Abstract
The repair of DNA single-strand breaks in mammalian cells is mediated by poly(ADP-ribose) polymerase 1 (PARP-1), DNA ligase IIIα, and XRCC1. Since these proteins are not found in lower eukaryotes, this DNA repair pathway plays a unique role in maintaining genome stability in more complex organisms. XRCC1 not only forms a stable complex with DNA ligase IIIα but also interacts with several other DNA repair factors. Here we have used affinity chromatography to identify proteins that associate with DNA ligase III. PARP-1 binds directly to an N-terminal region of DNA ligase III immediately adjacent to its zinc finger. In further studies, we have shown that DNA ligase III also binds directly to poly(ADP-ribose) and preferentially associates with poly(ADP-ribosyl)ated PARP-1 in vitro and in vivo. Our biochemical studies have revealed that the zinc finger of DNA ligase III increases DNA joining in the presence of either poly(ADP-ribosyl)ated PARP-1 or poly(ADP-ribose). This provides a mechanism for the recruitment of the DNA ligase IIIα-XRCC1 complex to in vivo DNA single-strand breaks and suggests that the zinc finger of DNA ligase III enables this complex and associated repair factors to locate the strand break in the presence of the negatively charged poly(ADP-ribose) polymer.
Three human genes, LIG1, LIG3, and LIG4, that encode DNA ligases have been identified (30). Unlike the LIG1 and LIG4 genes, which appear to be conserved among all eukaryotes, the LIG3 gene has been found only in the genomes of mammals and of the amphibian Xenopus laevis (6, 22, 32). Intriguingly, the LIG3 gene is more closely related to poxvirus DNA ligase genes than to those for the other eukaryotic DNA ligases (6, 10). Furthermore, the LIG3 gene is more complex than the other LIG genes in that it encodes multiple products that appear to have distinct biological functions.
Alternative splicing of the LIG3 gene transcript generates two species of mRNA, designated α and β, that encode polypeptides with different C termini (17, 22). DNA ligase IIIα mRNA is ubiquitously expressed, whereas DNA ligase IIIβ mRNA has been detected only in germ cells (17, 22). The unique C terminus of DNA ligase IIIα, which exhibits homology with the BRCT motif initially identified in the product of the breast cancer susceptibility gene BRCA1 (5, 11), mediates formation of a stable complex with the DNA repair protein XRCC1 (3, 4, 17, 21, 29). In contrast, no protein partner or biochemical activity has been ascribed to the unique C terminus of DNA ligase IIIβ. Further heterogeneity of products from the LIG3 gene is generated by translation initiation at different ATG codons within DNA ligase III mRNA, generating mitochondrial and nuclear forms of DNA ligase III (14, 15, 22).
A unique feature of the DNA ligases encoded by the LIG3 gene is the zinc finger motif situated at the N termini of these polypeptides (32). Interestingly, this motif is closely related to the two tandem-arrayed zinc fingers that constitute the DNA binding domain of poly(ADP-ribose) polymerase 1 (PARP-1), a nuclear protein that binds avidly to DNA strand breaks and catalyzes ADP-ribosylation of itself and other proteins by using NAD as a cofactor (7, 32). Previous studies have shown that the zinc finger of DNA ligase III enables this enzyme to bind to DNA strand breaks, in particular single-strand breaks, and to efficiently ligate DNA nicks at physiological salt concentrations (16). However, the zinc finger is not required either for catalytic activity in vitro or for in vivo function in a heterologous organism (16).
Although Lig3 mutant mammalian cell lines are not currently available, the xrcc1 mutant Chinese hamster cell lines EM9 and EMC11 are functionally DNA ligase III deficient because, in the absence of XRCC1 protein, the levels of nuclear DNA ligase IIIα protein are significantly reduced (3, 4, 28, 36). Genetic and biochemical studies with these mutant cells have implicated XRCC1 in the short-patch subpathway of DNA base excision repair and DNA single-strand break repair (8, 28, 29). XRCC1 itself has no known catalytic activity, but this protein binds to nicked DNA (18) and to several other DNA repair proteins, including PARP-1 (19), PARP-2 (26), DNA polymerase β (Pol β) (2, 12), polynucleotide kinase (33), and apurinic-apyrimidinic (AP) endonuclease (31), in addition to DNA ligase IIIα (3, 17, 21). Based on these observations, it has been suggested that XRCC1 acts as a scaffolding factor in the assembly of multiprotein DNA repair complexes. Recent studies demonstrating that XRCC1 may also function independently of DNA ligase IIIα (20, 27) and that the mitochondrial form of DNA ligase IIIα functions independently of XRCC1 (14, 15) indicate that the roles of DNA ligase IIIα in somatic cells cannot be deduced solely on the basis of its interaction with XRCC1.
Using DNA ligase IIIβ as the ligand, we fractionated a HeLa extract by affinity chromatography and identified a specific association between DNA ligase III and the DNA strand break binding factor, PARP-1. In subsequent studies, we show that DNA ligase III not only directly interacts with PARP-1 but preferentially binds to poly(ADP-ribosyl)ated PARP-1, providing a mechanism for the recruitment of the DNA ligase IIIα-XRCC1 complex to DNA single-strand breaks in somatic cells. Finally, we demonstrate that the zinc finger of DNA ligase III is required for efficient ligation in the presence of poly(ADP-ribosyl)ated PARP-1, suggesting that this DNA binding activity allows the ligase to locate DNA nicks in the presence of the negatively charged poly(ADP-ribose) polymer (PAR).
MATERIALS AND METHODS
Plasmids.
The plasmid pTG-LigIII that encodes full-length DNA ligase IIIβ as a glutathione S-transferase (GST) fusion protein has been described previously (17). Removal of an XhoI fragment corresponding to nucleotides 459 to 2589 of DNA ligase III cDNA followed by religation generated the plasmid pTG-LigIII1-152 that encodes the N-terminal 152 amino acids of DNA ligase III as a GST fusion protein. The DNA sequence encoding the last 692 amino acids of DNA ligase IIIβ was amplified by PCR and then subcloned into the pGSTag vector (24) to generate the plasmid pTG-LigIIIβ170-862 that encodes a C-terminal fragment of DNA ligase IIIβ containing the catalytic domain as a GST fusion protein. Plasmids encoding His-tagged versions of DNA ligase III have been described previously (16).
Expression and purification of recombinant DNA ligase III.
