Abstract
Genome integrity is protected by Cds1 (Chk2), a checkpoint kinase that stabilizes arrested replication forks. How Cds1 accomplishes this task is unknown. We report that Cds1 interacts with Rad60, a protein required for recombinational repair in fission yeast. Cds1 activation triggers Rad60 phosphorylation and nuclear delocalization. A Rad60 mutant that inhibits regulation by Cds1 renders cells specifically sensitive to replication fork arrest. Genetic and biochemical studies indicate that Rad60 functions codependently with Smc5 and Smc6, subunits of an SMC (structural maintenance of chromosomes) complex required for recombinational repair. These studies indicate that regulation of Rad60 is an important part of the replication checkpoint response controlled by Cds1. We propose that control of Rad60 regulates recombination events at stalled forks.
Genome replication is fraught with danger. Among the many hazards are cross-linked DNA, modified bases, and protein complexes bound to DNA, all of which can halt DNA polymerases. Preserving stalled replication forks while these roadblocks are removed is of paramount importance for genome integrity and cell survival. Indeed, collapsed replication forks are a major source of genetic instability in eukaryotic cells (23). Deciphering how cells deal with these problems is therefore vital for understanding mechanisms of genome maintenance.
Studies of budding and fission yeasts have pioneered efforts to elucidate how eukaryotic organisms cope with replication fork arrest. Hydroxyurea (HU), the most widely used fork-stalling agent, causes deoxynucleoside triphosphate (dNTP) starvation by inactivating ribonucleotide reductase. HU-induced fork arrest triggers a replication checkpoint response that leads to the effector protein kinase Cds1 in fission yeast and Rad53 in budding yeast, homologs of human Chk2 (Cds1) (34, 36, 49). One of Cds1's functions is to enforce the cell cycle checkpoint that prevents mitosis during a replication arrest, but arguably its most important activity is to stabilize stalled forks (8, 33). In budding yeast, stalled forks collapse, regress, and form other abnormal structures in mutants that lack Rad53 activity (9, 27, 41, 43). Rad53 is required to suppress gross chromosomal rearrangements induced by replication difficulties in budding yeast (23).
How Cds1 stabilizes stalled forks is unknown. Speculation has focused on the possibilities that Cds1 controls core replication and recombination proteins. A paused or regressed fork should be a tempting substrate for DNA endonucleases and recombination enzymes. In bacteria, active collapse of regressed forks by a Holliday junction (HJ) resolvase can be a successful strategy for bypassing irreparable DNA lesions (30). Whether the same is true in eukaryotes is unknown, but Cds1 is known to associate with and control the phosphorylation of Mus81, a subunit of an HJ resolvase in fission yeast (5, 7). Rad53 controls the phosphorylation of at least two recombination enzymes, and it associates with Asf1, a chromatin assembly and silencing factor (2, 15, 21). The functional consequences of these interactions are unknown.
Proteins in the SMC (structural maintenance of chromosomes) family have recently been recognized as central players in recombinational repair of DNA (19). SMC proteins contain N-terminal and C-terminal nucleotide-binding motifs separated by an extensive coiled-coil region (protein-protein interface) that contains a central hinge region (19). SMC proteins form heterodimeric structures that also contain essential non-SMC subunits. These subunits control the association of SMC complexes with chromosomes, perhaps by regulating the opening and closing of a loop-shaped SMC complex that can encircle chromosomes (18). Although best known for their roles in chromatid cohesion (Smc1-Smc3 cohesin complex) and condensation (Smc2-Smc4 condensin complex), hypomorphic mutations of these essential proteins cause DNA damage sensitivity (1, 3, 22, 39). The importance of chromatid cohesion in maintaining the physical proximity of a template for repair of double-strand breaks (DSBs) by homologous recombination is readily understood, but the damage-sensitive phenotypes of condensin mutants were not anticipated. In fact, very little is known about how chromosome structure influences DNA repair.
Analysis of radiation-sensitive mutants in fission yeast led to the discovery of a third class of SMC complex that contains Spr18 and Rad18, more generally known as Smc5 and Smc6, respectively (16, 24, 47). Like condensin and cohesin, the Smc5-Smc6 complex is essential for viability and appears to control chromosome architecture, although its essential function and role in DSB repair are obscure. The Smc5-Smc6 complex has additional subunits (16), one of which is the essential protein Nse1 in budding yeast (17).
Here we report studies aimed at understanding how Cds1 promotes recovery from replication fork arrest. We describe how Cds1 regulates Rad60, a newly described protein proposed to function with Smc5-Smc6 in recombinational repair (32). Replication arrest leads to delocalization of Rad60 from the nucleus by a Cds1-dependent process. Analysis of a unique Rad60 mutant that is insensitive to Cds1 regulation strongly suggests that control of Rad60 is a vital part of the mechanism by which Cds1 promotes recovery from replication fork arrest.
MATERIALS AND METHODS
General techniques.
