Abstract
The Rac members of the Rho family GTPases control signaling pathways that regulate diverse cellular activities, including cytoskeletal organization, gene transcription, and cell transformation. Rac is implicated in apoptosis, but little is known about the mechanism by which it responds to apoptotic stimuli. Here we demonstrate that endogenous Rac GTPases are caspase 3 substrates that are cleaved in human lymphoma cells during drug-induced apoptosis. Cleavage of Rac1 occurs at two unconventional caspase 3 sites, VVGD11/G and VMVD47/G, and results in inactivation of the GTPase and effector functions of the protein (binding to the p21-activated protein kinase PAK1). Expression of caspase 3-resistant Rac1 mutants in the cells suppresses drug-induced apoptosis. Thus, proteolytic inactivation of Rac GTPases represents a novel, irreversible mechanism of Rac downregulation that allows maximal cell death following drug treatment.
Rac proteins constitute a subgroup of the Rho family of small GTPases and include the ubiquitously expressed Rac1, the myeloid lineage-specific Rac2, and the recently cloned Rac3 and Rac1b proteins found in breast cancer cells (27, 39). Activation of Rac induces reorganization of the actin cytoskeleton (16), gene transcription (34), cell transformation (26), and formation of endogenous reactive oxygen species (1). Rac has also been shown to modulate induction of apoptosis, giving either proapoptosis or survival signals, depending on the cell type and extracellular stimulus employed (2, 8). In fibroblasts and hematopoietic cells, constitutively active Rac protects cells from apoptosis induced by various stimuli, including Ras activation (21), antitumor drugs (20), UV radiation (29), death receptor activation (9), and factor deprivation (32, 33, 37). Paradoxically, Rac can also sensitize cells to induction of apoptosis by a variety of stimuli. Overexpression of active Rac1 induced apoptosis in fibroblasts (11, 42), neurons (17), and epithelial cells (13). In keeping with these observations, transgenic mice carrying a dominant active version of Rac2 showed increased apoptosis in the thymus (25). The molecular mechanisms that account for these strikingly different biological activities are poorly understood.
In general, apoptosis involves the activation of a cascade of cysteine-dependent aspartyl proteases, known as caspases, which catalyze the cleavage of cellular substrates at specific amino acid sequences (e.g., DXXD for caspase 3) and cause the biochemical and morphological changes that are characteristic of apoptotic death (7). The cytotoxicity of most cancer chemotherapy drugs is thought to derive from their ability to induce caspase activation and apoptosis (7, 28, 31). Fourteen mammalian caspases have been identified, among which caspase 3 is believed to play a pivotal role. Interestingly, a number of Rac-related regulatory proteins, including the D4 isoform of guanine nucleotide dissociation inhibitors (D4-GDI) (30), PAK2 (36), and Tiam1 (35), have been determined to be caspase substrates, and their cleavage is accompanied by the onset of apoptosis. This suggests that caspases may modulate Rho GTPase signaling pathways in addition to destroying structural components of the cell. Therefore, it is of significance to determine how the Rho GTPases are regulated in response to apoptotic stimuli and how the perturbation of Rac-related signaling pathways may contribute to cell death.
The goal of these studies was to determine whether Rac GTPases are susceptible to caspase-mediated degradation and whether such cleavage contributes to the progression of apoptosis. The results indicate that both Rac1 and Rac2 are caspase 3 substrates that are cleaved during apoptosis to generate a C-terminal fragment of 20 kDa. Proteolysis of Rac1 occurs at two unconventional caspase 3 cleavage sequences, VVGD11/G and VMVD47/G, removing the N-terminal guanine nucleotide-binding loop of the protein. Cleavage of Rac1 by caspase 3 inactivates the protein, as defined by failure to bind either to GTP or to the downstream effector protein PAK1. Caspase-mediated inactivation of Rac represents a novel, irreversible mechanism for downregulation of Rac-dependent signaling pathways and is required for maximal apoptosis in response to cytotoxic drugs.
MATERIALS AND METHODS
Reagents.
Human recombinant caspase 3 and caspase 8 and a monoclonal antibody against human Rac1 were purchased from BD Biosciences (Lexington, Ky.). Tetrapeptide inhibitors of caspase 3 (Z-DEVD-fmk), caspase 8 (Z-IETD-fmk), caspase 9 (Z-LEHD-fmk), and a general caspase inhibitor (Z-VAD-fmk) were obtained from ICN Biomedicals (Aurora, Ohio). Polyclonal antibodies against a human Rac1 C-terminal peptide (amino acids 179 to188; Rac1 C-11), a human Rac2 C-terminal peptide, (Rac2 C-11), the polyhistidine domain of the pET expression vector (His-probe G-18), and a monoclonal antibody against human N-Ras were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). Monoclonal antibody against poly(ADP-ribose) polymerase (PARP) was from Oncogene Research Products (Boston, Mass.). Monoclonal antihemagglutinin (HA) antibody was purchased from Covance Research Products (Berkeley, Calif.). VP-16 (etoposide), staurosporine, GDP, GTP, and nonhydrolyzable GTP analog guanine 5′-O-(3-thiotriphosphate) (GTP-γS) were purchased from Sigma. All radiolabeled nucleotides were from Perkin-Elmer Life Sciences.
Cell culture, transfection, and induction of apoptosis.
Burkitt's lymphoma JLP-119 and BL-41 cells were grown in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and 50 μM β-mercaptoethanol at 37°C in 5% CO2. Transfections were performed by electroporation as described (24). Transfected clones were selected by G418 resistance and tested for expression of Rac1 by immunoblotting analysis with antibodies to the HA epitope or to the human Rac1 protein. To induce apoptosis, cells were seeded at 5 × 105 cells/ml and treated with VP-16 or staurosporine at the indicated concentrations for various times. Where indicated, caspase inhibitors were added 1 h before induction of apoptosis. Apoptotic cells were assessed by nuclear morphology with Hoechst 33342-propidium iodide staining and fluorescence microscopy as described previously (24).
DNA plasmids and constructs.
The pET28a-Rac1 and pET28a-Rac2 constructs, encoding human Rac1 and Rac2 proteins, respectively, fused to (His)6, were prepared as previously described (45). A pcDNA3.1(+)-Rac1 plasmid encoding an HA-Rac1 protein was provided by Guthrie Research Institute (Sayre, Pa.). pGEX-KG-PAK1, encoding amino acids 51 to 135 of human PAK1 fused to glutathione S-transferase (GST) (GST-PBD), was a kind gift from Yi Zheng (Cincinnati Children's Hospital Research Foundation, Cincinnati, Ohio). Rac1 mutants described in this work were generated by site-directed mutagenesis with the QuickChange kit (Stratagene) as recommended by the manufacturer with primers containing the desired mutations and pET28a-Rac1 or pcDNA3.1(+)-Rac1 as a template. The His-tagged Rac1 fragments His-G12 and His-G48 were generated by PCR with internally derived oligonucleotides encoding the desired sequences and containing BamHI and EcoRI sites, followed by digestion and ligation into the pET28a vector. All constructs were confirmed by automated DNA sequencing.
Expression and purification of recombinant proteins.