Plasmids encoding the GST fusion derivatives of DNA ligase III were transformed into Escherichia coli BL21. One-liter cultures were grown at 37°C in Terrific Broth medium containing 100 μg of ampicillin/ml. At an optical density at 600 nm of 0.6, isopropyl thiogalactoside was added to a final concentration of 1 mM and incubation was continued for 4 h. Cells were collected by centrifugation and then resuspended in a 100-ml solution of 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 1 mM benzamidine-HCl. After clarification by sonication, GST fusion proteins were purified from the lysate by stepwise elution from a phosphocellulose column and then further purified to near homogeneity by glutathione-Sepharose affinity chromatography. His-tagged versions of human DNA ligase IIIβ were expressed and purified as described previously (16). Unless specifically noted, the experiments were carried out with DNA ligase IIIβ translated from the preferred internal ATG site (6, 14). This form of the protein and its truncated derivatives, which lack the mitochondrial leader sequence (14), are referred to as DNA ligase III. Recombinant human PARP-1 purified from baculovirus-infected insect SF9 cells (9) was a gift from Gilbert de Murcia.
Fractionation of a HeLa nuclear extract by DNA ligase IIIβ affinity chromatography.
Purified GST (2 mg) and GST-DNA ligase IIIβ (2 mg) were each covalently attached to 1 ml of Affigel-10 beads (Bio-Rad) according to the manufacturer's instructions. Nuclear extracts were prepared from a frozen pellet of HeLa S3 cells (5 × 109 cells) as described previously (34) and dialyzed against a solution containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.5 mM EDTA, 10 μg of leupeptin/ml, 1 mM PMSF, and 1 mM benzamidine-HCl. Nuclear extract (20 mg) was loaded onto Affigel beads liganded by either GST or GST-DNA ligase IIIβ that had been preequilibrated with buffer A (100 mM morpholinepropanesulfonic acid [MOPS, pH 7.5], 50 mM NaCl, 10 μg of leupeptin/ml, 1 mM PMSF, 1 mM benzamidine-HCl). After a wash with buffer A, bound proteins were eluted stepwise with buffer A containing 0.15 M NaCl and then 0.3 M NaCl. Proteins in equivalent column fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13) and then detected by using the Silver Stain Plus kit (Bio-Rad).
Mass spectrometry.
Proteins that were preferentially retained by the GST-DNA ligase IIIβ beads were separated by preparative SDS-PAGE and stained with Coomassie blue. Bands were cut out of the gel and diced into small pieces. The gel pieces were vortexed with 100 μl of 25 mM ammonium bicarbonate-50% acetonitrile three times and then dried in a SpeedVac SC100 for 20 min. The dried gel pieces were resuspended in 20 μl of 25 mM ammonium bicarbonate containing 5 μg of sequencing-grade trypsin (Promega)/ml, vortexed for 10 min, and then incubated at 4°C for 30 min. The supernatant was discarded and 20 μl of 25 mM ammonium bicarbonate was added to the gel pieces, which were then incubated at 37°C for 16 h. After the addition of 100 μl of H2O, samples were vortexed for 10 min. The supernatant was collected and added to 5 μl of 50% acetonitrile-5% formic acid in a 0.65-ml siliconized tube. The gel pieces were washed two more times with 5 μl of 50% acetonitrile-5% formic acid, and the supernatants were all combined. The pooled supernatants were reduced to 10 μl in a SpeedVac and then diluted by the addition of 100 μl of H2O. After concentration to 5 μl, an equal volume of 50% acetonitrile-5% formic acid was added, and an aliquot (1 μl) was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Voyager-DE Pro; Applied Biosystems). Peptides in the sample were compared against those in the University of California, San Francisco, MS-FIT database (ProteinProspector package) for identification.
Immunoblotting.
Proteins were separated by SDS-PAGE and then transferred onto nitrocellulose membranes. The membranes were incubated with antibodies against DNA ligase III (GeneTex), Ku70 and Ku80 (NeoMarkers), PARP-1 (Trevigen), PAR (Trevigen), or XRCC1 (NeoMarkers) in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.05% Tween 20 containing 2% dried milk overnight at 4°C. After incubation with the appropriate secondary antibody linked to horseradish peroxidase, antigen-antibody complexes were detected by enhanced chemiluminescence (Pierce).
Yeast two-hybrid genetic screen.
PARP-1 and DNA ligase IIIα cDNAs were cloned into the two-hybrid vectors pGADT7 and pGBKT7 (Clontech), respectively. Plasmids were transformed individually into the isogenic haploid Saccharomyces cerevisiae strains PJ69-4a and PJ69-4α. Diploids were selected on synthetic medium lacking tryptophan and leucine and replica plated onto plates of synthetic complete medium supplemented with 30 mM 3-amino-1,2,4-triazole but lacking tryptophan, leucine, and histidine. Colonies were incubated at 30°C for 3 days and then assayed for β-galactosidase activity (1). To confirm the protein-protein interaction, the colonies were transferred onto synthetic medium lacking tryptophan, leucine, and adenine.
Pull-down assays.
Nickel-agarose beads (10 μl) liganded by either His-tagged DNA ligase IIIβ (2 μg) or DNA ligase IIIβ lacking the zinc finger (2 μg) were washed with buffer B (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 20 imidazole) and then resuspended in 0.5 ml of buffer B containing 5 mg of bovine serum albumin/ml and purified PARP-1 (1 μg). After incubation at 4°C with constant rotation for 1 h, the beads were washed extensively with buffer B. Proteins were eluted from the beads with SDS sample buffer. After separation by SDS-PAGE, PARP-1 was detected by immunoblotting.
Surface plasmon resonance.
A 40-bp hairpin duplex containing a single-strand break was biotinylated at the 3′ terminus and coupled to a streptavidin-coated Biacore CM-5 sensor chip. Full-length DNA ligase IIIβ, DNA ligase IIIβ lacking the zinc finger, or PARP-1 dialyzed against 10 mM HEPES (pH 7.4)-150 mM NaCl-3 mM EDTA was injected (60 μl at 5 μl/min) at various concentrations and monitored for association and dissociation. Dissociation constants were calculated using BIAevaluation software (version 3.1) in which experimental data are fitted with kinetic models.
DNase I footprinting.
An 80-mer oligonucleotide (50 pmol) was end labeled with [γ-32P]ATP (6,000 Ci/mmol; NEN) and polynucleotide kinase (New England Biolabs) and then annealed with two complementary oligonucleotides (50 pmol of each) to generate a labeled 80-bp duplex with a single nick. Radiolabeled nicked duplexes (800 fmol) and DNA ligase IIIβ were incubated on ice for 10 min in a solution containing 50 mM Tris-HCl (pH 8.0), 0.1 M KCl, 12.5 mM MgCl2, 1.0 mM EDTA, 20% glycerol, and 1.0 mM dithiothreitol. The DNase I cleavage reaction was started with the addition of 50 μl of 5 mM CaCl2-10 mM MgCl2 and 75 μl of DNase I (50 U/ml; Promega) at room temperature. Reactions were terminated after 1 min by the addition of 90 μl of stop solution (0.05% bromophenol blue, 0.05% xylene cyanol, 0.2 M NaCl, 0.03 M EDTA, 1% SDS, and 100 μg of yeast RNA/ml). After phenol-chloroform extraction and ethanol precipitation, DNA was electrophoresed through a 15% polyacrylamide gel containing 8 M urea. Labeled DNA products in the dried gel were detected by autoradiography.