Standard fission yeast methods and media were used in these studies (31). Yeast two-hybrid screens were performed with previously described forkhead-associated (FHA) domain bait plasmids, libraries, and methods (7). UV, ionizing radiation (IR), and HU sensitivity assays were performed as described previously (7). Error-prone PCR conditions were used to generate the rad60-4 allele.
Immunoblotting and microscopy techniques.
Immunoblotting was performed as described previously with extracts made from cells lysed in a bead beater (7). Briefly, cells were lysed using in buffer A and resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% polyacrylamide gels. Proteins were transferred to Immobilon membrane, blocked in 5% milk in Tris-buffered saline (TBS) and 0.3% Tween 20, and probed with antibodies to the epitope. Rad60-myc was detected with anti-myc antibody (9E10 at 1:5,000 dilution; Santa Cruz Biotechnology). Rad60 tagged with a tandem affinity purification tag (Rad60-TAP) was detected with peroxidase-antiperoxidase reagent (PAP at 1:2,000 dilution; Sigma). Phosphatase treatments were carried out with lambda phosphatase according to the guidelines in the New England Biolabs catalog. Glutathione S-transferase (GST) precipitations were performed as described previously (38). For GST pull-down experiments, GST-Cds1 expression was induced from the nmt1 promoter for 18 h (28a). Cells were lysed in buffer A (50 mM Tris [pH 8]; 150 mM NaCl; 2 mM EDTA; 10% glycerol; 0.2% Nonidet P-40; 5 μg each of leupeptin, pepstatin, and aprotinin per ml; 1 mM phenylmethylsulfonyl fluoride [PMSF]), and glutathione-Sepharose (Pharmacia) was added to the lysates followed by incubation at 4°C for 1.5 h with rotation. Complexes were collected by centrifugation and washed three times with buffer A before resuspension in SDS-PAGE loading buffer.
Indirect immunofluorescence microscopy was performed by established methods (28). Cells were stained with primary anti-myc antibody (9E10 at 1:2,000) and secondary antibody (CY3 conjugated antimouse antibody at 1:1,000). DAPI (4′,6′-diamidino-2-phenylindole) was used at 0.5 μg/ml. Cells were photographed with Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera.
Identification of Rad60-interacting proteins.
Proteins that associated with Rad60-TAP were identified by multidimensional protein identification technology (MudPIT) by established methods (5, 48). Briefly, cells (∼40 g, wet weight) expressing Rad60-TAP at the genomic locus were frozen in liquid nitrogen and lysed with a motorized mortar and pestle (Retsch) in buffer A (50 mM Tris [pH 8]; 150 mM NaCl; 2 mM EDTA; 10% glycerol; 0.2% Nonidet P-40; 5 μg each of leupeptin, pepstatin, and aprotinin per ml; 1 mM PMSF). Rad60-TAP was purified from clarified lysate as described previously (35). The final eluate was precipitated with trichloroacetic acid (25% [vol/vol]) for 1 h on ice. The precipitate was pelleted in a bench top microcentrifuge (Eppendorf) at a relative centrifugal force of 16. The pellet was washed twice with acetone (−20°C) and air dried. The sample was reduced and alkylated with dithothreitol and iodoacetamide and then sequentially digested with endonuclease lyse-C (Roche) and trypsin (Perceptive Biosystems) (29). The resulting peptide mixture was analyzed by MudPIT (26, 48) with modifications described by W. H. McDonald et al. (submitted for publication). Tandem mass spectra were searched against the latest version of the pompep database to which common contaminants such as keratin and trypsin were added (These sequence data were produced by the S. pombe Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/yeast/Pombe/Protein_data/.) Search results were filtered and grouped by using the DTASelect program, and identifications were confirmed through manual evaluation of spectra. Common background proteins were also excluded by comparing the Rad60-TAP data set to the large number of other data sets obtained by purification of unrelated proteins in the laboratory.
Strains.
The following strains were used in this study (all ura4-D18 leu1-32): NB3156 (rad60:13myc:kanMx6), NB3157 (rad60:13myc:kanMx6 cds1-fha1), NB3158 (rad60:13myc:kanMx6 cds1::ura4+), NB3159 (rad60:TAP:kanMx6), NB3160 (rad60-3), NB3161 (rad60-3 cds1::ura4+), NB3162 (rad60-3 chk1::ura4+), NB3163 (rad60-3 rad3::ura4+), NB3164 (rad60-3 rhp51::ura4+), NB3165 (rad60-4:13myc:kanMx6), NB3166 (spr18:13myc:kanMx6), NB3167 (rqh1::ura4+), NB3168 (rad2::ura4+), EN3169 (brc1::kanMx6), and NB3170 (rad18-X).
RESULTS
Cds1 associates with Rad60.
