Abstract
Endothelial cells differentiate from mesoderm-derived precursors to initiate the earliest events in vascular development. Although the signaling events that regulate the successive steps of vascular development are known in some detail, the transcriptional processes that regulate the first steps in vasculogenesis are not well defined. We have studied the regulatory mechanisms of flk1 expression as a model to understand the upstream events in endothelial cell differentiation, since flk1 is the earliest marker of endothelial precursors. Using a variety of biochemical approaches, we identified a cis-acting element in the first intron of the flk1 gene that is required for endothelium-dependent expression in transgenic reporter gene assays. Using the yeast one-hybrid system, we identified HoxB5 as the transcription factor that binds this cis-acting element, the HoxB5-binding element (HBE). HoxB5 mRNA colocalized with flk1 expression in differentiating embryoid bodies, and HoxB5 potently transactivated the flk1 promoter in an HBE-dependent fashion in transient-transfection assays. Overexpression of HoxB5 led to expansion of flk1+ angioblasts in differentiating embryoid bodies and increased the number of PECAM (platelet-endothelial cell adhesion molecule)-positive primitive blood vessels. HoxB5 is necessary and sufficient to activate the cell-intrinsic events that regulate the differentiation of angioblasts and mature endothelial cells from their mesoderm-derived precursors.
The vascular endothelium is a continuous surface that forms a boundary between circulating hematopoietic cells and the underlying mural vascular cells. In addition to its barrier function, the endothelium integrates signals from the circulation to modulate vascular tone and is a major determinant of the inflammatory process by regulating activation and adhesion of white blood cells. Appropriate activation of the endothelium is also the necessary proximate event for growth of new blood vessels from existing vessels during development and in physiologic and pathological conditions via the process of angiogenesis. Even earlier in development, the first blood vessels form in situ by migration and differentiation of endothelial precursors, or angioblasts, via the process of vasculogenesis. Although angiogenesis and vasculogenesis have distinguishing features, the signaling events that regulate these processes are, in most cases, overlapping.
The cellular and developmental processes that coordinate endothelial cell differentiation are only now being well explained. The earliest marker of cells fated to the endothelial lineage is the receptor for vascular endothelial growth factor (VEGF), flk1 (VEGFR-2; the human homolog is also known as KDR). flk1 is first expressed at day 7.0 postconception (p.c.) in the mouse embryo in the yolk sac mesoderm and in the lateral plate mesoderm of the embryo proper (50). Deletion of flk1 by homologous recombination results in absence of both endothelial and hematopoietic cells, suggesting that early populations of flk1-expressing cells define a bipotential precursor for these two lineages (41). This conclusion has been supported by the identification of flk1+ blast colony-forming cells that give rise to hematopoietic and endothelial lineage progeny in in vitro assays (11). These early bipotential cells have been termed hemangioblasts. Identification of hemangioblasts in vivo, however, has proven to be an elusive task, and their existence remains controversial (14, 26). The weight of evidence seems to be in favor of a hemangioblast at some point during development, although the extent of its spatiotemporal distribution is unclear. In any event, it is accepted that flk1+ cells define a major population of angioblasts and that definitive endothelial cells maintain flk1 expression, whereas hematopoietic populations do not.
In contrast to other lineages, our appreciation of the transcriptional events that determine commitment down the endothelial developmental pathway is limited. The basic helix-loop-helix protein SCL/tal-1 is the transcription factor most closely linked to the earliest stages of endothelial cell differentiation. flk1 and SCL/tal-1 are coexpressed in presumptive angioblasts within the lateral plate mesoderm (14), although temporally SCL/tal-1 expression follows that of flk1 (17), and targeted deletion of SCL/tal-1 affects early hematopoietic differentiation and angiogenic remodeling of the yolk sac but not endothelial differentiation (35, 39, 42). However, expression of SCL/tal-1 under the flk1 promoter in flk1-null embryos and embryonic stem cells results in partial rescue of endothelial cell development in vivo and in vitro, suggesting that a combinatorial genetic effect of flk1 and SCL/tal-1 in establishing the endothelial lineage after flk1 expression is initiated in mesoderm-derived progenitors (16). VEZF1/DB1 is a zinc finger transcription factor that is also specifically expressed during early stages of endothelial differentiation (49). However, VEZF1/DB1 does not regulate the earliest set of angioblast markers (such as flk1), at least in transient-transfection assays (1). VEZF1/DB1 more likely participates in later events in vascular development, although a complete analysis of its function—including its deletion by homologous recombination—has yet to be reported.
We have used formal analysis of the flk1 promoter as a model to understand the proximate events in the commitment of lateral mesoderm precursors to the endothelial lineage. The 5′-flanking sequence of the human flk1 gene supports high-level reporter gene expression in cell culture (32), and the transcription factors Sp1, TFII-I, and GATA-2 are implicated in regulation via the flk1 promoter (30, 33, 48). However, large fragments of the flk1 5′-flanking sequence from either the mouse or human genes do not support reporter gene expression in transgenic mice (C. Patterson, unpublished observations), indicating that other elements are necessary for expression within the endothelium. Recently, the Breier laboratory has identified a 510-bp enhancer-like element in the distal portion of the first intron of the mouse flk1 gene that confers endothelium-specific expression in vivo (23), indicating that proteins regulating flk1 expression through this element are likely bona fide upstream contributors to the determination of the endothelial cell lineage. Binding sites for GATA and Ets family members have been tested deductively based on sequence analysis of this fragment and are indeed necessary for its activity (24). However, proteins from these families are likely to be only ancillary factors in flk1 expression, since genetic studies have not implicated any of them as being clearly upstream of angioblast determination. (Interestingly, putative SCL/tal-1 sites are present in the flk1 element defined by these studies, but they are dispensable for reporter gene expression in transgenic mice [24].)
As an alternative approach, we have searched for potential upstream transcriptional regulators of endothelial cell differentiation by using an unbiased, inductive approach. Using a combination of molecular, biochemical, and genetic approaches, we identified HoxB5 (a member of the classical and highly conserved family of homeobox proteins) as a potent transcriptional regulator of flk1 expression. In addition, we have found that the HoxB5-binding site in the flk1 gene is required for expression within the vascular endothelium during development. Most importantly, we show here that overexpression of HoxB5 increases the number of angioblasts during embryonic stem cell differentiation (as determined by measuring the number of flk1+ cells at early stages of angioblast differentiation in this system) and also the number of mature endothelial cells, which form primitive blood vessels and express platelet-endothelial cell adhesion molecule (PECAM). These studies unexpectedly place HoxB5 at a very early step in the process of angioblast differentiation and indicate that HoxB5 is both necessary and sufficient to regulate flk1 expression.
MATERIALS AND METHODS
In vitro DNase I footprinting.
In vitro DNase I footprinting was performed as described previously (33). Overlapping DNA fragments containing bp 1 to 256 and bp 138 to 511 of the mouse flk1 first intronic enhancer were labeled with [α-32P]dGTP by using the Klenow fragment of DNA polymerase I. Approximately 10,000 cpm of the labeled DNA fragment was incubated with 25 μg of nuclear extract or bovine serum albumin (BSA) and 1 μg of poly(dI-dC)-poly(dI-dC) for 25 min on ice and then for 2 min at room temperature. Samples were treated with increasing doses of DNase I (0.0005 to 0.005 Kunitz units with BSA and 0.005 to 0.05 Kunitz units with nuclear extract) at room temperature for 2 min. Samples were analyzed on 6% (wt/vol) denaturing polyacrylamide-urea gels as described previously (47).