Wild-type Rac1 and mutants were expressed in BL21(DE3)-Gold cells as (His)6-tagged fusions in the pET28a expression system (46). GST-PBD was expressed in Escherichia coli DH5α cells and purified by glutathione-coupled agarose chromatography as described (47). The quality of the proteins was judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentrations were determined with BCA protein assay reagents (Pierce) and confirmed visually with Coomassie blue-stained gels.
Detection of Rac cleavage.
Drug-treated cells were washed twice in ice-cold phosphate-buffered saline and lysed in lysis buffer (25 mM Tris-HCl [pH 6.8], 2% SDS, and 10% glycerol). Total protein was estimated with Coomassie Plus protein assay reagent (Pierce). Equal amounts of protein (20 μg per lane) were separated by electrophoresis with a 4 to 12% NuPAGE Bis-Tris gel (Invitrogen), and transferred to nitrocellulose membranes (Millipore) for immunoblotting. When necessary, the membrane was stripped by Restore Western blot stripping buffer (Pierce) and reprobed with appropriate antibodies. For in vitro cleavage assays, recombinant His-Rac1 protein (100 ng/μl) was incubated with purified caspase 3 or 8 at a concentration of 5 ng/μl at 37°C in caspase reaction buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EDTA, 10% [wt/vol] sucrose, 0.1% CHAPS, 10 mM dithiothreitol) (BD Biosciences). At the indicated times, the reaction was terminated by the addition of SDS sample buffer. Samples were analyzed by Western blotting. In the experiment in which we examined the cleavage of endogenous Rac1 by recombinant caspase 3, normal JLP-119 cells were lysed in caspase reaction buffer supplemented with 1 μg of leupeptin, 1 μg of aprotinin, and 40 μg of phenylmethylsulfonyl fluoride per ml, and 10 mM dithiothreitol. Cells were disrupted by nitrogen cavitation with a Parr bomb (150 lb/in2 for 2 min to 5 × 108 cells) (41). The resulting supernatant was used as endogenous Rac1 to perform the caspase cleavage assays.
Filter binding assay.
The radioactive filter binding assays measuring the retention of [3H]GDP or [γ-32P]GTP-bound Rac proteins were carried out as described (47). Briefly, [3H]GDP or [γ-32P]GTP at a specific activity of 6,000 cpm/μmol was incubated with Rac1 or mutant proteins (100 nM each) at 25°C for 1 h in 50 mM HEPES (pH 7.6)-100 mM NaCl-0.2 mg of bovine serum albumin per ml-1 mM EDTA-1 mM dithiothreitol, followed by addition of MgCl2 to a final concentration of 5 mM. The equilibrium mixture was filtered through a nitrocellulose filter (0.45 μm; Millipore), and washed with 5 ml of ice-cold buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, and 5 mM MgCl2). The amount of [3H]GDP or [γ-32P]GTP bound to the protein was measured by scintillation counting. To measure the intrinsic GTPase activity, Rac1 or mutant protein (250 nM) was first loaded with [γ-32P]GTP as described above. An aliquot of the Rac1-[γ-32P]GTP was mixed with a reaction buffer containing 50 mM HEPES (pH 7.6), 0.2 mg of bovine serum albumin per ml, and 5 mM MgCl2. At the indicated times, aliquots of the mixtures were withdrawn, and the [γ-32P]GTP that remained bound to Rac1 was subjected to nitrocellulose filtration and scintillation counting.
Rac pulldown assay.
The activity of Rac1 was assayed based on its specific binding to the p21-binding domain (PBD) of p21-activated kinase (PAK1) (3). JLP-119 cells were induced to undergo apoptosis with VP-16 as indicated. Cells were lysed in the presence of purified GST-PBD in Mg2+ lysis buffer (25 mM HEPES [pH 7.5], 1 mM EDTA, 1% NP-40, 10% glycerol, 10 mM MgCl2) supplemented with protease inhibitors and incubated at 4°C for 30 min. After broken cells and nuclei were removed by centrifugation, the supernatant was precipitated with glutathione-agarose beads (Sigma) for 30 min. The bead pellets were then washed three times with the same buffer without NP-40. To examine the interactions of Rac1 fragments with PAK1, recombinant Rac proteins (100 ng) were loaded with GDP or GTP-γS and incubated at 4°C for 30 min with 10 μg of GST-PBD immobilized on glutathione-Sepharose beads (Qiagen, Valencia, Calif.) as described (46). The resulting pellets were subjected to Western blot analysis with anti-Rac1 antibody.
RESULTS
Cleavage of Rac GTPases in lymphoma cells undergoing apoptosis.
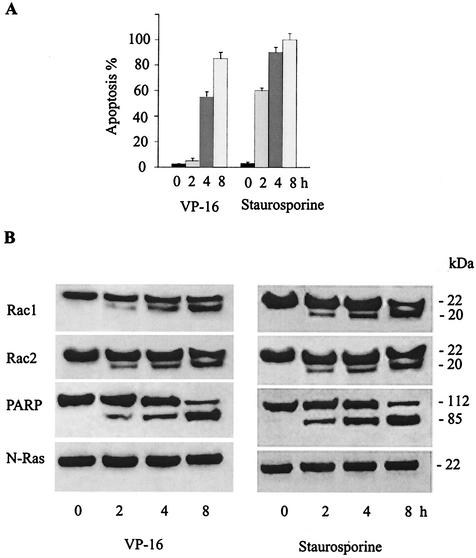
To determine whether endogenous Rac1 and Rac2 become cleaved during apoptosis, JLP-119 Burkitt's lymphoma cells were induced to undergo apoptosis with two different agents, the chemotherapy drug VP-16 (etoposide) and the protein kinase inhibitor staurosporine. Apoptosis was quantified by examining nuclear morphology following staining with Hoechst 33342 and propidium iodide. Protein cleavage was examined by Western blot immunoassay on whole-cell extracts collected at different time points after drug addition with antibodies raised against the C-terminal peptides from Rac1 and Rac2. The results (Fig. 1) show that both VP-16 and staurosporine induced extensive apoptosis in the cells. In healthy cells, Rac1 and Rac2 migrated at a molecular mass of approximately 22-kDa. However, upon treatment with either apoptosis-inducing agent, the intensity of the 22-kDa band decreased and a C-terminal cleavage product of approximately 20-kDa was formed. The accumulation of Rac cleavage product correlated with the extent of apoptosis. As expected, cleavage of the 116-kDa poly(ADP-ribose) polymerase (PARP) protein, a well-known substrate for caspase 3 (6), also occurred. In contrast, N-Ras remained intact throughout the course of apoptosis. Similar results were obtained with Burkitt's lymphoma BL-41 cells and with human breast cancer SKRB3 cells treated with VP-16 (data not shown).
FIG. 1.