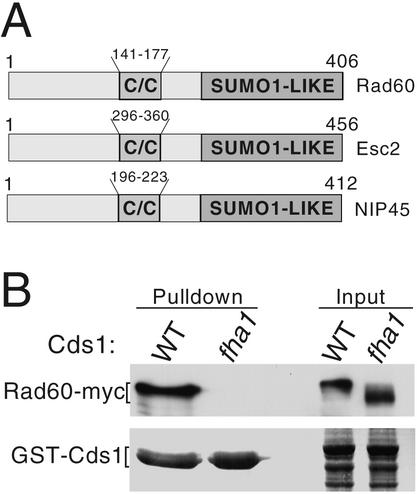
Cds1 homologs contain an N-terminal FHA domain that mediates binding to target proteins (13, 14). Fission yeast Mus81 was previously identified in a yeast two-hybrid screen using the FHA domain of Cds1 as bait (7). The fission yeast gene SPBC1921.02, recently described under the name rad60+ as a gene required for DSB repair (32), was also identified in this screen. Rad60 has a central coiled-coil motif and a C-terminal domain related to the ubiquitin-like modifier PIC1/SUMO-1 (6, 37). Rad60 shares this domain composition with Saccharomyces cerevisiae ESC2p and mammalian NIP45 (11, 20) (Fig. 1A).
FIG. 1.
Rad60 associates with Cds1. (A) Members of the Rad60 family. Shown are Rad60 of fission yeast (406 amino acids), Esc2 of budding yeast (456 amino acids), and Nip45 of humans (412 amino acids). The C terminus of each protein contains a ubiquitin-like domain related to SUMO-1 (none has the C-terminal motifs for covalent attachment to other proteins). All contain a central coiled-coil domain (C/C). (B) Confirmation of the Rad60-Cds1 interaction in vivo. GST fusions of wild-type (WT) or mutant Cds1 (fha1) were expressed in cells that express Rad60-myc from the rad60 genomic locus. Rad60-myc coprecipitates with the wild-type but not mutant Cds1 (fha1). Approximately 1% of the total Rad60-myc coprecipitated with GST-Cds1.
Coprecipitation studies confirmed that Rad60 and Cds1 associate in vivo. Full-length Cds1 was expressed as a GST fusion in a strain that expressed 13myc-epitope-tagged Rad60 from its genomic locus. Rad60-myc coprecipitated with wild-type Cds1 (Fig. 1B). The cds1-fha1 allele (previously named cds1-fha*) encodes proteins altered at two highly conserved residues in the FHA domain that are required for FHA-mediated protein interactions (7). Rad60-myc did not coprecipitate with Cds1-fha1 (Fig. 1B). We observed that Rad60-myc coprecipitates with a GST fusion protein that contains region 1 to 190 of Cds1 that includes the FHA domain, but failed to coprecipitate with the fha1 form of this construct (our unpublished data). Therefore, Cds1 interacted with Rad60 in an FHA domain-specific manner.
Cds1 controls Rad60 phosphorylation.
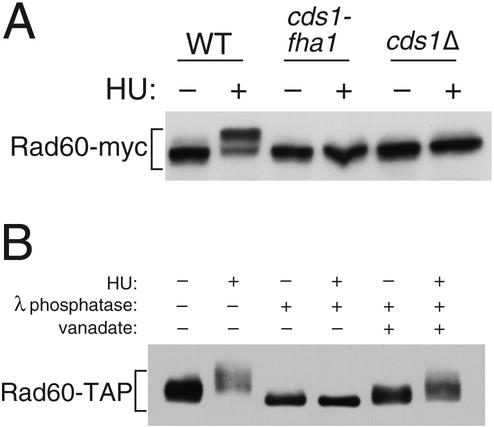
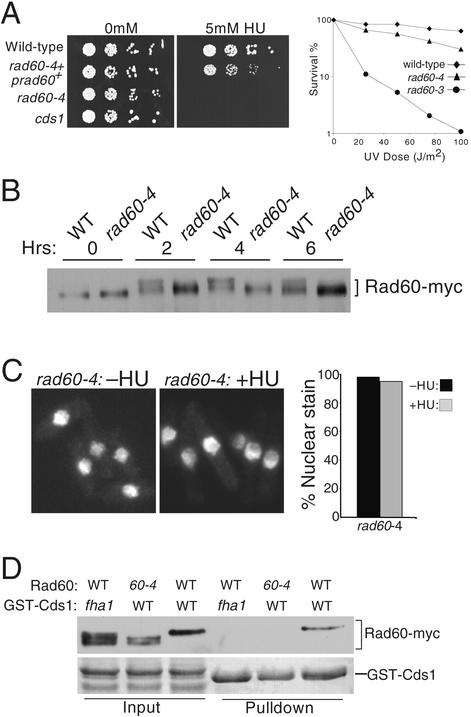
GST-Cds1 expression led to the appearance of a species of Rad60 with slower electrophoretic mobility, whereas expression of GST-Cds1-fha1 had no effect on Rad60 electrophoretic mobility (Fig. 1B). These findings suggested that Cds1 controls Rad60 phosphorylation. Indeed, a slower-electrophoretic-mobility form of Rad60-myc appeared in cells exposed to HU (Fig. 2A), a treatment that activates Cds1 (4, 25). Phosphatase treatment enhanced the electrophoretic mobility of Rad60-TAP expressed at endogenous levels, confirming that Rad60 electrophoretic retardation was caused by phosphorylation (Fig. 2B). Importantly, HU-induced phosphorylation of Rad60 was eliminated in cds1Δ and cds1-fha1 cells (Fig. 2A). Therefore, Cds1 activation leads to Rad60 phosphorylation.