EMSA.
Mouse myocardial endothelial cells (MECs), Py-4-1 mouse hemangioma endothelial cells, C166 mouse embryonic yolk sac endothelial cells, bEnd.3 mouse endothelial cells, and C2C12 mouse myoblast cells were grown in Dulbecco modified Eagle medium with 10% fetal calf serum. (MECs and C166 cells were generous gifts from Robert Auerbach.) Preparation of nuclear extract and electrophoretic mobility shift assay (EMSA) were performed as previously described (33). The probe consisted of annealed synthetic 46-bp complementary oligonucleotides corresponding to bp 150 to 195 of the mouse flk1 first-intronic enhancer or mutated oligonucleotides as indicated. Prior to annealing, the oligonucleotides were labeled with [γ-32P]ATP by using polynucleotide kinase. A typical binding reaction contained 20,000 cpm of DNA probe, 0.5 μg of poly(dI-dC)-poly(dI-dC), 25 mM HEPES (pH 7.9), 40 mM KCl, 3 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 5 μg of nuclear extract in a final volume of 20 μl. The reaction was incubated at room temperature for 20 min and fractionated on a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. To determine the specificity of the DNA-protein complexes, we performed competition assays by using a molar excess of unlabeled wild-type oligonucleotides. To characterize specific DNA-binding proteins, a recombinant glutathione S-transferase (GST)-HoxB5 fusion protein was used in place of nuclear extract, with or without preincubation with anti-GST antibody (or an isotype-specific control antibody) for 3 h at 4°C before adding probe.
Generation of transgenic mice.
The mouse flk1 promoter-reporter transgene consisted of the mouse 940-bp (−640 to +299) flk1 promoter fragment followed by the LacZ reporter gene, and 510 bp of the first intronic enhancer, with or without the M4 mutation, as depicted in Fig. 1A. Transgenic mice were generated by microinjection of purified transgene DNA into pronuclei of single-cell fertilized eggs as described previously (22). Transgenic positive embryos were identified by PCR analysis of yolk sac genomic DNA by using lacZ-specific primers (sense, 5′-CAACTTAATCGCCTTGCCTTGCAGCAC-3′; antisense, 5′-CTTCCAGATAACTGCCGTCACT-3′) to generate a 500-bp PCR product. Whole-mount lacZ staining of embryos derived from injected eggs was performed as described previously (22).
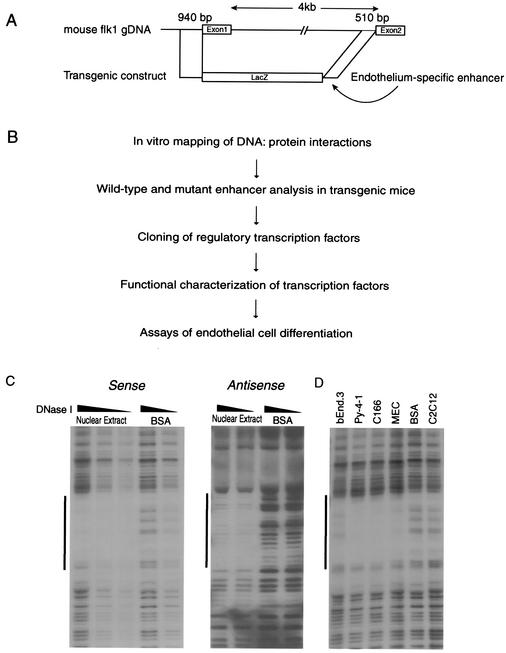
FIG. 1.
Identification of cis-acting elements in the mouse flk1 first-intronic enhancer. (A) Scheme of mouse flk1 genomic locus and transgenic construct. A total of 940 bp (positions from −640 to +299) of the 5′-flanking sequence of the mouse flk1 gene and 510 bp of the first intronic enhancer are sufficient to target lacZ expression to the vascular endothelium in transgenic mice. (B) Diagram of the general approach undertaken to identify cis-acting elements in the mouse flk1 first intronic enhancer. (C) Protein-binding site identification by in vitro DNase I footprinting. A single footprint, located between bp 150 and 195 in the intronic enhancer, was detected on both sense and antisense strands. The dark line denotes the putative binding site for DNA-binding proteins. (D) Analysis of cell type differences in DNase I footprinting patterns. The labeled probe was incubated with nuclear extracts from MECs, Py-4-1, C166, bEnd.3, and C2C12 cells, or BSA prior to digestion with DNase I.
Yeast one-hybrid screening.
The one-hybrid library screen was performed according to the protocol of the manufacturer (Clontech). The pHISi-HBE and pLacZi/HBE bait plasmids were constructed by using synthetic DNA oligomers containing three tandem repeats of the flk1 intronic enhancer from bp 164 to 183, defined by in vitro DNase I footprinting and EMSA. Linear bait plasmids were integrated into the genome of the yeast YM4271 by homologous recombination. The yeast strain was then transformed with a pACT2 vector-based mouse embryo day 10.5 p.c. cDNA library (Clontech). About 2 × 106 yeast transformants were plated on synthetic complete medium lacking histidine and leucine and supplemented with 45 mM 3-amino-1,2,4-triazole. Selected clones were subjected to the β-galactosidase assay using the colony-lift filter method. Several negative control plasmids were used in the selection procedure, including the mutant constructs pHISi-HBEmt and pLacZi-HBEmt, p53Blue containing the consensus p53 binding site, and p53 containing the murine p53 gene fused to the GAL4 activation domain. Plasmids from putative positive clones were recovered from yeast and individually transformed into Escherichia coli DH10B cells for amplification. To eliminate false-positive results, these plasmids were separately introduced into yeast cells containing either bait or the p53 binding site, and transformants were tested for β-galactosidase assay. Inserts were sequenced, and the DNA sequences were used to query the GenBank database.
Transient-transfection assays.
MECs were used for transient transfection assays because they can be reproducibly transfected with high efficiency. Cells were grown to 40 to 60% confluence in six-well plates and then transfected with Lipofectamine (Life Technologies), as described previously (1). The cells were transfected with 1.0 μg of pflk1/en3 (which contains sequences from positions −640 to +299 of the mouse flk1 promoter upstream of the luciferase gene and the 510-bp intronic enhancer downstream) or pflk1/en3mt (containing a 5-bp mutation within the HoxB5-binding site) with different concentrations (indicated in Fig. 5) of pECH/HoxB5, pcDNA3/HoxB6, or control vectors. (Hox expression constructs were generous gifts from Craig Hauser.) To correct for variability in transfection efficiency, 0.25 μg of pCMV-βgal was cotransfected in all experiments. At 48 h after transfection, cells extracts were prepared by a detergent lysis method (Promega). Luciferase activity was measured in duplicates for all samples. β-Galactosidase assay was performed as previously described (32). The ratio of luciferase activity to β-galactosidase activity in each sample served as a measure of the normalized luciferase activity. The normalized luciferase activity was expressed as the fold induction. Each construct was transfected at least four times, and the data for each construct are presented as the mean ± the standard error.
