AMS is membrane-impermeant under the assay conditions, and derivatization may be taken as evidence for cysteine translocation.
Comparison of Translocation and Elongation.
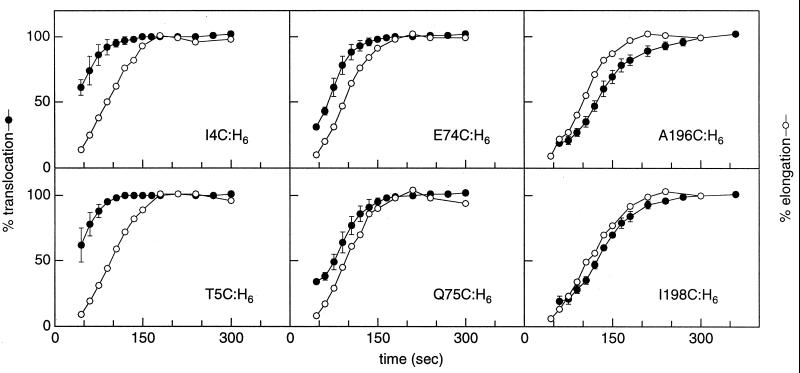
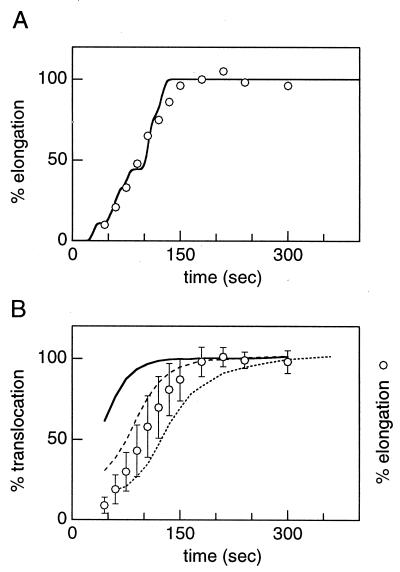
To compare the translocation assays, the final extent of derivatization for each data set was normalized to 100% (Fig. 4, filled circles). The high extent of derivatization and the excellent reproducibility of the data justify the normalization. The translocation data were plotted with the elongation time course of each protein (Fig. 4, open circles), which was determined by examining the incorporation of 35S-Met into the full-length His-tagged protein as a function of time after pulse–chase radiolabeling. The translocation and elongation time courses produce similarly shaped curves, consistent with the idea that the cysteines are translocated and the polypeptide is completed at fixed times during biogenesis. However, the translocation time courses are shifted along the time axis relative to the elongation time courses. Both the N-terminal domain and the BC loop are translocated before elongation is complete. In contrast, the FG loop is translocated after elongation is complete.
Figure 4.
Comparison of translocation and elongation data. Translocation time courses for I4C:H6, T5C:H6, E74C:H6, Q75C:H6, A196C:H6, and I198C:H6 were obtained as described in Fig. 3. The extent of derivatization was quantified as described in Materials and Methods and the plateau value was set to 100%. The average of three experiments is plotted as a function of time after the pulse (filled circles) with error bars that represent one SD. To determine the elongation time course (open circles), cells expressing the cysteine mutants were preincubated for 1 min with DMSO, pulse-radiolabeled with 35S-Met for 20 sec, then chased with nonradioactive Met. Aliquots were removed at various times and mixed with lysis solution (20) containing FM-derivatized T5C:H6 to normalize recovery. The full-length protein was Ni2+-affinity-purified and electrophoresed on a 5% SDS/PAGE gel. The radioactivity in each BO band was quantified as described in Materials and Methods, normalized to a plateau value of 100% elongation, and plotted as a function of time after initiation of the pulse. The average of seven experiments is shown.
Both cysteine mutants in each extracellular domain yielded similar results, suggesting that the observed translocation time courses reflect changes in cysteine accessibility and are not influenced by differences in the intrinsic reactivity of individual cysteines. At the concentration of 2.5 mM AMS used in the assay, the time course of T5C:H6, E74C:H6, and A196C:H6 derivatization proceeded at a maximal rate (data not shown). This finding suggests that cysteine translocation is rate-limiting for derivatization.
Order and Timing of Translocation.