FIG. 2.
Cds1 controls Rad60 phosphorylation. (A) Wild-type (WT), cds1-fha1, and cds1Δ cells were treated or left untreated with 12 mM HU for 4 h. The electrophoretic mobility of Rad60-myc was analyzed in each strain. HU caused the appearance of a reduced-mobility form of Rad60-myc in wild-type but not cds1-fha1 and cds1Δ cells. (B) Precipitates of Rad60-TAP from wild-type cells, treated or not with HU, were subjected to lambda phosphatase treatment. The slow-migrating forms of Rad60-TAP induced by HU were converted to a single faster-migrating species, showing that Rad60 is phosphorylated in response to HU treatment. The phosphatase inhibitor vanadate largely blocks the conversion of Rad60-TAP to the high-mobility species.
Replication arrest survival defect of rad60-3 mutant.
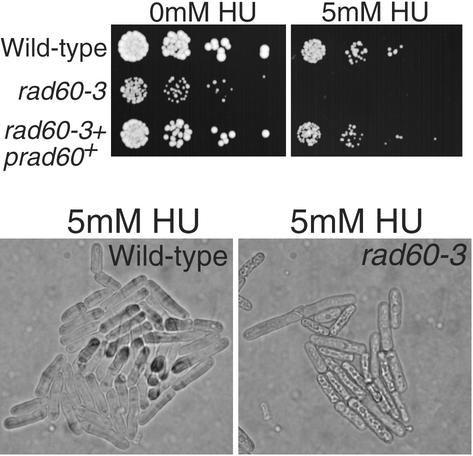
The evidence linking Cds1 and Rad60 suggested that Rad60 might be involved in tolerance of replication arrest. We isolated a rad60 temperature-sensitive allele to address this question. The rad60-3 allele changed codon 272 from phenylalanine to valine (F272V). This mutation is located very near the site mutated in the temperature-sensitive rad60-1 allele described by Morashita et al. (32), which changed codon 263 from lysine to glutamic acid (K263E). In common with rad60-1 cells, rad60-3 cells incubated at permissive temperature (25°C) were sensitive to DNA-damaging agents, such as IR (data not shown) and UV light (UV) (described below). Importantly, rad60-3 cells were also hypersensitive to the replication inhibitor HU (Fig. 3). Treatment with 5 mM HU led to the rapid arrest and death of rad60-3 cells, whereas wild-type cells continued division (Fig. 3, lower panel). These findings indicated that Rad60 plays an important role in survival of replication arrest.
FIG. 3.
Mutant rad60-3 cells are hypersensitive to HU. (A) The indicated strains were serially diluted (∼2,500, 500, 100, and 20 cells per spot) and plated on medium supplemented or not with 5 mM HU followed by incubation at 25°C. (B) Cells of the indicated strains were photographed on agar medium supplemented with 5 mM HU.
Rad60 nuclear delocalization provoked by replication arrest.
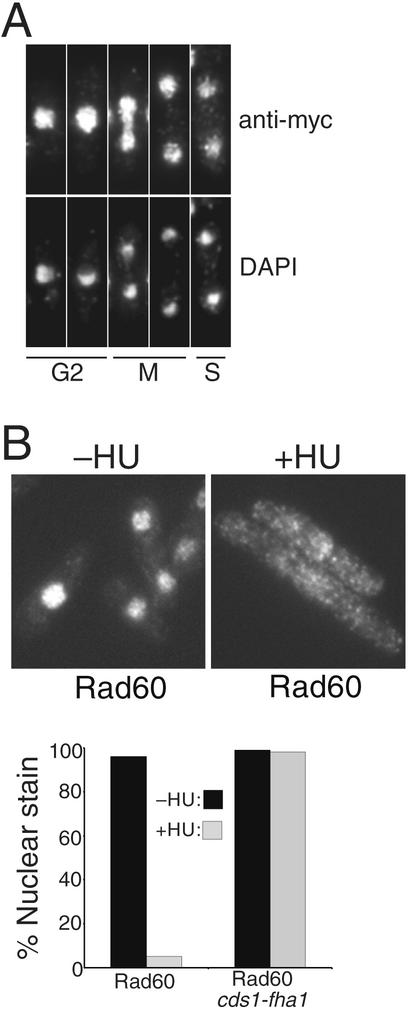
Our findings suggested that Cds1 regulates Rad60. We explored this possibility by examining the localization of 13myc-tagged Rad60 in cells exposed to HU. Rad60-myc, expressed at endogenous levels, was detected in the nucleus of untreated cells at all stages of the cell cycle (Fig. 4A). Strikingly, HU treatment provoked dispersal of Rad60 from the nucleus, resulting in a pan-cellular signal (Fig. 4B). This change occurred very soon after cells completed their first division following addition of HU. The G1 phase is very short in fission yeast grown in rich medium; thus, nuclear dispersal of Rad60 coincided with cells encountering the HU-induced S-phase block. We examined whether Rad60 delocalization was dependent on Cds1. In cds1-fha1 cells, Rad60 remained in the nucleus after exposure to HU (Fig. 4B, right panel). Thus, Cds1 controls localization of Rad60. The nuclear dispersal of Rad60 was specific for S-phase arrest, because it was not observed in G2 cells treated with IR (our unpublished observations). IR is not a potent activator of Cds1 (25).