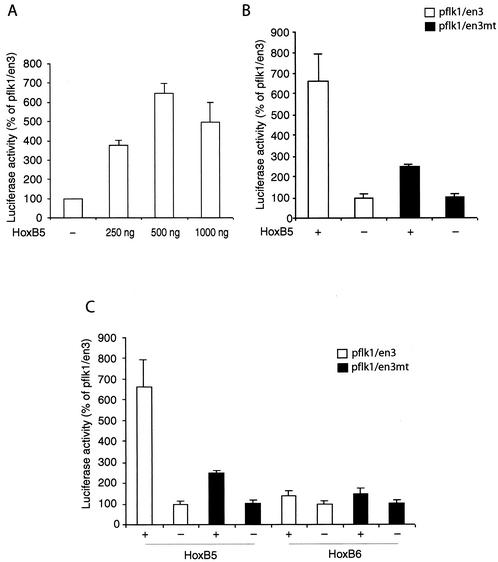
FIG. 5.
Transactivation of the mouse flk1 promoter-enhancer by HoxB5 in transient-transfection assays. (A) Dose-response analysis of transactivation of the flk1 promoter-enhancer construct pflk1/en3 by HoxB5. pflk1/en3 (1 μg) was transiently transfected with the indicated doses of pECH/HoxB5 or vector pECH in MECs. pCMV-βgal was cotransfected to normalize transfection efficiency. The ratio of luciferase activity to β-galactosidase activity in each sample served as a measure of the normalized luciferase activity. The normalized luciferase activity was expressed as a percentage of pflk1/en3. (B) The ability of HoxB5 to transactivate the flk1 promoter was examined in the presence or absence of an intact HBE. HoxB5 (500 ng) was transfected along with wild-type pflk1/en3 (□) or with a mutant containing a 4-bp mutation in the HBE (▪). The results were normalized by cotransfection with pCMV-βgal. (C) Comparison of transactivation of the flk1 promoter-enhancer by HoxB5 and its adjacent cluster mate HoxB6.
Embryonic stem cells growth and stable transfection.
R1 mouse embryonic stem cells were maintained by growth and passage on gelatinized tissue culture dishes in maintenance medium containing conditioned medium from human 5637 bladder carcinoma cells. Differentiation was initiated by treating embryonic stem cell aggregates with dispase and culturing embryoid bodies in differentiation medium (Dulbecco modified Eagle medium-HEPES with 15% lot selected fetal calf serum, 5 μg of gentamicin/ml, and 150 μM monothioglycerol). Cells were grown in bacterial dishes to allow for the formation of cystic embryoid bodies and then plated on tissue culture plates after 60 h for attachment. Attached embryoid bodies were grown under these conditions for the indicated times (5). To construct the corresponding stably transfected cell lines, the cDNAs for HoxB5 and HoxB6 were inserted into pcDNA3 (which contains the neomycin resistance cassette) downstream of the cytomegalovirus (CMV) promoter. Linear plasmids were electroporated into R1 mouse embryonic stem cells. Stable clones were selected with G418 for 12 days, were screened for transgene expression by reverse transcription-PCR (RT-PCR), and confirmed by Northern blotting. The specific primers used for HoxB5 were as follows: sense, 5′-TCCTCGGAGCCTGAGGAAGCGGCAAG-3′, and antisense, 5′-CCCGTCCGGCCCGGTCATATCATG-3′.
RNA preparation and RT-PCR.
Total RNA was isolated from cultured cells by using the High-Pure MiniRNA kit (Roche). All RNA samples were treated with DNase I before cDNA synthesis to eliminate contaminating genomic DNA. Equal amounts of RNA were converted to first-strand cDNA with reverse transcriptase and were then amplified with Taq DNA polymerase. (The DNA amplification reaction was also carried out without reverse transcriptase to ensure that amplified bands were not due to contamination by genomic DNA in the RNA preparations.) The oligonucleotides used for detecting specific cDNA were as follows: mouse HoxB5 (sense, 5′-TCCTCTGAGCCCGAGGAAGCGGCGAG-3′; antisense, 5′-CCACTTCATGCGACGGTTCTG-3′), flk1 (sense, 5′-GGAACCTGACTATCCGCAGG-3′; antisense, 5′-CCTCAACAAAGCCTGAGCTGG-3′); or GAPDH (glyceraldehyde-3-phosphate dehydrogenase; sense, 5′-ACCACAGTCCATGCCATCAC-3′; antisense: 5′-TCCACCACCCTGTTGCTGTA-3′). GAPDH served as an RNA integrity and normalization control.
Flow cytometry, cell sorting, and immunostaining.
Phycoerythrin-conjugated rat anti-mouse flk1 antibody and rat anti-mouse CD31 (PECAM-1) antibody were purchased from Becton Dickinson. Alexafluor 488-conjugated goat anti-rat immunoglobulin G antibody was from Molecular Probes. Differentiated embryonic stem cells were trypsinized (when sorted for flk1) or subjected to collagenase digestion (when sorted for PECAM-1) to produce a single-cell suspension. Cells were counted, and equal numbers were stained with an appropriate antibody in 0.1% BSA-phosphate-buffered saline for 30 min, washed twice with 0.1% BSA-phosphate-buffered saline, and then stained with secondary antibody or analyzed directly on a Becton Dickinson FACSscan by using the Cytomation Summit software or, alternatively, sorted on a MoFlo instrument (Cytomation). Dead cells were excluded from the analysis. Immunostaining of cultured cells with the anti-PECAM-1 antibody was performed as described previously (5). Images were collected with a Qimaging Retica 1300 digital camera by using Qcapture and IPlab software.
RESULTS
Mapping of DNA-protein interactions in the flk1 intronic element.
Previous studies have indicated that a 510-bp positive regulatory element in the 3′ end of the mouse flk1 first intron confers endothelial cell-specific expression on the minimal 5′-flanking sequence in transgenic reporter gene assays (Fig. 1A) and that binding sites for GATA and Ets family members contribute to this activity (23, 24). We have confirmed the activity of this regulatory element by using similar constructs (data not shown). Because this element has been incompletely characterized and because neither GATA nor Ets proteins acting alone are likely to account for the restricted pattern of flk1 expression, we used a variety of biochemical, molecular, and genetic approaches to determine the upstream factors regulating flk1 expression by using an unbiased approach (Fig. 1B). As a first step, we performed in vitro DNase I footprinting with extracts from C166 embryonic endothelial cells to search for nuclear protein-binding sites. Although we sequenced the entire element on both strands, only a single region, located 340 bp upstream of exon 2, was clearly footprinted (Fig. 1C). In particular, we did not observe nuclear protein binding at consensus GATA, Ets, or SCL/tal-1 sites (23). The binding activity identified here was present in nuclear extracts from a wide panel of endothelial cells but, interestingly, nuclear extracts from C2C12 myoblasts did not footprint this region (Fig. 1D), suggesting that endothelial cell-specific factor(s) may be responsible for the footprint.