Comparison of the translocation data for each cysteine mutant is possible only if the average elongation rates of the proteins do not differ significantly from each other and from the wild-type His-tagged protein (BO:H6). To determine the average elongation rates, elongation time courses were obtained as described in Fig. 4 and fit to a theoretical curve as in Fig. 5A. The average elongation rates of the cysteine mutants range from 1.6 to 1.9 amino acids per sec (Table 2) and do not vary significantly from each other or from BO:H6, as determined by statistical analysis (ANOVA; graphpad; Prism Software, San Diego). Thus, the cysteine substitutions do not perturb translation of the protein, allowing direct comparison of the translocation data.
Figure 5.
Fitting of elongation data and order of translocation. (A) Elongation time course of BO:H6 (open circles) obtained and analyzed as in Fig. 4. The data were fit with a theoretical elongation curve as described in Materials and Methods. (B) Averaged translocation time courses for the N-terminal domain (solid line), the BC loop (dashed line), and the FG loop (dotted line). Elongation time courses obtained as in A were averaged for the entire set of mutants (open circles). Error bars represent one SD.
Table 2.
Translocation and elongation times
| Protein | Elongation rate*†, aa/sec | Elongation time‡, sec | Tshift§, sec | Translocated chain length¶, aa |
|---|---|---|---|---|
| BR:H6 | 1.8 ± 0.4 | 140 ± 30 | ||
| N-terminal domain | 67 | 130 | ||
| I4C:H6 | 1.9 ± 0.4 | 130 ± 30 | ||
| T5C:H6 | 1.8 ± 0.4 | 140 ± 30 | ||
| BC loop | 25 | 210 | ||
| E74C:H6 | 1.9 ± 0.4 | 130 ± 30 | ||
| Q75C:H6 | 1.8 ± 0.3 | 140 ± 20 | ||
| FG loop | −28‖ | >251 | ||
| A196C:H6 | 1.7 ± 0.2 | 150 ± 20 | ||
| I198C:H6 | 1.6 ± 0.2 | 160 ± 20 |
Values reported are the average of seven experiments with one SD.
Elongation rates were calculated by fitting elongation time courses (see Materials and Methods).
Elongation times were calculated by dividing the number of amino acids in the mature protein by the elongation rate.
Tshift values were estimated as described in Results.
The translocated chain length was calculated by (elongation time − Tshift) × 1.8, where 1.8 is the average elongation rate.
A negative value indicates that translocation occurs after elongation.
Normalized derivatization data from the two cysteine mutants in each extracellular domain were averaged and compared with elongation data averaged from the entire set of mutants and wild-type BO (Fig. 5B, open circles). The data show that the N-terminal domain translocates first (Fig. 5B, solid line), followed by the BC loop (Fig. 5B, dashed line), and finally by the FG loop (Fig. 5B, dotted line). Thus, these domains are translocated sequentially, from the N terminus to the C terminus. The first two extracellular domains become accessible to the extracellular environment cotranslationally, whereas the last domain becomes accessible after elongation is complete.
To relate translation and translocation quantitatively, we estimated the length of the polypeptide chain upon translocation of the cysteine. The separation of the curves in Fig. 5B was evaluated at time points in the rising portion of the translocation curves corresponding to values of 30–80% translocation. By this analysis, the N-terminal domain and BC loop were found to translocate ≈70 and 25 sec before completion of the polypeptide chain, respectively, whereas the FG loop was translocated ≈30 sec after the polypeptide was completed (Table 2, Tshift). Assuming a uniform elongation rate, cysteines in the N-terminal domain or BC loop are accessible to derivatization 125–130 amino acids after leaving the ribosomal peptidyl transfer site.
Translocation Order of Two Cysteines in a Single Polypeptide.