FIG. 4.
Cds1 controls nuclear delocalization of Rad60 in HU-treated cells. (A) Rad60 is a nuclear protein throughout the cell cycle. The localization of endogenous Rad60 was determined by indirect immunofluorescence of 13myc-tagged protein. The Rad60-myc signal was strongest in the chromatin (DAPI staining) region of the nucleus. Cell cycle position was determined by DAPI stain and by a phase-contrast photo (data not shown) showing whether a septum was present in binucleate cells. (B) Rad60-myc delocalizes from the nucleus during replication arrest. Rad60-myc cells were treated or not with 10 mM HU for 4 h and fixed, and Rad60-myc was detected by indirect immunofluorescence. The percentage of cells with exclusively nuclear staining was determined and is shown at the bottom. Rad60-myc delocalization was abrogated by the cds1-fha1 mutation (right panel).
A Rad60 mutant insensitive to Cds1 control.
These findings suggested that Rad60 was an important target of regulation by Cds1. We sought to test this proposition by identifying a Rad60 mutant that was functional but uncoupled from regulation by Cds1. We hypothesized that such a mutant would be defective in survival of replication arrest induced by HU but proficient for survival of DNA damage caused by UV. A library of rad60 mutants was made by gene conversion of genomic rad60+ with copies of rad60 mutagenized in vitro. Extensive screening identified one mutant gene, rad60-4, that had the desired properties. Sequencing of rad60-4 revealed that it contains four mutations: T72A, I232S, Q250R, and K312N. Survival of HU treatment was profoundly impaired in rad60-4 cells, whereas their UV resistance was almost the same as that of the wild type (Fig. 5A). The levels of Rad60 abundance were approximately equal in the wild type and rad60-4 mutants, but electrophoretic retardation of Rad60 caused by HU-induced phosphorylation was almost abolished in rad60-4 cells (Fig. 5B). Strikingly, these properties of rad60-4 cells were accompanied by a failure to delocalize Rad60 from the nucleus in response to HU treatment (Fig. 5C).
FIG. 5.
A rad60 mutant insensitive to control by Cds1. (A) Mutant rad60-4 cells are sensitive to HU but relatively insensitive to UV. Cells were serially diluted (∼2,500, 500, 100, and 20 cells per spot) and plated in the presence or absence of 5 mM HU (left panel). A single integrated copy of wild-type rad60+ (prad60+) allowed rad60-4 cells to form colonies in the presence of 5 mM HU. Survival analysis of wild-type, rad60-3, and rad60-4 cells irradiated with UV is shown in the right panel. Cells were maintained at 25°C. (B) Electrophoretic retardation of Rad60 that induced by HU was largely eliminated in rad60-4 cells. Rad60-myc (wild type [WT]) and rad60-4-myc (rad60-4) cells were treated with 12 mM HU from 0 to 6 h. Samples were taken every 2 h and analyzed by SDS-PAGE. (C) Nuclear delocalization of Rad60 induced by 10 mM HU at 4 h was abrogated in rad60-4 cells. (D) The interaction between Rad60 and Cds1 was abolished by rad60-4. GST fusions of wild-type or mutant Cds1 (fha1) were expressed in cells that express myc epitope-tagged wild type Rad60 or rad60-4 from the rad60 genomic locus. Wild-type Rad60 coprecipitated with GST-Cds1 but not GST-Cds1-fha1. Rad60-4 did not coprecipitate with GST-Cds1. In addition, Rad60 but not Rad60-4 displayed maximum electrophoretic retardation caused by phosphorylation in response to expression of GST-Cds1 but not GST-Cds1-fha1.