Kinase protection assay.
A 50-bp duplex oligonucleotide containing a single-nucleotide gap with only the 5′ terminus at the gap unphosphorylated (0.5 pmol) was preincubated in kinase buffer with 3 pmol of either full-length DNA ligase IIIβ or PARP-1 in the presence or absence of 1 mM NAD. After 10 min at room temperature, 10 U of T4 polynucleotide kinase was added and incubation was continued for 5 min. Reactions were stopped by phenol-chloroform extraction. DNA was precipitated with ethanol and then electrophoresed through a 6% denaturing polyacrylamide gel for 1.5 h. Labeled DNA products in the dried gel were detected and quantitated by phosphorimager analysis.
DNA ligation assays.
The substrate used in DNA joining assays was a 50-bp duplex oligonucleotide containing a single nick. The 5′ terminus of the 30-mer at the nick was end labeled with polynucleotide kinase and [γ-32P]ATP. DNA substrate (1 pmol) was preincubated in ligation buffer with either 4 or 2 pmol of full-length PARP-1 for 10 min at room temperature in the presence or absence of 1 mM NAD as indicated. Subsequently, DNA ligase IIIβ, truncated DNA ligase IIIβ lacking the zinc finger, or DNA ligase I (50 or 100 fmol as indicated) was added and incubation was continued for 10 min at room temperature. Reactions were stopped with 5 μl of gel loading dye (0.05% bromophenol blue, 0.05% xylene cyanol, and 20 mM EDTA in 100% formamide), and then DNA was electrophoresed through a 6% denaturing polyacrylamide gel for 1.5 h. Labeled DNA products in the dried gel were detected and quantitated by phosphorimager analysis.
Immunoprecipitation.
For immunoprecipitation of purified proteins, protein A/G Sepharose beads (10 μl) were incubated with monoclonal anti-PARP-1 or anti-PAR antibody and then washed twice with 100 mM MOPS (pH 7.5)-50 mM NaCl. The beads were resuspended in 0.5 ml of the same buffer containing 5 mg of bovine serum albumin/ml and either purified PARP-1 (1 μg) or PAR (0.2 μg; Trevigen) and then incubated at 4°C with constant rotation for 1 h. Purified DNA ligase IIIβ (1 μg) or truncated DNA ligase IIIβ lacking the zinc finger (1 μg) was added to the reaction mixture, which was incubated at 4°C with constant rotation for 1 h. The beads were washed extensively, and the proteins were eluted with SDS sample buffer. After separation by SDS-PAGE, proteins were detected by immunoblotting with the appropriate antibody.
HeLa cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum prior to incubation with 5 mM hydrogen peroxide (H2O2) in serum-free media for 1 h at 37°C. Following H2O2 treatment or mock treatment, cells were harvested and lysed in a solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet-P40, 10 μg of leupeptin/ml, 1 mM PMSF, and 1 mM benzamidine-HCl. The lysate was then cleared by centrifugation for 10 min at 16,000 × g at 4°C. PARP-1 polyclonal antibody (3 μl; Serotec) was combined with 2 mg of cleared lysate and protein A/G Sepharose beads (15-μl bed volume) preequilibrated with lysis buffer and then incubated with constant rotation at 4°C for 16 h. After extensive washing, proteins were eluted from the beads with SDS sample buffer and then separated by SDS-PAGE. Proteins were detected by immunoblotting.
RESULTS
DNA ligase III interacts directly with PARP-1.
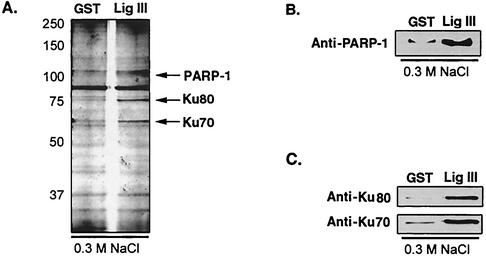
To identify proteins in somatic cells that associate with DNA ligase IIIα independently of XRCC1, we fractionated a HeLa nuclear extract by affinity chromatography by using GST-DNA ligase IIIβ, which lacks the C-terminal XRCC1-interacting domain of DNA ligase IIIα (6, 21) as the ligand. Analysis of proteins in the column eluates by silver staining after SDS-PAGE revealed the presence of three polypeptides with molecular masses of 110, 80, and 70 kDa that were much more abundant in the 0.3 M NaCl eluate from the DNA ligase column than in that from the GST column (Fig. 1A). To identify these polypeptides, the bands were excised from the gel and, after in situ digestion by trypsin, the masses of peptides were determined by MALDI-TOF and compared against those in a public database. This initial identification was confirmed by immunoblotting with antibodies against PARP-1 (Fig. 1B) and Ku80 and Ku70 (Fig. 1C).
FIG. 1.
Identification of DNA ligase III-associated proteins by affinity chromatography. A HeLa nuclear extract was fractionated by using a DNA ligase III affinity chromatography column as described in Materials and Methods. (A) Proteins in comparable fractions eluted with 0.3 M NaCl from the beads liganded by GST and by GST-DNA ligase III (Lig III) were detected by silver staining after separation by SDS-PAGE. The polypeptides indicated were identified by MALDI-TOF mass spectrometry. The identities of PARP-1, Ku70, and Ku80 were verified by immunoblotting with PARP-1 (B) and Ku70 and Ku80 (C) monoclonal antibodies.
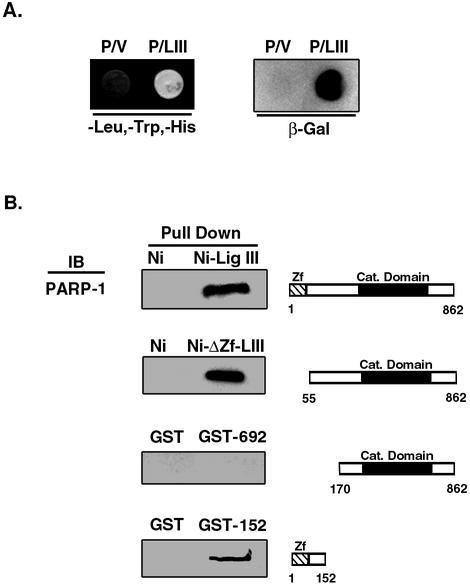
Since both PARP-1 and the Ku70/Ku80 heterodimer are abundant nuclear proteins that bind avidly to DNA strand breaks, the preferential binding of these factors to the DNA ligase III column could have been mediated by DNA. Because of genetic and biochemical studies linking PARP-1 and DNA ligase IIIα with the repair of DNA single-strand breaks (2, 19, 29, 36), we chose to further characterize the association between these proteins by using different approaches. PARP-1 and DNA ligase III were found to specifically interact both in the yeast two-hybrid in vivo assay (Fig. 2A) and in pull-down assays with purified proteins (Fig. 2B). By deletion analysis, the PARP-1 binding site was localized to residues 55 to 152 of the nuclear form of DNA ligase III (6, 14). Together, these results provide compelling evidence that PARP-1 and DNA ligase III interact directly in a reaction that is dependent on the N-terminal region of DNA ligase III adjacent to the zinc finger motif.