Proteolytic cleavage of Rac GTPases during drug-induced apoptosis in human B lymphoma cells. (A) JLP-119 Burkitt's lymphoma cells were treated with VP-16 (8.5 μM) or staurosporine (2.0 μM) for the indicated times. Apoptosis was quantified by staining cells with Hoechst 33342 and propidium iodide and assessing nuclear morphology by fluorescence microscopy. Values are given as the percentage of total cells that are apoptotic. The results represent the means ± standard deviation from three separate experiments. (B) Extracts from the drug-treated JLP-119 cells described in the legend for panel A were subjected to Western blot immunoassay with antibodies against Rac1, Rac2, PARP, and N-Ras as described under Materials and Methods. Each lane was loaded with 20 μg of total cellular protein. Sizes are indicated to the right of the blots. The results shown are representative of three independent experiments.
Rac GTPases are substrates for caspase 3.
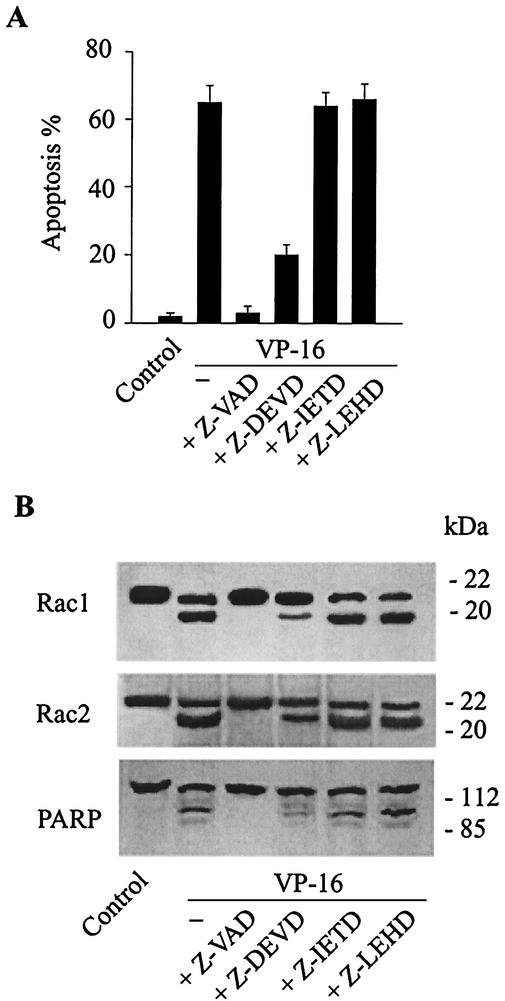
Both VP-16 and staurosporine are known to be able to activate cellular caspases (10, 15). Our observation that Rac GTPases are proteolyzed during apoptosis prompted us to investigate whether Rac was cleaved by endogenous caspases. JLP-119 cells were treated with peptide inhibitors of several different caspases prior to VP-16 treatment. As shown in Fig. 2, VP-16-induced apoptosis was blocked by a broad-spectrum caspase inhibitor, z-VAD, and was partially inhibited by the caspase 3 inhibitor z-DEVD. Peptide inhibitors of caspases 8 and 9 (z-IETD and z-LEHD, respectively) had no effect on apoptosis. Consistent with the effects on apoptosis, cleavage of Rac1, Rac2, and PARP was blocked by z-VAD, partially inhibited by z-DEVD, and unaffected by z-IETD and z-LEHD.
FIG. 2.
Proteolytic cleavage of Rac1 and Rac2 during apoptosis is inhibited by caspase 3 inhibitors. JLP-119 cells were pretreated with different concentrations of tetrapeptide inhibitors of caspase 3 (z-DEVD), caspase 8 (z-IETD), caspase 9 (z-LEHD), or a broad-spectrum caspase inhibitor (z-VAD) for 1 h and then incubated with 8.5 μM VP-16 for an additional 6 h. The extent of apoptosis (A) and cleavage of Rac1, Rac2, and PARP (B) were measured as described in the legend to Fig. 1.
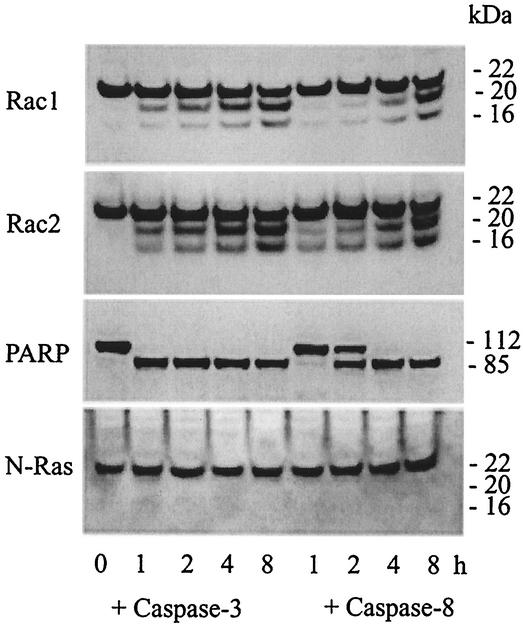
The ability of caspase 3 to cleave endogenous Rac1 and Rac2 in the intracellular milieu was confirmed by adding purified active caspase 3 to whole-cell extracts isolated from healthy lymphoma cells and measuring Rac cleavage by immunoblotting (Fig. 3). In addition to the 20-kDa fragment formed during apoptosis, a proteolytic product of approximately 16 kDa was also observed, suggesting that a second caspase site in Rac1 or Rac2 is recognized in vitro. As expected, PARP was also cleaved in these incubations, while N-Ras was not. Purified active caspase 8 also led to cleavage of Rac1, Rac2, and PARP, but the rate was significantly slower and likely resulted from cleavage and activation of endogenous pro-caspase 3 in the cell extracts (see below).
FIG. 3.
Cleavage of endogenous Rac GTPases by recombinant caspase 3 in vitro. Whole-cell extracts from healthy JLP-119 cells containing endogenous Rac GTPases were incubated for different times at 37°C with purified recombinant caspase 3 or 8 at final concentrations of 5 ng/μl in a caspase reaction buffer containing 50 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, 10% (wt/vol) sucrose, and 10 mM dithiothreitol. Proteins were examined by Western blot immunoassay with the indicated antibodies.
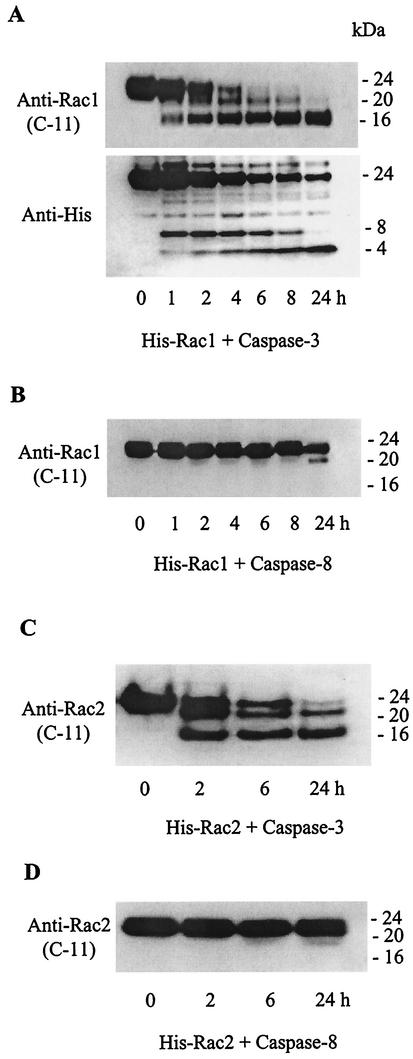
Exposure of purified samples of recombinant (His-tagged) Rac1 and Rac2 proteins (≈24 kDa) to purified active caspase 3 resulted in similar patterns of cleavage (Fig. 4), forming two C-terminal fragments of 16 and 20 kDa and two N-terminal fragments of approximately 8 and 4 kDa (detected with an antibody to the His tag on the N terminus of the recombinant proteins). Consistent with the results with the intracellular Rac proteins (see Fig. 3), caspase 8 had very little direct catalytic activity on either recombinant Rac1 or Rac2. None of the caspase 3 cleavage products were formed when the caspase 3 inhibitor z-DEVD-fmk was included in the reaction mixture (data not shown).