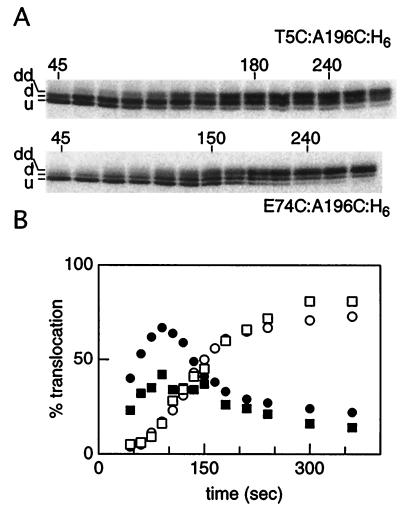
A potential concern of the above analysis is that the cysteine substitutions may change the elongation rate in different regions of the polypeptide without altering the average rate, making it difficult to interpret the translocation data. To address this concern, we examined T5C:A196C:H6 and E74C:A196C:H6, which contain cysteines in two external domains of the same polypeptide. These proteins showed larger gel shifts than did single cysteine mutants (Fig. 2, samples 5/196 and 74/196), consistent with derivatization by two AMS molecules. Translocation assays yielded time courses that were a composite of the corresponding single cysteine mutants, showing a singly derivatized band at early times (Fig. 6A, d) and a doubly derivatized band at later times (Fig. 6A, dd). The doubly derivatized band was generated with the same kinetics in both T5C:A196C:H6 and E74C:A196C:H6 (Fig. 6B, open symbols) and thus corresponds to the common cysteine at position 196. The singly derivatized band, which formed with different kinetics in the two mutants (Fig. 6B, filled symbols), corresponds to derivatization of the cysteine at positions 5 or 74. From these studies, the N-terminal domain and the BC loop were shown to translocate before the FG loop, supporting the results obtained from single cysteine mutants. In addition, the translocation time course of the singly derivatized band in the two mutants (Fig. 6B, filled symbols) supports the finding that the N-terminal domain translocates before the BC loop does.
Figure 6.
Ordered translocation of two domains in double cysteine mutants. (A) Autoradiograms of T5C:A196C:H6 and E74C:A196C:H6 translocation time courses. Cells expressing the mutant proteins were treated and analyzed as in Figs. 3 and 4. u, underivatized; d, singly derivatized; dd, doubly derivatized. (B) The percentage of singly derivatized (filled symbols) and doubly derivatized (open symbols) protein for both T5C:A196C:H6 (circles) and E74C:A196C:H6 (squares) is plotted as a function of time after initiation of the pulse.
Discussion
We have applied an in vivo kinetic assay of membrane protein translocation to determine the translocation order of three extracellular domains of the archaeal membrane protein BO. Our results indicate that the N-terminal domain, the BC loop, and the FG loop are translocated in the same order as in the primary sequence. In addition, our results confirm that the N-terminal domain is translocated cotranslationally (20) and establish that this finding also is true of the BC loop. In contrast, the FG loop is translocated posttranslationally, as expected from its proximity to the C terminus. This study provides a description of the insertion order and timing under physiological conditions where translation is active.
The translocation data support the sequential insertion of transmembrane segments in BO. Nearly all of the extracellular domains are shorter than 13 amino acids, which is the minimum length required to span the lipid bilayer in extended conformation. Thus, the translocation of these domains must be coupled to the insertion of the adjacent transmembrane segments, possibly as a helical hairpin. However, the BC loop is longer than 13 amino acids, which might span the membrane and permit the insertion of helix C before helix B. Our data argue against this possibility, because the cysteine mutants at positions 74, 75, and 68 (data not shown) showed similar translocation time courses. This finding suggests that much of the BC loop is translocated at the same time and that helices B and C are inserted at close to the same time. We infer from these data that insertion of helix A occurs first, presumably by pairing with the presequence, followed by insertion of helices B and C, and finally by helices F and G. Because most of the protein is inserted cotranslationally, the D and E helices are likely to insert after helices B and C but before helices F and G. However, our data do not rule out the possibility that helices D and E are inserted posttranslationally.
Because translocation of the N-terminal domain, BC loop, and FG loop occurs sequentially, we can eliminate a concerted mechanism of transmembrane segment insertion, as proposed for bacterial toxins (25). We also can eliminate a mechanism in which a large fraction of BO is synthesized before insertion, as proposed for translocation of E. coli secreted proteins (26), because the N-terminal domain and the BC loop are clearly translocated before elongation is complete. Our results are consistent with the model that polytopic membrane proteins are inserted cotranslationally and sequentially by the secretory translocase (4, 5). H. salinarum contains SecY (C.M.A. and M.P.K., unpublished results) and other components of the secretory machinery (S. DasSarma, personal communication) that may mediate BO insertion. However, a direct demonstration of an interaction between nascent BO and the translocase is needed to test this hypothesis.