FIG. 2.
(A) The yeast two-hybrid genetic screen was employed to determine whether DNA ligase III interacts directly with PARP-1. Yeast strains were constructed as described in Materials and Methods. Colonies were tested for growth on triple dropout medium (-Leu,-Trp,-His) and subjected to a β-galactosidase (β-Gal) filter assay to detect protein-protein interactions as indicated. P/LIII, yeast strain expressing PARP-1 fused to the GAL4 activation domain (P) and DNA ligase III fused to the GAL4 DNA binding domain (LIII); P/V, yeast strain expressing the GAL4 binding domain (V) and PARP-1 fused to the GAL4 activation domain (P). (B) Mapping of the region of DNA ligase III that interacts with PARP-1. Pull-down assays with glutathione Sepharose and nickel beads were performed as described in Materials and Methods with purified PARP-1 (10 nM) and tagged versions of DNA ligase III. Ni, nickel beads only; Ni-Lig III, nickel beads liganded by His-tagged full-length DNA ligase III; Ni-ΔZf-LIII, nickel beads liganded by His-tagged DNA ligase III lacking the N-terminal 55 amino acids; GST, glutathione Sepharose beads liganded by GST; GST-692, glutathione Sepharose beads liganded by a GST fusion protein containing the C-terminal 692 residues of DNA ligase III; GST-152, glutathione Sepharose beads liganded by a GST fusion protein containing the N-terminal 152 residues of DNA ligase III; Zf, zinc finger; cat. domain, catalytic domain. The binding of PARP-1 to the beads was detected by immunoblotting (IB) with PARP-1 monoclonal antibody.
Binding of PARP-1 and DNA ligase III to DNA single-strand breaks.
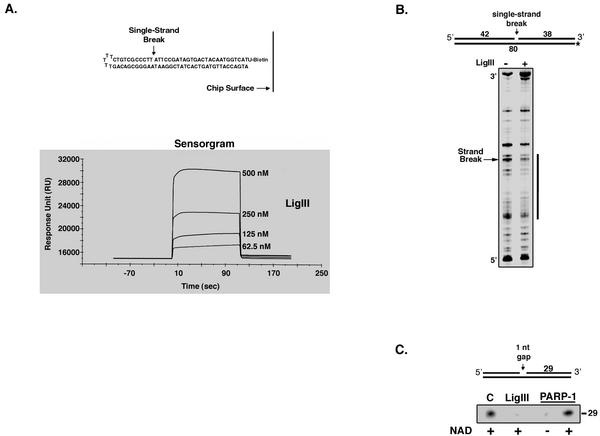
The direct interaction between PARP-1 and DNA ligase III, both of which possess similar zinc finger motifs at their N termini (32), suggests that these DNA binding domains may functionally interact in the repair of DNA single-strand breaks. To begin to examine this idea, we initially characterized the binding of DNA ligase III to DNA single-strand breaks. Using surface plasmon resonance, we monitored the binding and release (Fig. 3A) of DNA ligase III from an immobilized DNA substrate containing a single-strand break. From this data, the dissociation constant of DNA ligase III was calculated to be 5 nM. In similar experiments, the dissociation constant of truncated DNA ligase III lacking the zinc finger and that of PARP-1 were calculated to be 300 nM and 0.5 pM, respectively (data not shown). Thus, we conclude that the zinc finger is the major single-strand break binding activity in DNA ligase III and that the DNA binding domain of PARP-1 containing the two tandem-arrayed zinc fingers binds to DNA strand breaks with much higher affinity than does the single zinc finger of DNA ligase III.
FIG. 3.
Binding of DNA ligase III and PARP-1 to DNA single-strand interruptions. Effects of NAD on DNA binding by PARP-1 are shown. (A) Analysis of the binding of DNA ligase III to a DNA single-strand break by surface plasmon resonance. (Top panel) Schematic representation of the nicked hairpin oligonucleotide immobilized on the chip surface. (Bottom panel) Representative sensorgram showing the binding and release of untagged full-length DNA ligase III (LigIII), injected at the concentrations indicated, from the nicked hairpin oligonucleotide immobilized on the chip surface. (B) Visualization of DNA ligase III bound to a DNA single-strand break by DNase I footprinting. Intact DNA ligase III (LigIII; 20 nM) was preincubated with the labeled, nicked DNA substrate (16 nM) indicated and then incubated with DNase I as described in Materials and Methods. After separation by denaturing gel electrophoresis, labeled oligonucleotides were detected by autoradiography. −, no enzyme. The vertical line indicates the region protected from DNase I activity. (C) Effect of DNA strand break binding by DNA ligase III and PARP-1 on T4 polynucleotide kinase activity. Intact DNA ligase III (150 nM) or PARP-1 (150 nM) was preincubated with the indicated DNA substrate containing a single-nucleotide (1 nt) gap in the presence (+) or absence (−) of NAD as indicated prior to incubation with polynucleotide kinase and [γ-32P]ATP as described in Materials and Methods. After separation by denaturing gel electrophoresis, labeled 29-mer oligonucleotide was detected and quantitated by phosphorimager analysis. Lane C, no PARP-1 or DNA ligase III.
Next, the binding of DNA ligase III to a nonligatable DNA single-strand break was visualized by DNase I footprinting. Full-length DNA ligase III protected a region of 14 to 18 nucleotides in the intact strand that encompassed the strand break plus about 4 nucleotides on the 5′ side and about 14 nucleotides on the 3′ side of the break (Fig. 3B). The size and asymmetry of this footprint are similar to those observed with both PARP-1 and an N-terminal fragment of DNA ligase III that had been denatured by SDS-PAGE and then immobilized on a nitrocellulose membrane (16). These results suggest that binding of DNA ligase III to a DNA single-strand break will cover regions of the broken strand immediately adjacent to the break and block access to the termini at the break. To test this directly, we examined whether the complex formed by DNA ligase III at a DNA single-strand break prevented T4 polynucleotide kinase from reacting with the 5′ terminus of the DNA strand break. In accord with the footprinting results, preincubation of DNA ligase III with a DNA substrate containing a single-nucleotide gap effectively inhibited T4 polynucleotide kinase activity (Fig. 3C). As expected, purified PARP-1 also inhibited T4 polynucleotide kinase activity in the absence of NAD. However, in the presence of NAD, PARP-1 did not inhibit T4 polynucleotide kinase activity (Fig. 3C), indicating that, upon automodification, PARP-1 dissociates from the DNA strand break.
Effect of PARP-1 on DNA joining by DNA ligase III.