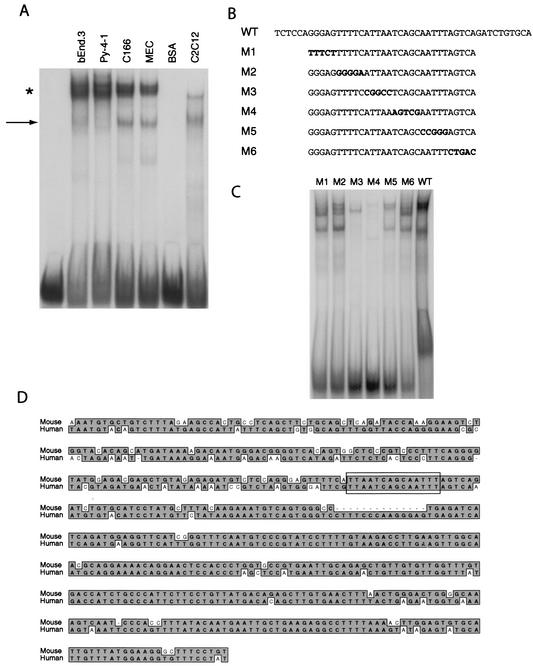
We performed EMSA to characterize this binding activity further. Two complexes were identified: a more rapidly migrating complex that was variably present in endothelial cell nuclear extracts, and a more intense, slowly migrating complex that was present in all endothelial cell nuclear extracts but was weak or absent in extracts from C2C12 cells (Fig. 2A). In order to define the binding determinants for this complex with greater resolution, we created serial mutations within our EMSA probe (Fig. 2B) and examined the effects of these mutations on binding activity using C166 nuclear extracts. The sequential mutations M3, M4, and M5 almost completely abolished binding, whereas other mutants had more subtle effects on DNA-protein interaction (Fig. 2C). The element identified by mutagenesis corresponds to a 14-bp sequence containing bipartite AT-rich stretches flanking an atypical AP1-like element that is conserved across species (Fig. 2D). Examination of this sequence by using the TRANSFAC 5.0 database indicated that it most closely corresponds to a binding site for homeodomain-containing proteins.
FIG. 2.
Identification of the minimal DNA sequences in the flk1 intronic enhancer necessary for nuclear protein interactions. (A) Nuclear protein interactions with the flk1 cis-acting element detected by EMSA. The probe corresponding to bp 150 to 195 of the mouse flk1 first-intronic enhancer was used to characterize nuclear protein interactions with endothelial and nonendothelial nuclear extracts. The arrow denotes a rapidly migrating complex, while the asterisk denotes a more intense, slowly migrating complex. (B) Mutant oligonucleotides used in EMSA. Residues that differ from the wild-type sequence are in boldface. (C) EMSA with C166 nuclear extract with mutant probes described in panel B. (D) Comparison of partial sequences of the mouse and human flk1 first introns. Identical residues are highlighted, and the nuclear protein-binding site identified by DNase I footprinting and EMSA is boxed.
Transgenic analysis of the flk1 intronic element.
To test the functional significance of this element in vivo, we created transgenic mice expressing β-galactosidase under the control of a 940-bp flk1 5′-flanking sequence and the 510-bp intronic element, with or without a 5-bp mutation (corresponding to the M4 mutation in Fig. 2B) in the protein-binding element identified in our footprinting experiments. Transgenic embryos were analyzed at day 11.0 p.c., i.e., during the critical window of embryonic blood vessel growth. Consistent with results from previous studies (24), about half of the transgenic mouse embryos containing the wild-type construct expressed β-galactosidase strongly in an endothelium-restricted pattern (Fig. 3A and B). In contrast, mutation within the footprinted region abolished reporter gene expression in almost all transgenic embryos; only 1 of 16 transgenic mice containing the mutated construct had endothelial cell expression, and in this lone embryo staining was weak and inhomogeneous within the endothelium. These experiments indicate that the footprinted sequence is critical for endothelium-specific activation of the flk1 promoter during embryonic development and that the nuclear protein(s) that interact with this element are likely to play a key role in the early steps in endothelial cell differentiation.
FIG. 3.
The nuclear protein-binding site is required for in vivo activity of the mouse flk1 promoter in transgenic mouse embryos. (A) Representative whole-mount β-galactosidase-stained E11.0 embryos. Transgenic mouse embryos expressing β-galactosidase under control of the flk1 promoter-enhancer stain the vascular endothelium diffusely and strongly. In contrast, a 5-bp mutation within the nuclear protein-binding site identified by footprinting resulted in the loss of β-galactosidase expression in most transgenic embryos. (B) Summary of in vivo activity of the wild-type and mutant transgenic constructs.
Identification of HoxB5 binding to the flk1 element in a genetic screen.
We performed a yeast one-hybrid screen to determine which nuclear protein(s) bound to our footprinted element. Four tandem integrated copies of this element were inserted upstream of selection markers in Saccharomyces cerevisiae strain YM4271, and a day 10.5 p.c. mouse embryo library was screened to identify proteins binding this element. We screened 2 × 106 clones, and of these five survived selection. After plasmid rescue and sequencing, three of the five clones were found to contain full-length cDNA sequence for the homeobox protein HoxB5 in frame with the Gal4 activation domain. The specificity of this binding was confirmed in two assays. First, we measured activation of the lacZ gene by activation domain fusions in yeast (Fig. 4A). β-Galactosidase activity was only detected when the HoxB5 activation domain fusion was expressed in yeast containing the wild-type flk1 element upstream of lacZ but not in yeast containing a mutated flk1 element or a p53 binding site. Interestingly, the strength of the HoxB5 interaction with its cognate element is greater than that of p53 with its well-characterized response element, as measured by relative expression of β-galactosidase. Similarly, we performed selection assays in which yeast cells grow in media lacking histidine only when activation domain fusions bind to their specific elements. In this assay, yeast cells containing the HoxB5 fusion only grew when they contained the wild-type flk1 element upstream of the selection marker and not when they contained a mutated element or a nonspecific binding site (Fig. 4B). These results confirm the specificity of interaction between HoxB5 and the intronic element in the yeast one-hybrid system.
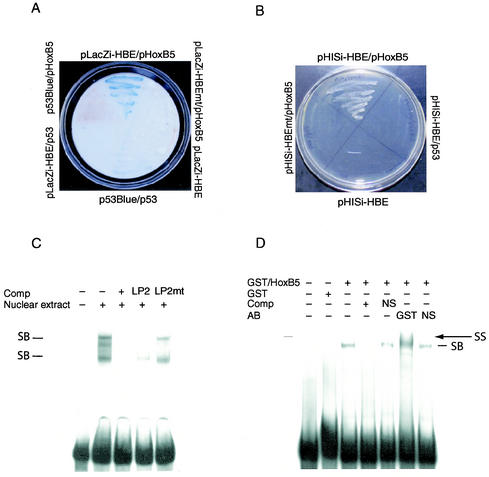
FIG. 4.
Identification of HoxB5 binding to the flk1 enhancer in a yeast one-hybrid screen. (A) β-Galactosidase assays of clones transfected with the indicated plasmids grown on synthetic complete medium lacking uracil to test for specific DNA-protein interactions. (B) Yeast transfectants were grown under growth-resistant conditions (−His, −Leu, +3-amino-1,2,4-triazole). Growth was detected only in yeast transfected with both the wild-type flk1 intronic enhancer motif (HBE) and the HoxB5 expression plasmid. (C) EMSA examining the binding of the labeled HBE with nuclear proteins in C166 cell extract in the presence of excess unlabeled HBE (+) or with mutant or wild-type HoxB5-binding element LP2 as marked. (D) EMSA with recombinant GST-HoxB5 (or GST alone) indicating specific binding to the HBE that is competed away by excess specific (+) competitor but not by a mutant sequence (NS). In addition, migration of this specific activity is shifted with a GST antibody and not by an isotypic control antibody (NS). SB, specific binding; SS, supershift.