We speculated that Rad60-4 might be refractory to Cds1 regulation because it failed to interact properly with Cds1. To test this possibility, we overexpressed GST-Cds1 in cells that expressed myc-tagged versions of rad60+ and rad60-4 from their genomic loci. Purification of GST-Cds1 resulted in coprecipitation of Rad60 but not Rad60-4 (Fig. 5D). The negative control GST-Cds1-fha1 failed to bind Rad60 (Fig. 5D), as previously observed (Fig. 1B). Interestingly, GST-Cds1 overexpression had different effects on the electrophoretic mobilities of Rad60 and Rad60-4. All of the Rad60-4 was detected in two faster-mobility species, whereas all of the Rad60 migrated at a slower position (Fig. 5D, lanes 2 and 3). These observations paralleled the results obtained by HU treatment of rad60+ and rad60-4 cells (Fig. 5B). The most straightforward interpretation of these observations is that Cds1 binds to and directly phosphorylates Rad60, with Rad60-4 being insensitive to Cds1 control because it fails to interact with Cds1. These findings provided compelling evidence for the model that regulation of Rad60 is a critical part of the replication checkpoint response controlled by Cds1.
Interactions involving Rad60 and Smc5-Smc6.
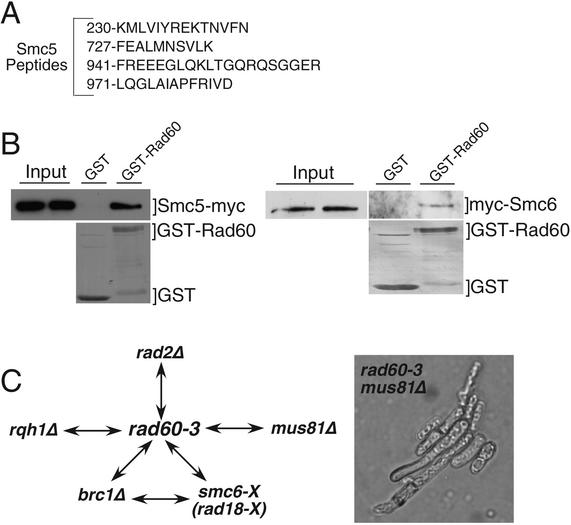
To gain more insight into the essential function of Rad60, as well as its role in recombinational repair and tolerance of replication arrest, we used MudPIT to identify proteins that coprecipitated with Rad60-TAP (35, 48). Excluding common contaminants, the best candidate uncovered by this method was Smc5 (Spr18), for which four peptides were identified (Fig. 6A). Smc5 is the binding partner of the Smc6 (Rad18) DNA repair protein (16). Smc6 was not detected in the MudPIT analysis of Rad60-TAP, but identification of Smc5 was near the limits of detection by this method. Coprecipitation of 13myc-tagged Smc5 with GST-Rad60 expressed in fission yeast confirmed the Rad60-Smc5 interaction (Fig. 6B). We estimated that ∼2% of the Smc5 coprecipitated with GST-Rad60, indicating that the interaction is substoichiometric and/or quite transient. Immunoblot analysis of GST-Rad60 preparations detected 3myc-tagged Smc6 (Fig. 6B; right panel), although again we estimated that only a very small fraction (∼0.5%) of the total Smc6 precipitated with GST-Rad60.
FIG. 6.
Rad60 interacts with Smc5 and Smc6. (A) Smc5 peptides identified in Rad60-TAP preparation. (B) GST or GST-Rad60 were expressed in strains that expressed 13myc-tagged Smc5 (Spr18) or 3myc-tagged Smc6 (Rad18) from their genomic loci (left and right panels, respectively). GST and GST-Rad60 were purified, and coprecipitating proteins were analyzed by anti-myc immunoblot (upper panels). The lower panels show Coomassie blue staining of purified GST-Rad60 and GST. Smc5 and Smc6 coprecipitated with GST-Rad60 but not GST. (C) Summary of synthetic lethal interactions involving rad60 and other genes. Double-headed arrows indicate synthetic lethality. The right-hand panel shows the rad60-3 mus81 synthetic lethal phenotype, following germination and formation of an approximately five-cell colony. Shown are cells on a tetrad dissection plate; wild-type cells had formed large colonies (data not shown).
The interactions involving Rad60, Smc5, and Smc6 were intriguing in light of studies showing that rad60-1 and smc6-X (rad18-X) have a synthetic lethal interaction (32). We observed a similar interaction for rad60-3 and smc6-X (Fig. 6C). The smc6-X allele is itself synthetic lethal with the brc1Δ mutation that eliminates Brc1, a BRCT domain protein required for optimum chromosome segregation and DNA damage tolerance (47). We found that rad60-3 and brc1Δ mutations were also synthetically lethal (Fig. 6C).
Interestingly, we also found that rad60-3 is synthetically lethal with mus81Δ and rqh1Δ (Fig. 6C). The rad60-3 mus81Δ double mutant germinated but arrested as elongated cells after one to three divisions (Fig. 6C, right panel). Mus81 is an HJ resolvase that is crucial for recovery from replication fork collapse, a process that requires HJ resolution (5, 7, 12). Mus81 was also proposed to play a backup role to the DNA helicase Rqh1 in processing stalled replication forks that regress to form HJs (5, 7, 12). The synthetic lethal interaction between rad60-3 and mus81Δ supports the suggestion that rad60-3 cells experience spontaneous DNA damage arising from fork collapse. Rqh1 has been implicated in the maintenance of replication forks and prevention of illegitimate recombination (12). These facts suggest that rqh1 mutants experience spontaneous fork collapse, a possibility consistent with the synthetic lethal interaction between mus81 and rqh1 mutations (7). The synthetic lethality of rad60-3 and rqh1Δ is consistent with a model in which Rad60 is required for recombinational repair of fork breaks that arise in an rqh1Δ background, although other explanations cannot be excluded.