FIG. 4.
Treatment of recombinant Rac GTPases with recombinant caspases 3 and 8 in vitro. (A) Purified Rac1 (100 ng/μl) containing a His tag at the N terminus was incubated with caspase 3 (5 ng/μl) under the conditions described for Fig. 3. Upper panel, Western blot probed with a polyclonal antibody to Rac1 (C-11) against a C-terminal peptide of Rac1 (amino acids 177 to 184). Bottom panel, following stripping of the blot in the upper panel, it was reprobed with an anti-(His)6 antibody to detect the N-terminal fragments. (B) Western blot immunoassay of Rac1 incubated with recombinant caspase 8. (C and D) Cleavage patterns of recombinant His-tagged Rac2 incubated with caspase 3 or 8, respectively, under the conditions described for His-tagged Rac1 and analyzed with an anti-Rac2 (C-11) antibody.
Identification of caspase cleavage sites in Rac1 by site-directed mutagenesis.
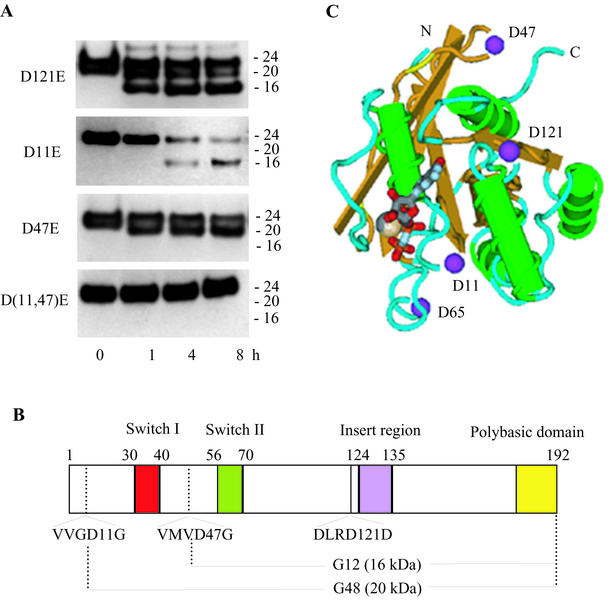
Caspase 3 has a preference for substrates having the consensus sequence DXXD (31). Knowing this, and based upon the fragment sizes estimated by Western blot immunoassay and the known amino acid sequences for the Rac proteins, we identified three potential caspase cleavage sites: Asp121, Asp47, and Asp11. None of these sequences is found in N-Ras, which is not a substrate for caspase 3. Of the putative Rac cleavage sequences, only DLRD121 represents a consensus sequence for caspase 3, while the other two sequences, VMVD47/G and VVGD11/G, would be noncanonical cleavage sites. Nonetheless, proteolytic cuts of Rac1 at Asp11 and Asp47 would lead to C-terminal fragments of 20 and 16 kDa, respectively.
To confirm whether these aspartyl residues represent the cleavage sites, each was mutated to a glutamyl residue, and the resulting mutants were tested for their susceptibility to cleavage by purified caspase 3. The results (Fig. 5A) show that the D11E and D47E point mutants prevented formation of one or the other cleavage fragment. A double mutant protein containing Glu substitutions at both Asp11 and Asp47 was completely resistant to caspase 3 cleavage. In contrast, the Glu substitution at Asp121 had no effect on cleavage of Rac1 by caspase 3. A schematic linear diagram showing the functional domains, the sites of cleavage of Rac1 by caspase 3, and the resulting 16- and 20-kDa polypeptide fragments (designated G48 and G12, respectively) is shown in Fig. 5B. A diagram of the three-dimensional structure of Rac1 with the locations of the critical Asp residues is shown in Fig. 5C.
FIG. 5.
Identification of potential and actual caspase 3 cleavage sites in Rac1. (A) Caspase 3 treatment (5 ng/μl for the indicated times) and Western blot immunoassay (anti-Rac1 C terminus antibody) of purified, recombinant His-tagged Rac1 mutant proteins (100 ng/μl) containing Asp→Glu mutations at amino acid position 121, 11, or 47, or both 11 and 47. (B) Schematic diagram showing the key functional domains and caspase 3 cleavage sites of Rac1. Also shown are the truncated 20- and 16-kDa C-terminal proteins (G12 and G48) that result from caspase 3 cleavage at Asp11 and Asp47, respectively. (C) Schematic diagram of the tertiary structure of Rac1 (derived from the crystal structure of human Rac1-GMPPNP) (18) showing the critical Asp11 and Asp47 residues and the nucleotide and magnesium binding sites.
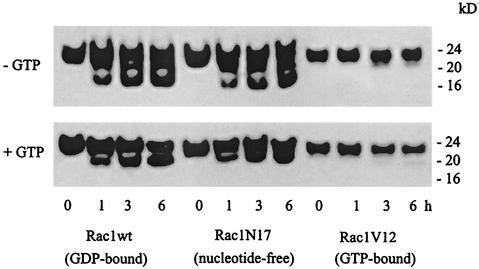
In apoptotic cells, Rac1 only underwent cleavage at the Asp11 site (see Fig. 1 and 2) to generate the 20-kDa band, but in vitro, recombinant Rac1 exposed to purified active caspase 3 was cleaved at both Asp11 and Asp47 (see Fig. 3 to 5). The in vitro assay was carried out in the absence of free nucleotides, but the cells are known to contain high levels (≈150 μM) of free GTP. To determine whether the presence of free nucleotides influenced the cleavage of Rac1 by caspase 3, experiments were carried out in the presence and absence of added GTP (100 μM). With wild-type Rac1 as the substrate, only the 20-kDa band was formed, mimicking the intracellular cleavage pattern (Fig. 6, lower panel). A similar effect was observed when GDP was added to the caspase reaction mixture (data not shown). Similar experiments were carried out with two cloned mutants of Rac1 that differ from the wild-type protein in their nucleotide binding capacities: a nucleotide-depleted, dominant-negative form of Rac 1 denoted Rac1N17, and a dominant-active, GTP-bound mutant denoted Rac1V12 (9, 33). As shown in Fig. 6, Rac1N17 was cleaved at essentially the same rate and with essentially the same pattern as wild-type Rac1. In contrast, Rac1V12 was relatively resistant to cleavage by caspase 3. These results show that caspase-mediated cleavage of Rac GTPases is somehow influenced by the presence of excess guanine nucleotides.