Although the identification of a homeodomain-containing protein in this assay was predicted by our TRANSFAC analysis, neither HoxB5 nor any of the other classical homeobox proteins have been previously linked with the earliest stages of endothelial cell development. However, HoxB5 is expressed in vascular endothelium (7, 8), and other Hox family members (in particular, HoxD3 and HoxB3 [8, 31]) have been implicated in later stages of vessel formation. We confirmed the interaction between HoxB5 and this element (which we call HBE, HoxB5-binding element) by EMSA. Unfortunately, a supershifting antibody for HoxB5 is not available; therefore, we first sought to determine whether a well-characterized HoxB5 binding sequence, LP2 (40), could compete for the binding of nuclear proteins to the HBE (Fig. 4C). Indeed, excess unlabeled LP2 could compete for nuclear protein binding to the HBE with nearly or virtually the same efficiency as that of HBE itself. In contrast, a 4-bp mutation within LP2 (LP2mut), which is known to weaken the affinity of HoxB5 with LP2, competed with nuclear protein binding to the HBE much less efficiently. In addition, we directly tested the ability of HoxB5 to bind to the HBE by expressing recombinant HoxB5 as a GST fusion protein. Whereas GST alone did not bind the HBE, the HoxB5-GST fusion produced a single complex that could be competed away with excess cold HBE but not with a nonspecific competitor (Fig. 4D). An antibody that recognized GST retarded the migration of this complex, whereas a nonspecific antibody had no effect. Therefore, we can confirm specific HoxB5-HBE interactions in two independent assays.
Trans-activation of the flk1 promoter by HoxB5.
To test the functional consequences of HoxB5 interactions with the HBE, we performed transient-transfection assays with a luciferase reporter gene under control of the flk1 promoter and minimal intronic enhancer in MECs. Expression of HoxB5 potently transactivated the flk1 promoter in a dose-dependent fashion (Fig. 5A). To determine whether this effect depends on interactions between HoxB5 and the HBE, we mutated the HBE within the intronic enhancer. The transactivation activity of HoxB5 was markedly diminished by mutation of the HBE (Fig. 5B), indicating that HoxB5 mediates the major portion of its effects on the flk1 promoter through direct interactions with its cognate element. We tested the specificity of this functional interaction by comparing the effects of HoxB5 with its cluster sibling, HoxB6. In contrast to the activity of HoxB5, HoxB6 did not affect flk1 promoter activity (Fig. 5C). Thus, the interactions of HoxB5 with HBE result in potent and specific transactivation of flk1, which suggested to us that HoxB5 plays an unanticipated role in the early stages of angioblast differentiation from precursors.
Coexpression of HoxB5 and flk1.
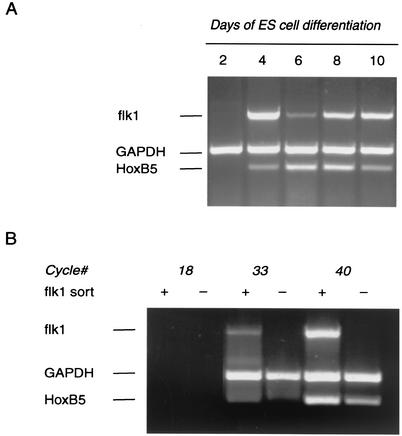
Although HoxB5 has not been clearly linked to angioblast differentiation, it is expressed in lateral plate mesoderm and in somites (28, 45), both of which are rich sources of endothelial precursors (2, 34). In addition, HoxB5 has been detected repeatedly in screens for Hox family members in cultured endothelial cells (7, 8). We have confirmed that HoxB5 mRNA is expressed in a panel of embryonic endothelial cell lines (data not shown). To further explore the developmental relationship between HoxB5 expression and angioblast development, we utilized differentiating mouse embryonic stem cell-derived embryoid bodies grown in attached cultures; the anatomic and molecular events in embryonic endothelial cell differentiation and blood vessel development are faithfully recapitulated in this model (4, 44, 46). The HoxB5 and flk1 mRNAs first appeared cosynchronously on day 4 of culture in this model (Fig. 6A). We confirmed coexpression of flk1 and HoxB5 by sorting cells on day 4 of culture with an flk1-specific antibody. As demonstrated by mRNA expression, this antibody sorted cells cleanly into flk1+ and flk1− populations (Fig. 6B). HoxB5 mRNA was coexpressed with flk1 and was indeed enriched in this cell population. Spatiotemporal colocalization of HoxB5 with flk1 provides additional support for a regulatory role for HoxB5 in angioblast differentiation.
FIG. 6.
Expression of flk1 and HoxB5 mRNA in differentiating embryoid bodies. (A) To assess the time course of HoxB5 and flk1 mRNA expression, embryoid bodies were differentiated for 2, 4, 6, 8, or 10 days, and total RNA was extracted from the cells at the indicated times. The specific genes were detected by RT-PCR. GAPDH served as an RNA integrity and normalization control. (B) To determine whether HoxB5 is expressed in flk1+ populations, embryoid bodies were differentiated for 4 days and then sorted into flk1+ and flk1− populations. Total RNA was isolated from 106 sorted cells from each population. RT-PCR analysis demonstrated expression and relative enrichment of HoxB5 mRNA in the flk1+ cell population.
Enhanced differentiation of angioblasts and definitive endothelial cells by HoxB5.
To test the physiologic function of HoxB5 in flk1 gene regulation and angioblast differentiation, we examined the effects of manipulating HoxB5 expression on the number of flk1+ cells differentiating in embryoid body cultures. Because of functional redundancy of Hox family genes both within paralogous groups and within each Hox family cluster (12, 38), we felt that underexpression studies would be less informative, and therefore we chose to stably overexpress HoxB5 under control of the CMV promoter in embryonic stem cells prior to differentiation. This approach has been used previously with success to define the ability of HoxB4 to expand hematopoietic stem cell populations (21). We tested at least three independent clones for each condition, and each clone was differentiated at least three times. Overexpression of HoxB5 mRNA in stable lines was confirmed by RT-PCR (data not shown). In a representative experiment after 4 days of differentiation, 17.7% of vector-transfected cells were detected by the flk1 antibody, whereas the flk1+ population was expanded to 52.5% in cells overexpressing HoxB5 (Fig. 7A). Similar to the results of our reporter gene assays, this effect was specific to HoxB5 when cells overexpressing HoxB5 and HoxB6 were compared (Fig. 7B).
FIG. 7.
Increased flk1+ cells in embryoid bodies stably expressing HoxB5. (A) Single-cell suspensions were prepared from wild-type or stably transfected embryoid bodies at day 4, and the expression of flk1 was analyzed by flow cytometry with phycoerythrin-conjugated rat anti-mouse flk1 antibody. The y axis represents the relative cell number; the x axis represents the fluorescence intensity. Gray lines indicate negative control, light lines represent results obtained with the addition of specific antibody, and dark lines represent the flk1+ population. The percentages of cells that fall within the indicated gate are noted. Values of <2% represent background staining. A significantly increased number of cells expressing the mouse flk1 gene were observed in the HoxB5-transfected embryoid bodies. (B) Comparison of flk1+ cells in HoxB5- and HoxB6-overexpressing embryoid bodies.