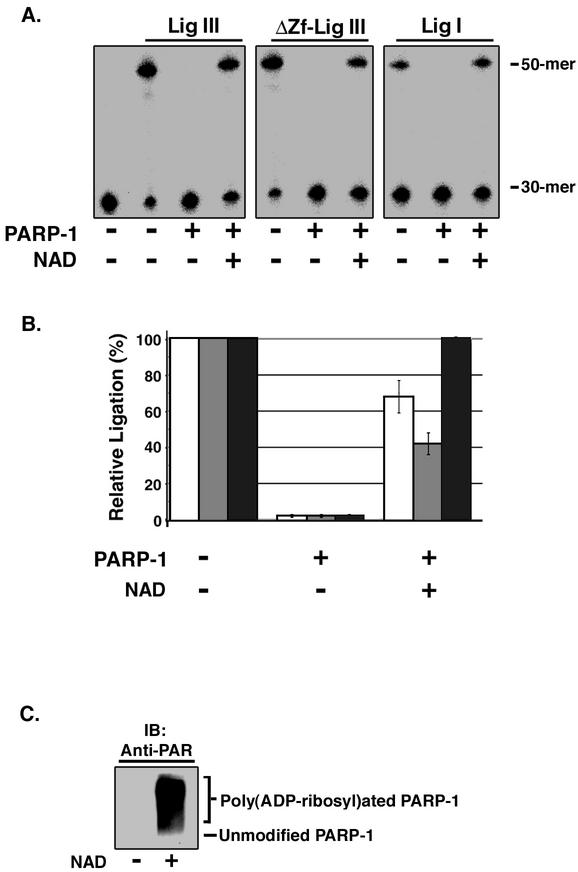
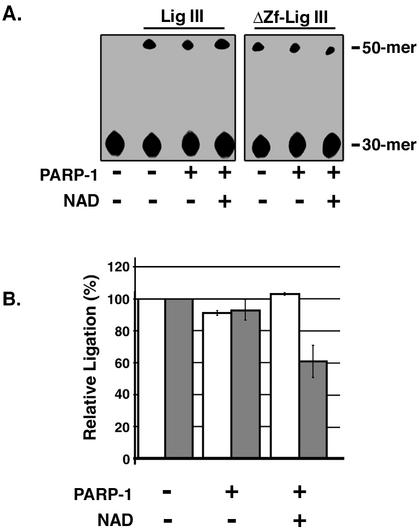
By quantitative immunoblotting of extracts from proliferating T24 and HeLa cells, we estimate that there are 2 × 105 PARP-1 molecules, 8 × 104 XRCC1 molecules, and 3 × 104 DNA ligase IIIα molecules per cell (Z. Dong and A. E. Tomkinson, unpublished results). Based on this numerical advantage and its higher affinity for DNA single-strand breaks, it seems likely that PARP-1 will bind to in vivo DNA single-strand breaks before DNA ligase IIIα does. In accord with the results of the kinase protection assays (Fig. 3C), DNA joining by DNA ligases I and III was almost completely inhibited by PARP-1 in the absence of NAD (Fig. 4A and B). However, in the presence of NAD when PARP-1 became poly(ADP-ribosyl)ated (Fig. 4C), DNA ligase I recovered maximum joining activity (Fig. 4A and B), providing further evidence that poly(ADP-ribosyl)ated PARP-1 dissociates from the DNA strand break. In contrast, both intact DNA ligase III and the truncated version of DNA ligase III lacking the zinc finger failed to recover maximum joining activity in the presence of poly(ADP-ribosyl)ated PARP-1 (Fig. 4A and B). Taken together, these results show that DNA joining by DNA ligase III but not DNA ligase I is partially inhibited by high levels of poly(ADP-ribosyl)ated PARP-1 and suggest that the interaction between DNA ligase III and PARP-1 modulates DNA joining.
FIG. 4.
Effect of DNA binding by PARP-1 on DNA joining. (A) Purified PARP-1 (200 nM) was preincubated with a labeled DNA duplex containing a single ligatable nick (50 nM) in the presence (+) or absence (−) of NAD. Subsequently, intact DNA ligase III (Lig III; 2.5 nM), a truncated version lacking the zinc finger (ΔZf-Lig III; 2.5 nM), or DNA ligase I (Lig I; 2.5 nM) was added to the reaction mixture as indicated. After incubation for 10 min at 25°C, labeled oligonucleotides were separated by denaturing gel electrophoresis. The labeled substrate (30-mer) and ligated product (50-mer) were detected and quantitated by phosphorimager analysis. (B) The results of three independent DNA joining assays are shown graphically. White bars, intact DNA ligase III; grey bars, truncated version of DNA ligase III lacking the zinc finger; black bars, DNA ligase I. (C) Purified PARP-1 (100 nM) was preincubated with a labeled DNA duplex containing a single ligatable nick (100 nM) in the presence (+) or absence (−) of NAD. After separation by SDS-PAGE, poly(ADP-ribosyl)ated PARP-1 was detected by immunoblotting (IB) with a monoclonal antibody against PAR. The positions of unmodified and poly(ADP-ribosyl)ated PARP-1 are indicated.
DNA ligase III binds directly to PAR.
With the use of a peptide binding assay, a putative PAR binding site (residues 12 to 34) has been identified within the zinc finger domain of DNA ligase III (23). To demonstrate that DNA ligase III does indeed interact with PAR, we performed coimmunoprecipitation experiments with PAR antibody. As expected, DNA ligase III was efficiently coimmunoprecipitated by the PAR antibody only in the presence of PAR (Fig. 5A). However, the truncated version of DNA ligase III lacking the zinc finger was also coimmunoprecipitated in a PAR-dependent manner, indicating that there are other PAR binding regions within DNA ligase III and that the previously identified PAR binding site within the zinc finger domain (23) may not be the predominant PAR binding site in DNA ligase III. In a similar experiment, DNA ligase I was not coimmunoprecipitated by the PAR antibody either in the presence or in the absence of PAR (data not shown).
FIG. 5.