FIG. 6.
Effect of guanine nucleotides on caspase 3-mediated cleavage of wild-type and mutant Rac1. Recombinant purified His-tagged wild-type Rac1 (Rac1wt), a constitutively active, GTP-bound Rac1 (Rac1V12), and a dominant-negative, nucleotide-depleted form of Rac1 (Rac1N17), all at 100 ng/μl, were treated with caspase 3 (5 ng/μl) in caspase buffer in the absence or presence of 100 μM GTP. The cleavage of Rac1 proteins was assessed at the indicated times by immunoblotting with anti-Rac1 (C-11) antibody, as described for Fig. 1.
Inactivation of Rac1 by caspase 3 cleavage.
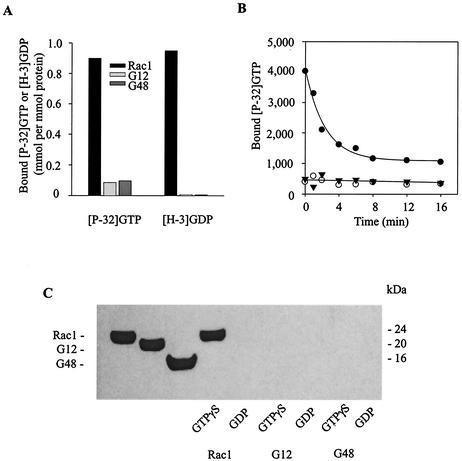
Rac GTPases are characterized functionally by their binding to and hydrolysis of guanine nucleotides and by their binding to and activation of effector proteins. Asp11 and Asp47 are located, respectively, in the nucleotide binding loop and the effector domain of Rac1 (see Fig. 5C). To determine how cleavage at these sites affects Rac1 activity, we assayed for GTPase activity in two Rac1 mutant proteins that represent the large caspase 3 cleavage products G12 (amino acids 12 to 192) and G48 (amino acids 48 to 192) (20 and 16 kDa, respectively) (Fig. 5B). Wild-type Rac1 bound to both GTP and GDP and hydrolyzed GTP effectively (Fig. 7A and B). In contrast, neither G12 nor G48 demonstrated either of these activities. As a result, only native Rac1 in the GTP-γS-bound form interacted with PBD; the truncated peptides did not bind measurably to the effector protein regardless of whether they were incubated with GTP-γS or GDP (Fig. 7C).
FIG. 7.
Inactivation of Rac1 by caspase 3 cleavage. (A) Recombinant purified His-tagged proteins representing wild-type Rac1 and the truncated proteins Rac1-G12 and Rac1-G48 were incubated with [γ-32P]GTP (left three bars) or [3H]GDP (right three bars) at room temperature for 30 min to achieve equilibrium. Bound nucleotide was quantified by a radioactive filter-binding assay as described under Materials and Methods. (B) Time courses of hydrolysis of [γ-P32]GTP by Rac1 (solid circles) and the truncated Rac1 G12 (open circles) and G48 (solid triangles) proteins. (C) Western blot immunoassay to test for binding of Rac1 and the G12 and G48 fragments to the p21-binding domain (PBD) of PAK1. Recombinant purified proteins (100 ng) (shown in the left three lanes) were loaded with GTP-γS or GDP and then incubated for 30 min at 4°C with 10 μg of GST-tagged PBD, followed by precipitation with glutathione-Sepharose beads. After each binding reaction, the proteins bound to the beads were eluted with SDS sample buffer, separated by SDS-PAGE, and visualized by anti-Rac1 (C-11) Western blot immunoassay.
Loss of Rac1 activity during apoptosis.
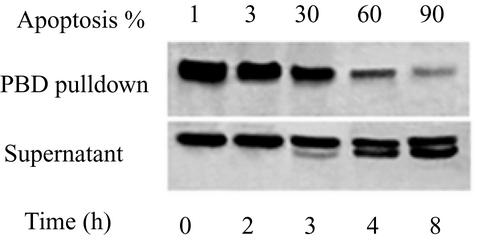
The same PBD binding assay was used to assess the activity of Rac1 in cells induced to undergo apoptosis with VP-16. As apoptosis proceeded, the amount of GTP-Rac1 bound to PBD gradually decreased (Fig. 8). At the same time, an increasing fraction of the Rac1 remaining in the supernatants showed proteolytic cleavage at the Asp11 site, resulting in the formation of an endogenous G12 protein. As shown in Fig. 7, generation of G12 can cause loss of binding to PBD.
FIG. 8.
Inactivation of Rac1 during apoptosis. JLP-119 cells were induced to undergo apoptosis by treatment with VP-16 (8.5 μM) for the indicated times. Cells were lysed by sonication in the presence of GST-tagged PBD and precipitated by mixing with glutathione-Sepharose beads. The presence of Rac1 was detected by immunoblotting with anti-Rac1 (C-11). Top panel, active Rac1 present in the GST-PBD precipitates; bottom panel, Rac1 remaining in the supernatant fractions after mixing with GST-PBD beads. The percentage of apoptotic cells at each time point is indicated above each lane. The results are representative of three independent experiments.
Expression of caspaseresistant Rac1 mutants suppresses apoptosis.
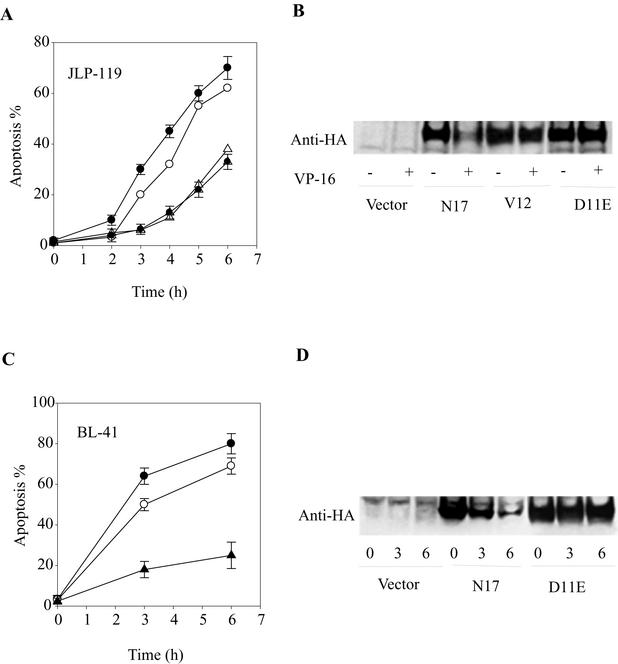
If the inactivation of Rac1 by caspase 3-like caspases is required for VP-16-induced apoptosis, then blocking Rac1 cleavage should suppress apoptosis. To test this hypothesis, JLP-119 and BL-41 lymphoma cells were stably transfected with vectors encoding different HA-tagged Rac1 mutants: the caspaseresistant point mutant D11E and double mutant D(11,47)E, Rac1N17, Rac1V12, or empty vector. Figure 9 shows the results obtained when the different cell lines were treated with VP-16. When expressed in JLP-119 cells (Fig. 9A and B), D11E and D(11,47)E (not shown) showed significant resistance (≈50%) to apoptosis induced by VP-16 compared to control cells expressing vector alone. Interestingly, cells expressing Rac1V12 showed essentially the same protection against apoptotic death. In contrast, expression of the dominant-negative Rac1N17 mutant showed slightly increased apoptosis in response to VP-16.