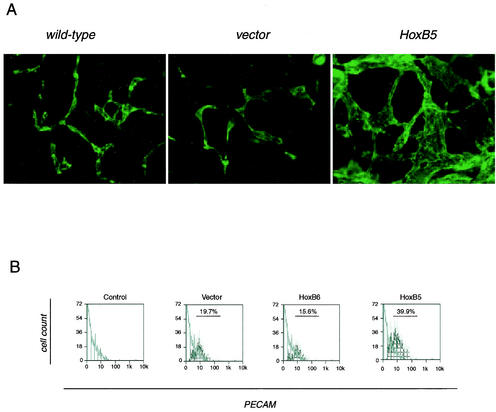
To determine whether activation of flk1 expression by HoxB5 is sufficient to expand the definitive endothelial population in differentiating embryoid bodies, we stained cells that had been differentiated for 8 days with an anti-PECAM antibody, which marks mature endothelial cells at this stage. Consistent with previous observations (46), endothelial cells arrange into vessel-like structures at this stage (Fig. 8A). In HoxB5-overexpressing cells, these structures were markedly hyperplastic, and the area of the culture covered by endothelial cells was greatly expanded. We sorted cultures at this stage with an anti-PECAM antibody to quantify the endothelial cell compartment. Overexpression of HoxB5 doubled the number of mature endothelial cells in these cultures, whereas HoxB6 again had no effect (Fig. 8B). Thus, HoxB5 alone is sufficient to induce angioblast differentiation and expansion of endothelial cell populations derived from differentiating embryonic stem cells.
FIG. 8.
HoxB5 is a positive regulator of endothelial cell differentiation and primitive blood vessel formation. (A) Immunostaining for PECAM. The indicated stably transfected embryonic stem cells were differentiated, fixed on day 8, and stained with antibody to PECAM, followed by secondary staining with an Alexafluor 488-conjugated antibody. Magnification, ×20. (B) PECAM sorting was performed to determine the absolute number of endothelial cells in embryoid bodies at day 8 of differentiation. The y axis represents relative cell number, and the x axis represents fluorescence intensity. Gray lines indicate negative control (secondary antibody only), light lines represent results obtained with the addition of the specific antibody, and dark lines represent the PECAM+ cell population. The percentages of cells that fall with the indicated gate are noted.
DISCUSSION
The transition from mesoderm to angioblasts to definitive endothelium is a highly ordered process, and extracellular signaling events that regulate this differentiation cascade are now being described in increasing detail—members of the transforming growth factor-β superfamily and Indian hedgehog regulate the earliest stages of endothelial development (9, 15), and the VEGF and angiopoietin families (among others) control later stages (19), culminating in the final appearance of a mature blood vessel. In contrast, the transcriptional events that direct the different steps in endothelial differentiation are much less well characterized. This is in contrast to other cell lineages of similar complexity, for which transcriptional networks are being described in detail. Several features of endothelial differentiation contribute to the difficulty in understanding key early transcriptional events. The endothelial lineage appears early in development and matures rapidly; important transcriptional regulators may appear transiently and in limited numbers of cells and yet still may have necessary effects on lineage determination. There is heterogeneity within the endothelial lineage that is likely determined early in development, and different endothelial populations may mature by somewhat different mechanisms. There is still no consensus on the developmental steps to maturity of the endothelium; for example, the extent to which the hemangioblast is a necessary stage for all endothelial cells during development is as yet unresolved. Finally, endothelial progenitors may have more plasticity than has been appreciated previously—under some circumstances, they retain potential for recommitment to smooth muscle and cardiomyocyte lineages (13, 51).
We have examined the mechanisms underlying flk1 expression to identify the earliest transcriptional events in endothelial cell differentiation. flk1 has several properties that make it especially well suited as a model of endothelial cell gene regulation. It is the earliest known marker of the endothelial cell lineage, its expression is exquisitely restricted to the endothelium and its precursor cells, and its pattern of expression during development and in embryonic stem cell differentiation models is well characterized (32, 50). Here, we have used an unbiased approach to determine the transcriptional events that regulate flk1 expression and endothelial cell differentiation by using both in vitro and in vivo models. A critical element, the HBE, is located in the 3′ portion of the first intron of the flk1 gene. The HBE is necessary for developmental activation of flk1, and HoxB5 binds and transactivates the flk1 promoter through this element. Given the size of the DNase I footprints observed in our studies (Fig. 1), it is possible that HoxB5 binds to the promoter cooperatively with other, as-yet-unidentified, transcription factors. Most remarkably, overexpression of HoxB5 is sufficient to increase the number of early endothelial precursors and to expand the endothelial population in differentiating embryoid bodies, which recapitulate the important molecular and morphological events of endothelial cell development with fidelity (4, 46).
Hox proteins are transcriptional regulators characterized by a 60-amino-acid DNA-binding domain, the homeodomain. The classical Hox proteins exist in four clusters (A through D) in mammals, with up to 13 Hox genes in each cluster. Their expression is dictated by temporal colinearity, with more 3′ genes expressed before the corresponding 5′ genes within a cluster. Based on studies in both mouse and Drosophila, Hox proteins are thought to generally regulate pattern formation and segmental identity along the anterior-posterior axis during development (36). However, overlapping spatiotemporal expression and functional redundancy have sometimes made the assignment of specific functions to individual Hox proteins problematic. In the case of HoxB5, specific target genes in mammals have not been clearly defined. The only defect in HoxB5−/− mice is a rostral shift in the shoulder girdle, but functional redundancy with other family members (possibly the paralogous Hox proteins A5 and C5) limits the utility of single-gene deletion for designating function to HoxB5 (38). HoxB5 has been identified in vascular endothelial cells in culture (7, 8), although a specific function in the endothelium (and in particular a developmental role) has not been previously established.
Nevertheless, there is reason to hypothesize a role for Hox proteins in vascular development and angiogenesis, given the necessity of pattern formation and tissue remodeling in these processes. HoxD3 is upregulated in endothelial cells by growth factor activation and induces a migratory, “invasive” endothelial cell phenotype but not well-formed blood vessels (8). In contrast, the paralogous gene HoxB3 promotes a later step in the angiogenic process: capillary morphogenesis (31). Given the spatiotemporal colinearity of expression and function of Hox family members, it is reasonable to speculate that other Hox proteins may participate in vascular developmental processes. The identification of HoxB5 as a necessary regulator of flk1 transcription in our studies provides support for a model in which the activation of Hox family members contributes to multiple steps in vascular development.
HoxB4 is the B-cluster member immediately upstream of HoxB5 in the chain of spatiotemporal expression, so it is informative to consider potential parallels in function between these two proteins. HoxB4 is expressed in primitive hematopoietic stem cells and enhances differentiation of hematopoietic lineages when it is overexpressed in embryonic stem cells (21), which is analogous to the effects of HoxB5 on endothelial lineages (Fig. 7 and 8). It is interesting that a role for HoxB4 in hematopoietic function is not suggested by studies of developmental expression (20) or deletion by homologous recombination (37). Recent reports suggest that HoxB4 facilitates the transition from primitive to definitive hematopoietic stem cells and is permissive for ex vivo expansion of adult hematopoietic stem cells by inducing a stable state of multipotency (3, 29). The downstream transcriptional targets mediating the effects of HoxB4 in hematopoietic stem cells are not yet defined. Taken together with the results presented here, it appears that HoxB5 acts as part of a pathway in early endothelial lineages (even possibly at the elusive hemangioblast commitment stage) that is parallel to that of HoxB4 in the hematopoietic system. Whether the effects of HoxB5 in our studies reflect increased differentiation or proliferation of endothelial precursors remains to be determined; such distinctions have often been difficult to make (18, 25).