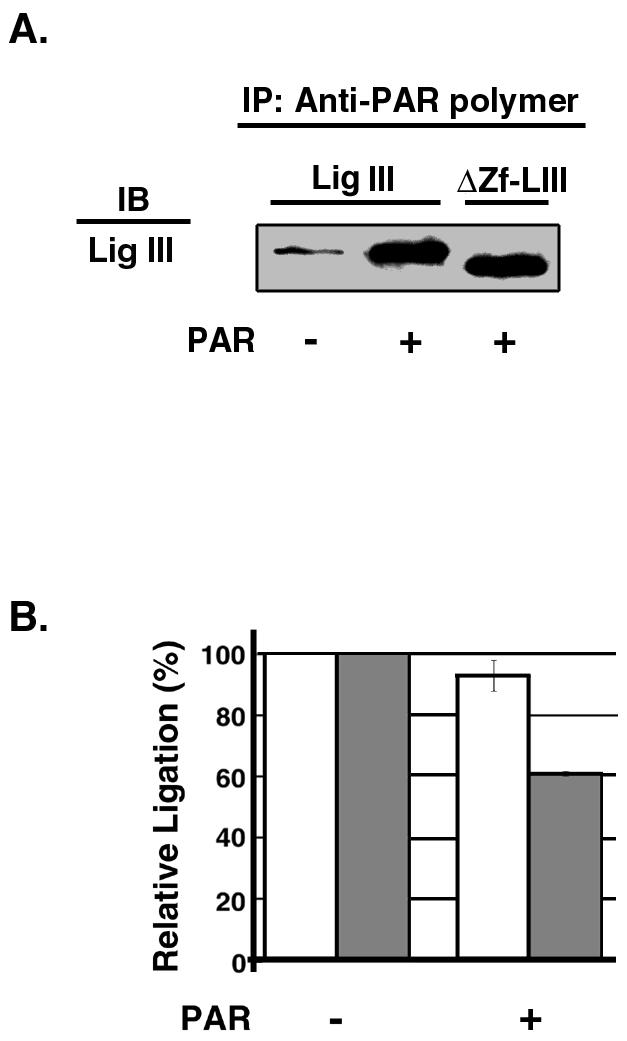
Interaction of DNA ligase III with PAR. Effects of PAR on DNA joining by DNA ligase III are shown. (A) Intact DNA ligase III (Lig III; 10 nM) or a truncated version lacking the zinc finger (ΔZf-LIII; 10 nM) was incubated with PAR antibody in the presence (+) or absence (−) of PAR. After separation of immunoprecipitated (IP) proteins by SDS-PAGE, DNA ligase III was detected by immunoblotting (IB). (B) DNA joining reactions with intact DNA ligase III (5 nM) or a truncated version lacking the zinc finger (5 nM) were carried out as described in Materials and Methods in the presence or absence of 0.5 nM PAR as indicated. The results of four independent DNA joining assays are shown graphically. White bars, intact DNA ligase III; grey bars, truncated version of DNA ligase III lacking the zinc finger.
Since the negatively charged PAR resembles the phosphodiester backbone of DNA, we examined the effect of PAR on DNA joining. Although both intact DNA ligase III and the truncated version of DNA ligase III lacking the zinc finger interact with PAR (Fig. 5A), PAR was significantly less effective at inhibiting DNA joining by intact DNA ligase III (<10% inhibition) (Fig. 5B) than that by the truncated version lacking the zinc finger (40% inhibition) (Fig. 5B). A similar effect was observed over a range of PAR concentrations from 0.5 to 50 nM (data not shown). These results suggest that the DNA ligase III zinc finger may facilitate recognition of the DNA nick in the context of the negatively charged PAR.
This model predicts that poly(ADP-ribosyl)ated PARP-1 should be more effective at inhibiting the truncated version lacking the zinc finger than it should be at inhibiting full-length DNA ligase III. Although we showed in Fig. 4 that the presence of the DNA ligase III zinc finger resulted in a greater recovery from inhibition by poly(ADP-ribosyl)ated PARP-1, the ratio of PARP-1 to DNA ligase III in these experiments was more than 10-fold higher than the in vivo ratio. Therefore, we examined the effect of PARP-1 on the DNA joining activity of DNA ligase III at a molar ratio that more closely reflects physiological conditions. In these reactions, unmodified PARP-1 inhibited DNA joining by less than 10% (Fig. 6). Formation of poly(ADP-ribosyl)ated PARP-1 by preincubation with the nicked DNA substrate and NAD resulted in a small increase in DNA joining by intact DNA ligase III (Fig. 6). In contrast, there was a decrease of about 30% in DNA joining by the truncated version of DNA ligase III lacking the zinc finger motif in similar reactions (Fig. 6). Taken together, these results strongly support the idea that the DNA ligase III zinc finger enhances the ability of DNA ligase III to detect DNA nicks in the presence of either PAR or poly(ADP-ribosyl)ated PARP-1.
FIG. 6.
Effect of poly(ADP-ribosyl)ated PARP-1 on DNA joining by DNA ligase III. (A) Purified PARP-1 (100 nM) was preincubated with a labeled DNA duplex containing a single ligatable nick (50 nM) in the presence (+) or absence (−) of NAD. Subsequently, intact DNA ligase III (Lig III; 5 nM) or a truncated version lacking the zinc finger (ΔZf-Lig III; 5 nM) was added to the reaction mixture as indicated. After incubation for 10 min at 25°C, labeled oligonucleotides were separated by denaturing gel electrophoresis. The labeled substrate (30-mer) and ligated product (50-mer) were detected and quantitated by phosphorimager analysis. (B) The results of three independent DNA joining assays are shown graphically. White bars, full-length DNA ligase IIIβ; grey bars, truncated version of DNA ligase IIIβ lacking the zinc finger.
DNA ligase III preferentially interacts with poly(ADP-ribosyl)ated PARP-1 in vitro and in vivo.
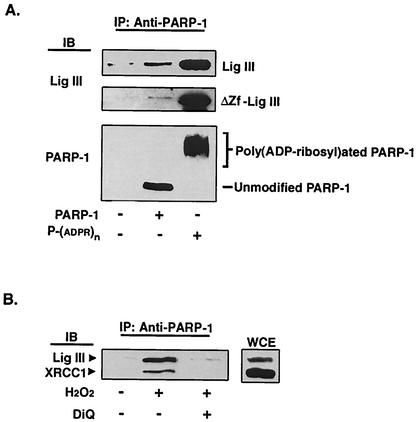
The direct binding of DNA ligase III to both PAR and PARP-1 suggests that this enzyme may preferentially bind to poly(ADP-ribosyl)ated PARP-1. In support of this idea, about fivefold more intact and truncated DNA ligase III was specifically coimmunoprecipitated by the PARP-1 antibody in the presence of automodified PARP-1 than in identical experiments with unmodified PARP-1 (Fig. 7A). The preferential interaction of DNA ligase III with poly(ADP-ribosyl)ated PARP-1 provides an attractive mechanism for the recruitment of DNA ligase IIIα to in vivo DNA single-strand breaks in the nuclei of somatic cells. To provide support for this hypothesis, extracts were prepared from undamaged cells and from cells treated with H2O2 to induce PAR synthesis and PARP-1 automodification. Although DNA damage did not cause a significant change in the cellular levels of DNA ligase IIIα or XRCC1, coimmunoprecipitation of the DNA ligase IIIα-XRCC1 complex by the PARP-1 antibody was DNA damage dependent (Fig. 7B). Pretreatment of the cells with the PARP-1 inhibitor 1,5-isoquinolinediol prevented the DNA damage-dependent association between PARP-1 and the DNA ligase IIIα-XRCC1 complex (Fig. 7B). Similar results were observed with another PARP-1 inhibitor, 3-aminobenzamide (data not shown). Since there are two- to threefold more XRCC1 molecules than DNA ligase IIIα molecules per cell, it is likely that there is a pool of free XRCC1 molecules. Notably, the ratio of DNA ligase IIIα to XRCC1 was significantly higher in the immunoprecipitates than in the cell extract (Fig. 7B), indicating that it is XRCC1 complexed with DNA ligase IIIα rather than free XRCC1 that preferentially associates with poly(ADP-ribosyl)ated PARP-1. Thus, our results indicate that the DNA ligase IIIα-XRCC1 complex is recruited to in vivo DNA strand breaks by its interaction with poly(ADP-ribosyl)ated PARP-1.