FIG. 9.
Expression of caspase 3-resistant Rac1 mutants in JLP-119 and BL-41 cells inhibits drug-induced apoptosis. Stable transfectants of JLP-119 cells (A and B) or BL-41 cells (C and D) were treated with 8.5 μM or 170 μM VP-16, respectively, for the indicated times. The cell lines expressed either empty pCDNA3.1(+) vector (open circles), HA-tagged dominant-negative Rac1N17 (solid circles), constitutively active Rac1V12 (open triangles), or caspaseresistant Rac1D11E (solid triangles). Apoptosis was assayed by nuclear morphology following Hoechst dye and propidium iodide staining and fluorescence microscopy (A and C). Expression of HA-tagged recombinant Rac1 protein was followed by Western blot immunoassay with an anti-HA antibody (B and D).
Despite their influence on drug-induced killing, neither the active nor the nonfunctional Rac1 mutants had any effect on cell death in the absence of apoptotic stimuli (data not shown). Immunoblotting analysis of JLP-119 cell extracts with anti-HA antibodies (Fig. 9B) revealed that Rac1N17, like the endogenous Rac1 protein (see Fig. 1 and 2) underwent significant degradation in the cells during the course of apoptosis. In contrast, Rac1V12 showed much less proteolysis, and, as expected, the D11E mutant was completely resistant to degradation during VP-16-induced apoptosis. These results are consistent with those obtained with the purified recombinant proteins (see Fig. 6). Similar results were obtained with BL-41 cells stably expressing HA-tagged Rac1D11E and Rac1N17 (Fig. 9C and D).
DISCUSSION
In these studies, we demonstrate that (i) endogenous Rac1 is cleaved by caspase 3 or a related enzyme during drug-induced apoptosis, (ii) the cleavage occurs at a noncanonical caspase cleavage site, (iii) this cleavage results in the inability of Rac1 to bind guanine nucleotides, catalyze GTP hydrolysis, and bind to downstream effector proteins such as p21-activated protein kinase (PAK1), and (iv) interference with Rac1 cleavage by expression of caspaseresistant mutants inhibits apoptosis. Overall, the results suggest that native Rac1 activity interferes with the apoptotic process and needs to be diminished in order to maximize cell killing by chemotherapy drugs. The results also provide a novel mechanism for the direct and irreversible downregulation of Rac GTPases in apoptotic cells.
Our finding that native endogenous Rac is antiapoptotic is consistent with several studies examining the effects of transfected Rac1 proteins. Thus, apoptosis induced by growth factor deprivation or treatment with tumor necrosis factor or antitumor drugs was inhibited by overexpression of a dominant-active version of Rac1 (Rac1-V12 or Rac1-L61) (4, 9, 20, 33, 37). The current work is the first to show that endogenous Rac1 is actually inactivated during apoptosis and that this inactivation stimulates apoptosis.
The molecular mechanism through which Rac inactivation promotes apoptosis has yet to be elucidated. One potential pathway would derive from the loss of PAK1 activity that occurs when Rac1 is inactivated. PAK1 is thought to promote cell survival by phosphorylating and inactivating Bad, a proapoptotic member of the Bcl-2 family of proteins, resulting in a reduced interaction between Bad and Bcl-2 or Bcl-x(L) and the increased association of Bad with 14-3-3 (40). Loss of PAK1 activity would thus relieve suppression of a proapoptotic factor and cause apoptosis. A similar mechanism would come from loss of phosphatidylinositol 3-kinase-Akt survival signaling by Rac inactivation, resulting in decreased Akt kinase activity and, again, decreased phosphorylation and inhibition of Bad activity (29, 40). Regardless of the molecular mechanisms involved, it may be expected that cleavage and inactivation of Rac1 will influence the cell morphology changes that are such a hallmark of apoptosis, since it is well established that Rac1 modulates the actin cytoskeleton (8, 16). Further experiments will be required to clarify the mechanism by which the caspasemediated inactivation of Rac1 promotes apoptotic cell death.
In contrast to the antiapoptotic function of Rac described above, there are several studies showing that Rac1, Rac2, and other members of the Rho family GTPases can induce apoptosis in cells (11-13, 17, 25, 42). For example, expression of Rac1-L61 in NIH 3T3 murine fibroblasts promoted apoptosis after serum deprivation (11). The apparent paradox in the pro- and antiapoptotic effects of Rac GTPases may be attributed to differences in the cell types, stimuli, and expression levels of the proteins being studied. In support of this notion, it has been shown that within a single cell type, Ras can induce either proliferation or apoptosis depending on its level of expression (21). Overall, however, additional studies are necessary to sort out the conditions under which Rac-dependent pathways are pro- or antiapoptotic.
Consistent with our finding of endogenous Rac1 cleavage by caspase 3, Tu and Cerione showed that transiently transfected, HA-tagged Cdc42 and Rac1 are cleaved during Fas-induced apoptosis in NIH 3T3 cells (44). The effects of this cleavage on GTPase activity and downstream effector functions were not examined, nor was it determined whether the cleavage actually plays a role in controlling apoptosis. Based upon their studies with Cdc42, those authors concluded that the cleavage occurs at the conventional caspase 3 cleavage site (DLRD121/D) in the protein (44). This potential cleavage site is present in most Rho GTPases (see Fig. 5 in this article). Our results differ and show through molecular analysis that in Rac1, this site is resistant to caspase 3 cleavage. Instead, cleavage of Rac1 by caspase 3 occurred at noncanonical sequences for this protease, XXXD/G (specifically, VVGD11/G and VMVD47/G) instead of DXXD, despite the presence of a caspase 3 consensus sequence (DLRD121). The reason for the discrepancy in the results is unclear but may be related to a difference in the conformation in Cdc42 compared to Rac1 or to the examination solely of transfected recombinant Cdc42 protein in the previous studies; we examined both endogenous (native) and recombinant Rac1.
Other examples where caspase 3 utilizes noncanonical sites are seen in the cleavage of protein kinase N at ATHD/G (43), human Rad51 at AQVD/G (14), topoisomerase I at EEED/G (38), and scaffold attachment factor A at SALD/G (22). A common feature of these noncanonical cleavage sites is that the P1 position is followed by a neutral residue (Gly). It has been suggested that protein structural elements other than primary sequence, such as protein conformation, can influence substrate recognition by caspases and that preferred caspase 3 cleavage sites are found in β-sheet conformations that are exposed and not in alpha helices. As shown in the tertiary structure of Rac1 (Fig. 5C), Asp121 is buried in an α-helix, while Asp11 and Asp47 are located in β-sheets and are relatively exposed. It should be noted that in the presence of free excess GTP or GDP, as occurs in cells, the cleavage at Asp47 is lost, presumably due to induction of a conformational change in the protein.