Early stages of endothelial cell differentiation from mesoderm-derived precursors are under strict control, and many factors—both intrinsic and extrinsic—coordinate this process. In particular, deletion of numerous growth factors can disrupt steps within this pathway. For example, lack of a single VEGF allele severely disrupts differentiation of the endothelium, leading to embryonic lethality before day 10 p.c. in mice (10) and arrest of endothelial precursor maturation in embryoid bodies (6). These and other studies suggest that endothelial precursors are under strict regulation that is difficult to override. Surprisingly, endothelium-associated transcription factors—including SCL/tal-1 and GATA-2—do not elicit expansion of angioblast-derived cells when overexpressed in mammalian systems, although they may be sufficient to amplify hematopoietic cell populations (27, 43). These studies indicate that hyperexpansion of endothelial precursors during development is restricted at multiple levels. The observation that HoxB5 is sufficient to increase flk1+ precursors and expand definitive endothelial populations in embryoid bodies is therefore a unique cell-intrinsic perturbation of the endothelial differentiation pathway. We show here that HoxB5 is a direct transcriptional activator of flk1 and that it is required for expression of flk1 in vivo. A careful characterization of whether the upregulation of flk1 alone is sufficient to trigger HoxB5-dependent differentiation down the endothelial lineage, whether or not there are other transcriptional targets of HoxB5 in endothelial precursors, and the cellular phenotypes of HoxB5-expressing angioblasts will provide a new window into the events of endothelial cell differentiation.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute grants HL03658, HL61656, and HL072347 (to C.P.) and HL43174 (to V.L.B.). M.M. was supported by Deutsche Forschungsgemeinschaft. C.P. is an Established Investigator of the American Heart Association.
We thank Daniel Hu for assistance with histology, Rebecka Rapaport and Joseph B. Kearney for technical advice with embryonic stem cell cultures, Robert Auerbach and Craig Hauser for providing reagents, and Mark Majesky for helpful commentary.
REFERENCES
- 1.Aitsebaomo, J., M. Kingsley-Kallesen, Y. Wu, T. Quertermous, and C. Patterson. 2001. Vezf1/DB1 is an endothelial cell-specific transcription factor that regulates expression of the endothelin-1 promoter. J. Biol. Chem. 276:39197-39205. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, C. A., J. L. Nowicki, A. C. Burke, and V. L. Bautch. 2001. Assembly of trunk and limb blood vessels involves extensive migration and vasculogenesis of somite-derived angioblasts. Dev. Biol. 234:352-364. [DOI] [PubMed] [Google Scholar]
- 3.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. [DOI] [PubMed] [Google Scholar]
- 4.Bautch, V., W. Stanford, R. Rapoport, S. Russell, R. Byrum, and T. Futch. 1996. Blood island formation in atached culture of murine embryonic stem cells. Dev. Dyn. 205:1-12. [DOI] [PubMed] [Google Scholar]
- 5.Bautch, V. L. 2002. Embryonic stem cell differentiation and the vascular lineage. Methods Mol. Biol. 185:117-125. [DOI] [PubMed] [Google Scholar]
- 6.Bautch, V. L., S. D. Redick, A. Scalia, M. Harmaty, P. Carmeliet, and R. Rapoport. 2000. Characterization of the vasculogenic block in the absence of vascular endothelial growth factor-A. Blood 95:1979-1987. [PubMed] [Google Scholar]
- 7.Belotti, D., N. Clausse, D. Flagiello, Y. Alami, M. Daukandt, C. Deroanne, B. Malfoy, E. Boncinelli, A. Faiella, and V. Castronovo. 1998. Expression and modulation of homeobox genes from cluster B in endothelial cells. Lab. Investig. 78:1291-1299. [PubMed] [Google Scholar]
- 8.Boudreau, N., C. Andrews, A. Srebrow, A. Ravanpay, and D. A. Cheresh. 1997. Induction of the angiogenic phenotype by Hox D3. J. Cell Biol. 139:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd, N., S. Becker, P. Maye, R. Narasimhaiah, B. St-Jacques, X. Zhang, J. McMahon, A. McMahon, and L. Grabel. 2002. Hedgehog is required for murine yolk sac angiogenesis. Development 129:361-372. [DOI] [PubMed] [Google Scholar]
- 10.Carmeliet, P., V. Ferriera, G. Breier, S. Pollefeyt, L. Nieckens, M. Gertsenstein, M. Fahrig, A. Vandenhoeck, K. Harpal, C. Eberhardt, C. Declercq, J. Pawling, L. Moons, D. Collen, W. Risau, and A. Nagy. 1996. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380:435-439. [DOI] [PubMed] [Google Scholar]
- 11.Choi, K., M. Kennedy, A. Kazarov, J. C. Papadimitriou, and G. Keller. 1998. A common precursor for hematopoietic and endothelial cells. Development 125:725-732. [DOI] [PubMed] [Google Scholar]
- 12.Condie, B. G., and M. R. Capecchi. 1994. Mice with targeted disruptions in the paralogous genes hoxa-3 and hoxd-3 reveal synergistic interactions. Nature 370:304-307. [DOI] [PubMed] [Google Scholar]
- 13.Condorelli, G., U. Borello, L. De Angelis, M. Latronico, D. Sirabella, M. Coletta, R. Galli, G. Balconi, A. Follenzi, G. Frati, M. G. Cusella De Angelis, L. Gioglio, S. Amuchastegui, L. Adorini, L. Naldini, A. Vescovi, E. Dejana, and G. Cossu. 2001. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: implications for myocardium regeneration. Proc. Natl. Acad. Sci. USA 98:10733-10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake, C. J., and P. A. Fleming. 2000. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood 95:1671-1679. [PubMed] [Google Scholar]
- 15.Dyer, M. A., S. M. Farrington, D. Mohn, J. R. Munday, and M. H. Baron. 2001. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development 128:1717-1730. [DOI] [PubMed] [Google Scholar]
- 16.Ema, M., P. Faloon, W. J. Zhang, M. Hirashima, T. Reid, W. L. Stanford, S. Orkin, K. Choi, and J. Rossant. 2003. Combinatorial effects of flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 17:380-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faloon, P., E. Arentson, A. Kazarov, C. X. Deng, C. Porcher, S. Orkin, and K. Choi. 2000. Basic fibroblast growth factor positively regulates hematopoietic development. Development 127:1931-1941. [DOI] [PubMed] [Google Scholar]
- 18.Fong, G. H., L. Zhang, D. M. Bryce, and J. Peng. 1999. Increased hemangioblast commitment, not vascular disorganization, is the primary defect in flt-1 knock-out mice. Development 126:3015-3025. [DOI] [PubMed] [Google Scholar]
- 19.Gale, N. W., and G. D. Yancopoulos. 1999. Growth factors acting via endothelial cell-specific receptor tyrosine kinases: VEGFs, angiopoietins, and ephrins in vascular development. Genes Dev. 13:1055-1066. [DOI] [PubMed] [Google Scholar]
- 20.Graham, A., N. Papalopulu, J. Lorimer, J. H. McVey, E. G. Tuddenham, and R. Krumlauf. 1988. Characterization of a murine homeo box gene, Hox-2.6, related to the Drosophila Deformed gene. Genes Dev. 2:1424-1438. [DOI] [PubMed] [Google Scholar]
- 21.Helgason, C. D., G. Sauvageau, H. J. Lawrence, C. Largman, and R. K. Humphries. 1996. Overexpression of HOXB4 enhances the hematopoietic potential of embryonic stem cells differentiated in vitro. Blood 87:2740-2749. [PubMed] [Google Scholar]
- 22.Hogan, B. L., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, vol. 2. Cold Spring Harbor Press, Plainview, N.Y.