The phenotypic similarities of rad60, smc5, and smc6 mutants, coupled with their parallel genetic interactions and the physical associations of their protein products, suggested strongly that Rad60 and Smc5-Smc6 have codependent functions in maintenance of chromosome structure and DNA repair.
DISCUSSION
This study has uncovered physical and functional interactions involving four proteins involved in genome maintenance: Cds1, Rad60, Smc5, and Smc6. The FHA module of Cds1 mediates an interaction with Rad60, controlling its phosphorylation and localization. Rad60 interacts with Smc5 and Smc6, subunits of an SMC complex essential for cell viability and DSB repair. Rad60 does not appear to be a stoichiometric subunit of the Smc5-Smc6 complex, but the similar phenotypes of rad60, smc5, and smc6 mutants, coupled with their synthetic lethal interactions, strongly suggest they have codependent functions. Replication arrest induced by HU leads to Cds1-dependent nuclear delocalization of Rad60, a protein that otherwise resides in the nucleus the entire cell cycle. These observations imply that Cds1 inhibits Rad60, because Rad60 presumably performs its DSB repair function in the nucleus. A failure of Cds1 to cause Rad60 nuclear delocalization, as observed in rad60-4 cells, correlates with a severe defect in survival of replication fork arrest. Therefore, regulation of Rad60 appears to be a significant part of the mechanism by which Cds1 promotes recovery from replication fork arrest.
Codependent functions of Rad60 and Smc5-Smc6.
Rad60 inactivation leads to a Chk1-dependent checkpoint arrest (our unpublished observation). This observation implies that DNA damage occurs when Rad60 function is impaired, most probably arising from defects in DNA replication. Synthetic lethal interactions involving rad60 and mus81 or rqh1 mutations support this model. The physical and genetic interactions involving Rad60, Smc5, and Smc6 strongly suggest codependent functions for these proteins. The simplest interpretation is that they function in a single pathway.
Rad60 interacts with Smc5 or Smc6, but these interactions are not as avid as that described between Smc5 and Smc6 (16). Indeed, the majority of Rad60 elutes as an apparent monomer in gel filtration columns (our unpublished observations). In this context, it is noteworthy that Psc3, which is required for cohesin (Smc1-Smc3) function in fission yeast, is not found in stable association with cohesin (44). We conclude that Rad60 is loosely or transiently associated with Smc5-Smc6 complex, but nevertheless is vital for its function.
The function of Rad60 may be comparable to those of Scc2 and Scc4 in budding yeast, which form a physically separate complex required to load cohesin onto chromosomes during S phase (10). Inactivation of Scc2 or Scc4 results in a cohesion defect equivalent to that of cohesin mutants. Another example is Eso1 and EcoI of fission and budding yeasts, which are required for the establishment of cohesion during S phase but are not required for its maintenance during G2 (40, 42, 46). It is tempting to speculate that Rad60 may have a comparable function, being required for Smc5-Smc6 loading onto chromosomes, although it has yet to be determined whether the Smc5-Smc6 complex stably associates with chromosomes.
Control of Rad60 localization.
Control of Rad60 by Cds1 is not a subtle effect. In response to HU treatment or overexpression of GST-Cds1, nearly all Rad60 becomes hyperphosphorylated and delocalized from the nucleus. Multiple electrophoretic forms of Rad60 were detected, suggesting that Rad60 is phosphorylated on several sites. The behavior of the rad60-4 mutant strongly suggests that Rad60's function in DNA repair does not require Cds1, but control of Rad60 phosphorylation and localization by Cds1 is important for survival of replication fork arrest.
Why does Cds1 control Rad60 localization? One interesting possibility is that Rad60 is removed from the nucleus precisely because it is required for recombinational repair. A stalled fork, with single-strand regions and DNA ends in close proximity to homologous sequences, would appear to be an ideal recombination substrate. DNA recombination is thought to be a choice of last resort in the resolution of stalled forks and may be useful only in rare circumstances when a fork has regressed to form a “chicken-foot,” an X-shaped DNA structure that can be cleaved by an HJ resolvase (5, 30, 41). The initial response to a stalled fork is probably to preserve the replisome and suppress recombination. Rad60 and Smc5-Smc6 appear to have central roles in repair of DNA damage by homologous recombination (24, 32, 47); thus, it is plausible that inactivation of these proteins by delocalization of Rad60 from the nucleus prevents counterproductive recombination events from occurring at stalled forks.
Potential Rad60 homologs in budding yeast and humans.