FIG. 7.
DNA ligase III preferentially binds to poly(ADP-ribosyl)ated PARP-1 in vitro. Effects of DNA damage on the association between PARP-1 and DNA ligase III-XRCC1 in vivo are shown. (A) Purified PARP-1 (500 nM) was preincubated with a labeled DNA duplex containing a single ligatable nick (300 nM) in the presence (+) or absence (−) of NAD. After treatment with DNase I, intact DNA ligase III (10 nM) or a truncated version lacking the zinc finger (10 nM) was added to the reaction mixture as indicated. Proteins immunoprecipitated (IP) by PARP-1 antibody were separated by SDS-PAGE and then detected by immunblotting (IB) with the indicated antibody. Upper panel, intact DNA ligase III (Lig III); middle panel, truncated version of DNA ligase III lacking the zinc finger (ΔZf-Lig III); lower panel, unmodified PARP-1 y(ADP-ribosyl)ated PARP-1 [P-(ADPR)n]. (B) Effect of H2O2 treatment on the association of PARP-1, DNA ligase IIIα, and XRCC1. Whole cell extracts (WCE) were prepared from undamaged (−) or damaged (+) HeLa cells as described in Materials and Methods. Equivalent aliquots of the cells to be damaged were pretreated with 1,5-isoquinolinediol (DiQ; 100 μM) as indicated for 1 h prior to and during H2O2 treatment. Proteins immunoprecipitated by PARP-1 antibody were separated by SDS-PAGE and then detected by immunoblotting with the indicated antibody. DNA ligase IIIα and XRCC1 in the extracts from undamaged cells were detected by direct immunoblotting.
DISCUSSION
The apparent absence of homologs of the LIG3 gene from the genomes of Drosophila, Caenorhabditis elegans, and S. cerevisiae indicates that this gene is restricted to mammals and amphibians and is presumably involved in DNA repair transactions that are unique to these higher eukaryotes (6, 16, 22, 27, 32). Because the nuclear form of DNA ligase IIIα is unstable without its partner protein XRCC1 (3, 4), it was assumed that DNA ligase IIIα functions together with XRCC1 in short-patch DNA base excision repair and the repair of DNA single-strand breaks (8, 28, 29, 36). However, recent studies have shown that the DNA ligase IIIα-XRCC1 complex is critical for repair for cells in the G1 phase of the cell cycle and in noncycling cells but that DNA ligase IIIα but not XRCC1 is dispensable for repair in cells in S phase (20, 27). To gain insights into the DNA repair events mediated by DNA ligase IIIα in the nuclei of somatic cells, we fractionated a HeLa nuclear extract by affinity chromatography and identified PARP-1 as a protein that specifically associates with DNA ligase III. In subsequent studies, we demonstrated that there is a direct physical interaction between PARP-1 and DNA ligase III that occurs via the region of amino acids immediately adjacent to the N-terminal zinc finger of DNA ligase III. Intriguingly, the DNA ligase III zinc finger is closely related to the tandem-arrayed N-terminal zinc fingers that constitute the DNA binding domain of PARP-1 (32). Our detection of a direct physical interaction between PARP-1 and the DNA binding region of DNA ligase III prompted us to determine whether these proteins functionally interact in DNA single-strand break repair.
Based on its relative abundance and high binding affinity, it seems likely that PARP-1 is the first factor to bind to DNA single-strand breaks in vivo (Fig. 8), resulting in activation of its polymerase activity and automodification since PARP-1 itself is the major acceptor for PAR (25). Although poly(ADP-ribosyl)ated PARP-1 dissociates from the DNA strand break (35), it is likely that PARP-1 molecules shuttle on and off the strand break, generating a network of PARs in the vicinity of the DNA strand break because of the relatively large number of PARP molecules per cell combined with the action of the poly(ADP-ribose) glycohydrolase (35). Presumably, this network of negatively charged polymers protects the DNA break by nonspecifically binding DNA metabolizing enzymes, such as nucleases, that recognize the negatively charged phosphodiester backbone of DNA (25, 35). The DNA damage-dependent association of DNA ligase IIIα with poly(ADP-ribosyl)ated PARP-1 in vivo suggests that the DNA ligase IIIα-XRCC1 complex is specifically recruited to the poly(ADP-ribosyl)ated PARP-1 molecules near the DNA strand break.
FIG. 8.
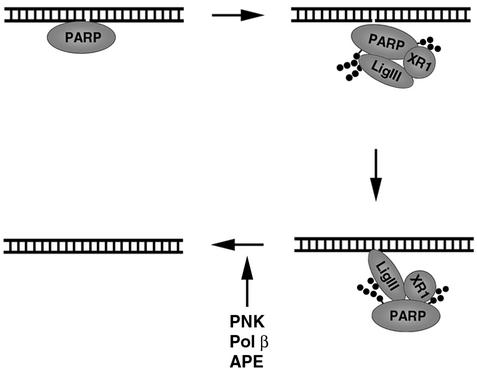
Model of the repair of DNA single-strand breaks by PARP-1 and DNA ligase III-XRCC1. The binding of PARP-1 to a DNA single-strand break activates PARP-1's polymerase activity that in turn results in automodification and dissociation of poly(ADP-ribosyl)ated PARP-1 from the strand break. DNA ligase IIIα (LigIII)-XRCC1 (XR1) associates with poly(ADP-ribosyl)ated PARP-1 in the vicinity of the DNA strand break. The zinc finger of DNA ligase IIIα enables this ternary complex to specifically recognize and bind to the DNA strand break despite the presence of negatively charged PARs. Additional repair factors such as polynucleotide kinase (PNK), Pol β, and AP endonuclease (APE) that process damaged termini are recruited via interactions with XRCC1. After the generation of ligatable termini, repair is completed by DNA ligase IIIα.