- 23.Kappel, A., V. Ronicke, A. Damert, I. Flamme, W. Risau, and G. Breier. 1999. Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood 93:4284-4292. [PubMed] [Google Scholar]
- 24.Kappel, A., T. M. Schlaeger, I. Flamme, S. H. Orkin, W. Risau, and G. Breier. 2000. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood 96:3078-3085. [PubMed] [Google Scholar]
- 25.Kearney, J. B., C. A. Ambler, K. A. Monaco, N. Johnson, R. G. Rapoport, and V. L. Bautch. 2002. Vascular endothelial growth factor receptor Flt-1 negatively regulates developmental blood vessel formation by modulating endothelial cell division. Blood 99:2397-2407. [DOI] [PubMed] [Google Scholar]
- 26.Kinder, S. J., D. A. Loebel, and P. P. Tam. 2001. Allocation and early differentiation of cardiovascular progenitors in the mouse embryo. Trends Cardiovasc. Med. 11:177-184. [DOI] [PubMed] [Google Scholar]
- 27.Kitajima, K., M. Masuhara, T. Era, T. Enver, and T. Nakano. 2002. GATA-2 and GATA-2/ER display opposing activities in the development and differentiation of blood progenitors. EMBO J. 21:3060-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumlauf, R., P. W. Holland, J. H. McVey, and B. L. Hogan. 1987. Developmental and spatial patterns of expression of the mouse homeobox gene, Hox 2.1. Development 99:603-617. [DOI] [PubMed] [Google Scholar]
- 29.Kyba, M., R. C. Perlingeiro, and G. Q. Daley. 2002. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109:29-37. [DOI] [PubMed] [Google Scholar]
- 30.Minami, T., R. D. Rosenberg, and W. C. Aird. 2001. Transforming growth factor-β1-mediated inhibition of the flk-1/KDR gene is mediated by a 5′-untranslated region palindromic GATA site. J. Biol. Chem. 276:5395-5402. [DOI] [PubMed] [Google Scholar]
- 31.Myers, C., A. Charboneau, and N. Boudreau. 2000. Homeobox B3 promotes capillary morphogenesis and angiogenesis. J. Cell Biol. 148:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson, C., M. A. Perrella, C.-M. Hsieh, M. Yoshizumi, M.-E. Lee, and E. Haber. 1995. Cloning and functional analysis of the promoter for flk1, a receptor for vascular endothelial growth factor. J. Biol. Chem. 270:23111-23118. [DOI] [PubMed] [Google Scholar]
- 33.Patterson, C., Y. Wu, M.-E. Lee, J. D. DeVault, M. S. Runge, and E. Haber. 1997. Nuclear protein interactions with the human flk1 promoter in vivo. J. Biol. Chem. 272:8410-8416. [DOI] [PubMed] [Google Scholar]
- 34.Poole, T. J., and J. D. Coffin. 1989. Vasculogenesis and angiogenesis: two distinct morphogenetic mechanisms establish embryonic vascular pattern. J. Exp. Zool. 251:224-231. [DOI] [PubMed] [Google Scholar]
- 35.Porcher, C., W. Swat, K. Rockwell, Y. Fujiwara, F. W. Alt, and S. H. Orkin. 1996. The T-cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47-57. [DOI] [PubMed] [Google Scholar]
- 36.Prince, V. E. 2002. The Hox paradox: more complex(es) than imagined. Dev. Biol. 249:1-15. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez-Solis, R., H. Zheng, J. Whiting, R. Krumlauf, and A. Bradley. 1993. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell 73:279-294. [DOI] [PubMed] [Google Scholar]
- 38.Rancourt, D. E., T. Tsuzuki, and M. R. Capecchi. 1995. Genetic interaction between hoxb-5 and hoxb-6 is revealed by nonallelic noncomplementation. Genes Dev. 9:108-122. [DOI] [PubMed] [Google Scholar]
- 39.Robb, L., I. Lyons, R. Li, L. Hartley, F. Kontgen, R. P. Harvey, D. Metcalf, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safaei, R. 1997. A target of the HoxB5 gene from the mouse nervous system. Dev. Brain Res. 100:5-12. [DOI] [PubMed] [Google Scholar]
- 41.Shalaby, F., J. Rossant, T. P. Yamaguchi, M. Gertsenstein, X.-F. Wu, M. Breitman, and A. Schuh. 1995. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376:62-66. [DOI] [PubMed] [Google Scholar]
- 42.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 43.Valtieri, M., A. Tocci, M. Gabbianelli, L. Luchetti, B. Masella, L. Vitelli, R. Botta, U. Testa, G. L. Condorelli, and C. Peschle. 1998. Enforced TAL-1 expression stimulates primitive, erythroid and megakaryocytic progenitors but blocks the granulopoietic differentiation program. Cancer Res. 58:562-569. [PubMed] [Google Scholar]
- 44.Vittet, D., M.-H. Prandini, R. Berthier, A. Schweitzer, H. Martin-Sisteron, G. Uzan, and E. Dejana. 1996. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood 88:3424-3431. [PubMed] [Google Scholar]
- 45.Wall, N. A., C. M. Jones, B. L. Hogan, and C. V. Wright. 1992. Expression and modification of Hox 2.1 protein in mouse embryos. Mech. Dev. 37:111-120. [DOI] [PubMed] [Google Scholar]
- 46.Wang, R., R. Clark, and V. L. Bautch. 1992. Embryonic stem cell-derived cystic embryoid bodies form vascular channels: an in vitro model of blood vessel development. Development 114:303-316. [DOI] [PubMed] [Google Scholar]
- 47.Wildeman, A., M. Zenke, C. Schatz, M. Wintzerith, T. Grundstrom, H. Matthes, K. Takahashi, and P. Chambon. 1986. Specific protein binding to the simian virus 40 enhancer in vitro. Mol. Cell. Biol. 6:2098-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, Y., and C. Patterson. 1999. The human flk1 gene contains a functional initiator element that is bound and transactivated by TFII-I. J. Biol. Chem. 274:3207-3214. [DOI] [PubMed] [Google Scholar]
- 49.Xiong, J. W., A. Leahy, H. H. Lee, and H. Stuhlmann. 1999. Vezf1: a Zn finger transcription factor restricted to endothelial cells and their precursors. Dev. Biol. 206:123-141. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi, T. P., D. J. Dumont, R. A. Conion, M. L. Breitman, and J. Rossant. 1993. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development 118:489-498. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita, J., H. Itoh, M. Hirashima, M. Ogawa, S. Nishikawa, T. Yurugi, M. Naito, and K. Nakao. 2000. flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408:92-96. [DOI] [PubMed] [Google Scholar]