Interestingly, Rac1V12 is not only constitutively active but also caspase 3 resistant, as demonstrated both in vitro and in the cells (Fig. 6 and 9). This most likely arises from the proximity of the mutated residue G12V to the Asp11 cleavage site in Rac1 and does not appear to depend on the nucleotide binding state of the protein (see Fig. 6). The relative resistance of Rac1V12 to caspase cleavage probably explains why this protein also inhibited apoptosis when transfected into cells.
Like the Ras family of proteins, Rac GTPases cycle between an active GTP-bound state and an inactive GDP-bound state. Only the active (GTP-bound) forms of the GTPases can transmit receptor-mediated signals by interacting with specific effectors, including a family of serine/threonine protein kinases termed PAKs. Rac GTPase activities are coordinately regulated by at least three classes of proteins — guanine nucleotide exchange factors, GTPase-activating proteins, and Rho guanine nucleotide dissociation inhibitors. Upon growth factor receptor stimulation, guanine nucleotide exchange factors stimulate the exchange of bound GDP for GTP, leading to the activation of GTPase activity. GTPase-activating proteins promote the inactivation of the GTPases by stimulating the intrinsic GTP hydrolysis rate of Rac-like proteins. Association of Rac proteins with guanine nucleotide dissociation inhibitors can block the exchange of GDP for GTP and are thought to keep the GTPases in an inactive form in the cytosol. Our finding that Rac GTPases serve as substrates for caspases provides an additional mechanism for regulation of Rac activity during apoptosis. Proteolysis of Rac1 abolished its GTP-binding/hydrolysis ability. Hence, the level of active Rac1 decreased progressively in the apoptotic cells, in parallel with apoptosis progression. Decreased Rac1 activity has also been observed in HMN1 motor neuron cells undergoing apoptosis after ceramide treatment (35). Thus, in addition to the conventional regulation of GTPase cycling, the proteolytic degradation of Rac provides an irreversible mechanism for downregulation of Rac-dependent signaling pathways.
Alternative mechanisms also exist for downregulating Rac family signaling pathways during apoptosis; for example, Tiam1 (a Rac-specific activator) also becomes cleaved and inactivated during apoptosis, leading to loss of Rac activity during apoptosis (35). D4-GDI inhibitor, a negative regulator of Rac GTPases, is also cleaved by caspases during apoptosis, but the effect of this cleavage on Rac activity is unclear (23, 30). Interestingly, transfection of Rac1 mutants did not affect background levels of apoptosis in the cells, consistent with observations made with Chinese hamster ovary cells (42). The finding that Rac inactivation is required for maximal drug-induced apoptosis is consistent with a large body of evidence indicating that Rac1 promotes cell growth (5, 19). Termination of Rac-mediated growth signals can thus lead to cell death.
Acknowledgments
We thank Yang-Ja Lee for help in setting up the electroporation transfections.
REFERENCES
- 1.Archer, H., and D. Bar-Sagi. 2002. Ras and Rac as activators of reactive oxygen species (ROS). Methods Mol. Biol. 189:67-73. [DOI] [PubMed] [Google Scholar]
- 2.Aznar, S., and J. C. Lacal. 2001. Rho signals to cell growth and apoptosis. Cancer Lett. 165:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Benard, V., B. P. Bohl, and G. M. Bokoch. 1999. Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils with a novel assay for active GTPases. J. Biol. Chem. 274:13198-13204. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, J. E., O. V. Chaika, and R. E. Lewis. 1999. Rac-dependent anti-apoptotic signaling by the insulin receptor cytoplasmic domain. J. Biol. Chem. 274:28632-28636. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch, G. M. 2000. Regulation of cell function by Rho family GTPases. Immunol. Res. 21:139-148. [DOI] [PubMed] [Google Scholar]
- 6.Boulares, A. H., A. G. Yakovlev, V. Ivanova, B. A. Stoica, G. Wang, S. Iyer, and M. Smulson. 1999. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 274:22932-22940. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, G. M. 1997. Caspases: the executioners of apoptosis. Biochem. J. 326:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, M. L., and M. F. Olson. 2002. Rho GTPase signalling pathways in the morphological changes associated with apoptosis. Cell Death Differ. 9:493-504. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, S. S., P. Angkeow, J. Huang, M. Ozaki, and K. Irani. 2000. Rac1 inhibits TNF-alpha-induced endothelial cell apoptosis: dual regulation by reactive oxygen species. FASEB J. 14:1705-1714. [DOI] [PubMed] [Google Scholar]
- 10.Droin, N., L. Dubrez, B. Eymin, C. Renvoize, J. Breard, M. T. Dimanche-Boitrel, and E. Solary. 1998. Upregulation of CASP genes in human tumor cells undergoing etoposide-induced apoptosis. Oncogene 16:2885-2894. [DOI] [PubMed] [Google Scholar]
- 11.Embade, N., P. F. Valeron, S. Aznar, E. Lopez-Collazo, and J. C. Lacal. 2000. Apoptosis induced by Rac GTPase correlates with induction of FasL and ceramide production. Mol. Biol. Cell 11:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteve, P., N. Embade, R. Perona, B. Jimenez, L. del Peso, J. Leon, M. Arends, T. Miki, and J. C. Lacal. 1998. Rho-regulated signals induce apoptosis in vitro and in vivo by a p53-independent, but BclII dependent pathway. Oncogene 17:1855-1869. [DOI] [PubMed] [Google Scholar]
- 13.Fiorentini, C., P. Matarrese, E. Straface, L. Falzano, G. Donelli, P. Boquet, and W. Malorni. 1998. Rho-dependent cell spreading activated by E. coli cytotoxic necrotizing factor 1 hinders apoptosis in epithelial cells. Cell Death Differ. 5:921-929. [DOI] [PubMed] [Google Scholar]
- 14.Flygare, J., D. Hellgren, and A. Wennborg. 2000. Caspase-3 mediated cleavage of HsRad51 at an unconventional site. Eur. J. Biochem. 267:5977-5982. [DOI] [PubMed] [Google Scholar]
- 15.Gescher, A. 2000. Staurosporine analogues—pharmacological toys or useful antitumour agents? Crit. Rev. Oncol. Hematol. 34:127-135. [DOI] [PubMed] [Google Scholar]
- 16.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 17.Harrington, A. W., J. Y. Kim, and S. O. Yoon. 2002. Activation of Rac GTPase by p75 is necessary for c-jun N-terminal kinase-mediated apoptosis. J. Neurosci. 22:156-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirshberg, M., R. W. Stockley, G. Dodson, and M. R. Webb. 1997. The crystal structure of human Rac1, a member of the Rho-family complexed with a GTP analogue. Nat. Struct. Biol. 4:147-152. [DOI] [PubMed] [Google Scholar]