Database searching has revealed a family of proteins sharing the size and domain structure of Rad60 (for examples, see Fig. 1A). Fission yeast Rad60, budding yeast Esc2p, and mammalian NIP45 are all ∼400 amino acids in length and share a C-terminal domain that is most closely related to the ubiquitin-like protein, SUMO-1. Unlike SUMO-1, the Rad60 family does not have the extreme C-terminal sequences required for covalent attachment to other proteins. Hence, the Rad60 SUMO-1-related domain is likely to function as a protein-protein interface. Genetic arguments support the notion that at least Rad60 and Esc2 may be functional homologs. We have observed that rad60 mutants are synthetically lethal with deletion of rqh1, a gene that encodes a RecQ-like DNA helicase (Fig. 4B). In addition, Morishita et al. (32) identified rad60-1 through its synthetic lethal interaction with deletion of rad2, which encodes the FEN-1 endonuclease homolog in fission yeast. Although Esc2 is not essential in budding yeast, esc2Δ mutations are synthetic lethal with mutations of sgs1 and rad27, which encode Rqh1 and Rad2 homologs, respectively (45). These similarities suggest that Rad60 and Esc2 perform related functions. It is unknown whether Esc2 interacts with the SMC5-SMC6 complex in budding yeast. Mammalian NIP45 appears to potentiate NF-AT-dependent transcription (20). Budding yeast Esc2 is so named for its ability to establish silent chromatin when targeted to a particular locus (11). Further, Esc2 mutants display a mild silencing defect, supporting a role for Esc2 in chromatin remodeling. These similarities suggest a tenuous but plausible link between NIP45 and Esc2 functions. Further exploration is required to establish the unifying functions of members of this new protein family.
Acknowledgments
We thank C. McGowan for comments on the manuscript and members of the Scripps Cell Cycle Groups for encouragement.
M.N.B. is a Research Special Fellow of the Leukemia & Lymphoma Society. W.H.M was supported by MERK-MGRI-241. E.N. was supported by the Human Frontiers Science Program. J.R.Y. was supported by RO1 EY1328801, MERK-MGRI-241, and CA81665 RR11823. This work was funded by NIH grants awarded to P.R.
REFERENCES
- 1.Aono, N., T. Sutani, T. Tomonaga, S. Mochida, and M. Yanagida. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197-202. [DOI] [PubMed] [Google Scholar]
- 2.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W.-D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkenbihl, R. P., and S. Subramani. 1995. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 270:7703-7711. [DOI] [PubMed] [Google Scholar]
- 4.Boddy, M. N., B. Furnari, O. Mondesert, and P. Russell. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280:909-912. [DOI] [PubMed] [Google Scholar]
- 5.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 6.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 7.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W.-D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddy, M. N., and P. Russell. 2001. DNA replication checkpoint. Curr. Biol. 11:R953-956. [DOI] [PubMed] [Google Scholar]
- 9.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 10.Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 11.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 12.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 13.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 14.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 15.Emili, A., D. M. Schieltz, J. R. Yates III, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 16.Fousteri, M. I., and A. R. Lehmann. 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe, and K. Kohno. 2002. Identification of a novel non-SMC component of the SMC5/SMC6 complex involved in DNA repair. J. Biol. Chem. 277:21585-21591. [DOI] [PubMed] [Google Scholar]
- 18.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 19.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 20.Hodge, M. R., H. J. Chun, J. Rengarajan, A. Alt, R. Lieberson, and L. H. Glimcher. 1996. NF-AT-driven interleukin-4 transcription potentiated by NIP45. Science 274:1903-1905. [DOI] [PubMed] [Google Scholar]
- 21.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, S. T., B. Xu, and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann, A. R., M. Walicka, D. J. F. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsay, H., D. Griffiths, R. Edwards, P. Christensen, J. Murray, F. Osman, N. Walworth, and A. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12:382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 27.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 regulated by DNA damage and 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 28a.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 29.McCormack, A. L., D. M. Schieltz, B. Goode, S. Yang, G. Barnes, D. Drubin, and J. R. Yates III. 1997. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69:767-776. [DOI] [PubMed] [Google Scholar]
- 30.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 31.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 32.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhind, N., and P. Russell. 2000. Checkpoints: it takes more than time to heal some wounds. Curr. Biol. 10:R908-R911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhind, N., and P. Russell. 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113:3889-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 36.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh, H., R. T. Pu, and M. Dasso. 1997. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22:374-376. [DOI] [PubMed] [Google Scholar]
- 38.Shiozaki, K., and P. Russell. 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 283:506-520. [DOI] [PubMed] [Google Scholar]
- 39.Sjogren, C., and K. Nasmyth. 2001. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11:991-995. [DOI] [PubMed] [Google Scholar]
- 40.Skibbens, R. V., L. B. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, K., Z. Hao, M. Kai, and H. Okayama. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 44.Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S. E. Kearsey, F. Uhlmann, K. Nasmyth, and M. Yanagida. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 46.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]