In vitro experiments have demonstrated that the ribosome forms a complex with the secretory translocase during insertion of polytopic membrane proteins into the eukaryotic endoplasmic reticulum (3). Our translocation results provide an estimate of the proximity of the ribosome to the membrane during cotranslational protein insertion in vivo. Previous studies have shown that ≈70 amino acids of a nascent polypeptide chain are protected by the ribosome–translocase complex (27). Cysteines in the N-terminal domain and the BC loop appear to be translocated when the polypeptide chain is extended past the cysteines by 125–135 amino acids, indicating that the ribosome is relatively near the membrane. It is possible that there is a lag between the synthesis of transmembrane segments and their insertion, perhaps to allow folding before insertion. Alternatively, the extracellular regions of BO may be translocated initially into a protected environment that does not permit access to AMS. For example, translocated regions trapped within the secretory translocase might be protected from derivatization but exposed on release into the lipid bilayer. Either model would support proximity of the ribosome to the membrane, which may be important for its recognition of transmembrane segments and regulation of the translocase, as suggested previously (28).
Acknowledgments
We thank A. Menon, T. Isenbarger, and R. Peck for comments on the manuscript. This work was supported in part by National Science Foundation Grant MCB-9514280 to M.P.K. and in part by a grant to the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools. C.M.A. was supported by a traineeship awarded through the Biotechnology Training Program of the National Institutes of Health (Grant 5T32 GM08349) to the University of Wisconsin.
Abbreviations
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid disodium salt
- BO
bacterioopsin
- BR
bacteriorhodopsin
- FM
fluorescein-5-maleimide
- Met
methionine
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140216497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140216497
References
- 1.Hegde R S, Lingappa V R. Cell. 1997;91:575–582. doi: 10.1016/s0092-8674(00)80445-6. [DOI] [PubMed] [Google Scholar]
- 2.Bibi E. Trends Biochem Sci. 1998;23:51–55. doi: 10.1016/s0968-0004(97)01134-1. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 4.Borel A C, Simon S M. Biochemistry. 1996;35:10587–10594. doi: 10.1021/bi960950q. [DOI] [PubMed] [Google Scholar]
- 5.Mothes W, Heinrich S U, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport T A. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 6.Wessels H P, Spiess M. Cell. 1988;55:61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- 7.Pohlschroder M, Prinz W A, Hartmann E, Beckwith J. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 8.Traxler B, Murphy C. J Biol Chem. 1996;271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]
- 9.Jander G, Cronan J E, Jr, Beckwith J. J Bacteriol. 1996;178:3049–3058. doi: 10.1128/jb.178.11.3049-3058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newitt J A, Bernstein H D. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
- 11.Ulbrandt N D, Newitt J A, Bernstein H D. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 12.Macfarlane J, Muller M. Eur J Biochem. 1995;233:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 13.de Gier J-W L, Scotti P A, Sääf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valent Q A, Scotti P A, High S, de Gier J W, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. J Mol Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 16.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 17.Seehra J S, Khorana H G. J Biol Chem. 1984;259:4187–4193. [PubMed] [Google Scholar]
- 18.Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 19.Gropp R, Gropp F, Betlach M C. Proc Natl Acad Sci USA. 1992;89:1204–1208. doi: 10.1073/pnas.89.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dale H, Krebs M P. J Biol Chem. 1999;274:22693–22698. doi: 10.1074/jbc.274.32.22693. [DOI] [PubMed] [Google Scholar]
- 21.Krebs M P, Behrens W, Mollaaghababa R, Khorana H G, Heyn M P. Biochemistry. 1993;32:12830–12834. doi: 10.1021/bi00210a035. [DOI] [PubMed] [Google Scholar]
- 22.Isenbarger T A, Krebs M P. Biochemistry. 1999;38:9023–9030. doi: 10.1021/bi9905563. [DOI] [PubMed] [Google Scholar]
- 23.Krebs M P, Mollaaghababa R, Khorana H G. Proc Natl Acad Sci USA. 1993;90:1987–1991. doi: 10.1073/pnas.90.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peck R F, DasSarma S, Krebs M P. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 25.Krishnasastry M, Walker B, Braha O, Bayley H. FEBS Lett. 1994;356:66–71. doi: 10.1016/0014-5793(94)01240-7. [DOI] [PubMed] [Google Scholar]
- 26.Randall L L. Cell. 1983;33:231–240. doi: 10.1016/0092-8674(83)90352-5. [DOI] [PubMed] [Google Scholar]
- 27.Matlack K E, Walter P. J Biol Chem. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- 28.Liao S R, Lin J L, Do H, Johnson A E. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]