- 19.Jaffe, A. B., and A. Hall. 2002. Rho GTPases in transformation and metastasis. Adv. Cancer Res. 84:57-80. [DOI] [PubMed] [Google Scholar]
- 20.Jeong, H. G., H. J. Cho, I. Y. Chang, S. P. Yoon, Y. J. Jeon, M. H. Chung, and H. J. You. 2002. Rac1 prevents cisplatin-induced apoptosis through downregulation of p38 activation in NIH 3T3 cells. FEBS Lett. 518:129-134. [DOI] [PubMed] [Google Scholar]
- 21.Joneson, T., and D. Bar-Sagi. 1999. Suppression of Ras-induced apoptosis by the Rac GTPase. Mol. Cell. Biol. 19:5892-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kipp, M., B. L. Schwab, M. Przybylski, P. Nicotera, and F. O. Fackelmayer. 2000. Apoptotic cleavage of scaffold attachment factor A (SAF-A) by caspase 3 occurs at a noncanonical cleavage site. J. Biol. Chem. 275:5031-5036. [DOI] [PubMed] [Google Scholar]
- 23.Krieser, R. J., and A. Eastman. 1999. Cleavage and nuclear translocation of the caspase 3 substrate Rho GDP-dissociation inhibitor, D4-guanine nucleotide dissociation inhibitors, during apoptosis. Cell Death Differ. 6:412-419. [DOI] [PubMed] [Google Scholar]
- 24.Lee, Y., and E. Shacter. 1997. Bcl-2 does not protect Burkitt's lymphoma cells from oxidant-induced cell death. Blood 89:4480-4492. [PubMed] [Google Scholar]
- 25.Lores, P., L. Morin, R. Luna, and G. Gacon. 1997. Enhanced apoptosis in the thymus of transgenic mice expressing constitutively activated forms of human Rac2GTPase. Oncogene 15:601-605. [DOI] [PubMed] [Google Scholar]
- 26.Meriane, M., S. Charrasse, F. Comunale, A. Mery, P. Fort, P. Roux, and C. Gauthier-Rouviere. 2002. Participation of small GTPases Rac1 and Cdc42Hs in myoblast transformation. Oncogene 21:2901-2907. [DOI] [PubMed] [Google Scholar]
- 27.Mira, J. P., V. Benard, J. Groffen, L. C. Sanders, and U. G. Knaus. 2000. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc. Natl. Acad. Sci. USA 97:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mow, B. M., A. L. Blajeski, J. Chandra, and S. H. Kaufmann. 2001. Apoptosis and the response to anticancer therapy. Curr. Opin. Oncol. 13:453-462. [DOI] [PubMed] [Google Scholar]
- 29.Murga, C., M. Zohar, H. Teramoto, and J. S. Gutkind. 2002. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-κB. Oncogene 21:207-216. [DOI] [PubMed] [Google Scholar]
- 30.Na, S., T. H. Chuang, A. Cunningham, T. G. Turi, J. H. Hanke, G. M. Bokoch, and D. E. Danley. 1996. D4-guanine nucleotide dissociation inhibitors, a substrate of CPP32, is proteolyzed during Fas-induced apoptosis. J. Biol. Chem. 271:11209-11213. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson, D. W., and N. A. Thornberry. 1997. Caspases: killer proteases. Trends Biochem. Sci. 22:299-306. [DOI] [PubMed] [Google Scholar]
- 32.Nishida, K., Y. Kaziro, and T. Satoh. 1999. Anti-apoptotic function of Rac in hematopoietic cells. Oncogene 18:407-415. [DOI] [PubMed] [Google Scholar]
- 33.Pervaiz, S., J. Cao, O. S. Chao, Y. Y. Chin, and M. V. Clement. 2001. Activation of the RacGTPase inhibits apoptosis in human tumor cells. Oncogene 20:6263-6268. [DOI] [PubMed] [Google Scholar]
- 34.Philips, A., P. Roux, V. Coulon, J. M. Bellanger, A. Vie, M. L. Vignais, and J. M. Blanchard. 2000. Differential effect of Rac and Cdc42 on p38 kinase activity and cell cycle progression of nonadherent primary mouse fibroblasts. J. Biol. Chem. 275:5911-5917. [DOI] [PubMed] [Google Scholar]
- 35.Qi, H., P. Juo, J. Masuda-Robens, M. J. Caloca, H. Zhou, N. Stone, M. G. Kazanietz, and M. M. Chou. 2001. Caspase-mediated cleavage of the TIAM1 guanine nucleotide exchange factor during apoptosis. Cell Growth Differ. 12:603-611. [PubMed] [Google Scholar]
- 36.Rudel, T., and G. M. Bokoch. 1997. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276:1571-1574. [DOI] [PubMed] [Google Scholar]
- 37.Ruggieri, R., Y. Y. Chuang, and M. Symons. 2001. The small GTPase Rac suppresses apoptosis caused by serum deprivation in fibroblasts. Mol. Med. 7:293-300. [PMC free article] [PubMed] [Google Scholar]
- 38.Samejima, K., P. A. Svingen, G. S. Basi, T. Kottke, P. W. Mesner, Jr., L. Stewart, F. Durrieu, G. G. Poirier, E. S. Alnemri, J. J. Champoux, S. H. Kaufmann, and W. C. Earnshaw. 1999. Caspase-mediated cleavage of DNA topoisomerase I at unconventional sites during apoptosis. J. Biol. Chem. 274:4335-4340. [DOI] [PubMed] [Google Scholar]
- 39.Schnelzer, A., D. Prechtel, U. Knaus, K. Dehne, M. Gerhard, H. Graeff, N. Harbeck, M. Schmitt, and E. Lengyel. 2000. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19:3013-3020. [DOI] [PubMed] [Google Scholar]
- 40.Schurmann, A., A. F. Mooney, L. C. Sanders, M. A. Sells, H. G. Wang, J. C. Reed, and G. M. Bokoch. 2000. p21-activated kinase 1 phosphorylates the death agonist Bad and protects cells from apoptosis. Mol. Cell. Biol. 20:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senturker, S., R. Tschirret-Guth, J. Morrow, R. Levine, and E. Shacter. 2002. Induction of apoptosis by chemotherapeutic drugs without generation of reactive oxygen species. Arch. Biochem. Biophys. 397:262-272. [DOI] [PubMed] [Google Scholar]
- 42.Subauste, M. C., M. Von Herrath, V. Benard, C. E. Chamberlain, T. H. Chuang, K. Chu, G. M. Bokoch, and K. M. Hahn. 2000. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 275:9725-9733. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, M., H. Mukai, M. Toshimori, M. Miyamoto, and Y. Ono. 1998. Proteolytic activation of PKN by caspase 3 or related protease during apoptosis. Proc. Natl. Acad. Sci. USA 95:11566-11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu, S., and R. A. Cerione. 2001. Cdc42 is a substrate for caspases and influences Fas-induced apoptosis. J. Biol. Chem. 276:19656-19663. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, B., J. Chernoff, and Y. Zheng. 1998. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J. Biol. Chem. 273:8776-8782. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, B., Y. Gao, S. Y. Moon, Y. Zhang, and Y. Zheng. 2001. Oligomerization of Rac1 GTPase mediated by the carboxyl-terminal polybasic domain. J. Biol. Chem. 276:8958-8967. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, B., Z. X. Wang, and Y. Zheng. 1997. Characterization of the interactions between the small GTPase Cdc42 and its GTPase-activating proteins and putative effectors. Comparison of kinetic properties of Cdc42 binding to the Cdc42-interactive domains. J. Biol. Chem. 272:21999-22007. [DOI] [PubMed] [Google Scholar]