Although the binding to poly(ADP-ribosyl)ated PARP-1 provides a molecular mechanism for the relocation of the DNA ligase IIIα-XRCC1 complex to the vicinity of DNA damage sites in vivo, the complex then has to recognize and interact with DNA single-strand breaks when it is bound to negatively charged, automodified PARP-1. It has been previously shown that the DNA ligase III zinc finger is not required either for DNA joining activity in vitro or for the in vivo complementation of an E. coli lig mutant (16). These observations suggest that the DNA ligase III zinc finger may play a critical role only in the presence of PARP-1 and PAR. In support of this idea, we found that intact DNA ligase III was more effective at DNA joining than a truncated version of DNA ligase III lacking the zinc finger in the presence of either PAR or poly(ADP-ribosyl)ated PARP-1. Taken together, these results suggest that the role of the DNA ligase III zinc finger is to enable this enzyme to functionally interact with DNA single-strand breaks when it is complexed with poly(ADP-ribosyl)ated PARP-1 (Fig. 8). Although the effect of the DNA ligase III zinc finger was relatively small in the in vitro assays with naked DNA substrates, it may be more critical in vivo when the DNA strand breaks occur in chromatin with the adjacent histones probably poly(ADP-ribosyl)ated by PARP-1 (7, 25). Since the majority of DNA single-strand breaks will not have ligatable termini, it is likely that end processing enzymes, such as polynucleotide kinase, AP endonuclease, and Pol β, are recruited to the DNA ligase IIIα-XRCC1-DNA strand break complex via their interaction with XRCC1 (2, 12, 31, 33). Further biochemical and molecular genetic studies are needed to elucidate the complex relationship between PARP-1, DNA ligase IIIα-XRCC1, and XRCC1 in the network of DNA repair pathways that maintain genome stability in higher eukaryotes.
Acknowledgments
We thank Gilbert de Murcia for reagents and insightful comments on the manuscript and Guikai Wu for the mass spectrometry analysis.
This work was supported by National Institutes of Health grant GM47251 (to A.E.T.) and the Cancer Center support grant CA54174 to the San Antonio Cancer Institute.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. Kingston, D. Morre, J. Seidman, A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 2.Caldecott, K. W., S. Aoufouchi, P. Johnson, and S. Shall. 1996. XRCC1 polypeptide interacts with DNA polymerase β and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular nick-sensor in vitro. Nucleic Acids Res. 24:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott, K. W., C. K. McKeown, J. D. Tucker, S. Ljunquist, and L. H. Thompson. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldecott, K. W., C. K. McKeown, J. D. Tucker, L. Stanker, and L. H. Thompson. 1996. Characterization of the Xrcc1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callebaut, I., and J. P. Mornon. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400:25-30. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., A. E. Tomkinson, W. Ramos, Z. B. Mackey, S. Danehower, C. A. Walter, R. A. Schultz, J. M. Besterman, and I. Husain. 1995. Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol. 15:5412-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Murcia, G., and J. Menissier-de Murcia. 1994. Poly(ADP) ribose polymerase: a molecular nick sensor. Trends Biochem. Sci. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 8.Frosina, G., P. Fortini, O. Rossi, F. Carrozzino, G. Raspaglio, L. S. Cox, D. P. Dane, A. Abbondandolo, and E. Dogliotti. 1996. Two pathways of base excision repair in mammalian cells. J. Biol. Chem. 271:9573-9578. [DOI] [PubMed] [Google Scholar]
- 9.Giner, H., F. Simonin, G. de Murcia, and J. Menissier-de Murcia. 1992. Overproduction and large scale purification of human poly (ADP-ribose) polymerase using a baculovirus expression system. Gene 11:279-283. [DOI] [PubMed] [Google Scholar]
- 10.Husain, I., A. E. Tomkinson, W. A. Burkhart, M. B. Moyer, W. Ramos, Z. B. Mackey, J. M. Besterman, and J. Chen. 1995. Purification and characterization of DNA ligase III from bovine testes. J. Biol. Chem. 270:9683-9690. [DOI] [PubMed] [Google Scholar]
- 11.Koonin, E. V., S. F. Alschul, and P. Bork. 1996. Functional motifs. Nat. Genet. 13:266-267. [DOI] [PubMed] [Google Scholar]
- 12.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Lakshmipathy, U., and C. Campbell. 1999. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 19:3869-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakshmipathy, U., and C. Campbell. 2000. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 28:3880-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackey, Z. B., C. Niedergang, J. M. Murcia, J. Leppard, K. Au, J. Chen, G. de Murcia, and A. E. Tomkinson. 1999. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 274:21679-21687. [DOI] [PubMed] [Google Scholar]
- 17.Mackey, Z. B., W. Ramos, D. S. Levin, C. A. Walter, J. R. McCarrey, and A. E. Tomkinson. 1996. An alternative splicing event, which occurs in mouse pachytene spermatocytes, generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 17:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marintchev, A., M. Mullen, M. W. Maciejewski, B. Pan, M. R. Gryk, and G. P. Mullen. 1999. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 6:884-893. [DOI] [PubMed] [Google Scholar]
- 19.Masson, M., C. Niedergang, V. Schreiebr, S. Muller, J. Menissier de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, D. J., R. M. Taylor, P. Clements, and K. W. Caldecott. 2000. Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 97:13649-13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash, R. A., K. Caldecott, D. E. Barnes, and T. Lindahl. 1997. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 36:5207-5211. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Jannotti, R. M., S. M. Klein, and D. F. Bogenhagen. 2001. Two forms of mitochondrial DNA ligase III are produced in Xenopus laevis oocytes. J. Biol. Chem. 276:48978-48987. [DOI] [PubMed] [Google Scholar]
- 23.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 24.Ron, D., and H. Dressler. 1992. pGSTag—a versatile bacterial expression plasmid for enzymatic labeling of recombinant proteins. BioTechniques 13:866-868. [PubMed] [Google Scholar]
- 25.Satoh, M., and T. Lindahl. 1992. Role of poly(ADP-ribose) formation in DNA repair. Nature 356:356-358. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028-23036. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, R. M., D. J. Moore, J. Whitehouse, P. Johnson, and K. W. Caldecott. 2000. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol. 20:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, L. H., K. W. Brookman, L. E. Dillehay, A. V. Carrano, J. A. Mazrimas, C. L. Mooney, and J. L. Minkler. 1982. A CHO-cell strain having hypersensitivity to mutagens, a defect in strand break repair, and an extraordinary baseline frequency of sister chromatid exchange. Mutat. Res. 95:247-254. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, L. H., K. W. Brookman, N. J. Jones, S. A. Allen, and A. V. Carrano. 1990. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 10:6160-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomkinson, A. E., and Z. B. Mackey. 1998. Structure and function of mammalian DNA ligases. Mutat. Res. 407:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Vidal, A. E., S. Boiteux, I. D. Hickson, and J. P. Radicella. 2001. XRCC1 coordinates the initial and late stages of DNA abasic site repair though protein-protein interactions. EMBO J. 20:6530-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, Y.-F., P. Robins, K. Carter, K. Caldecott, D. J. C. Pappin, G.-L. Yu, R.-P. Wang, B. K. Shell, R. A. Nash, P. Schar, D. E. Barnes, W. A. Haseltine, and T. Lindahl. 1995. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and genetic recombination. Mol. Cell. Biol. 15:3206-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehouse, C. J., R. M. Taylor, A. Thistlethwaite, H. Zhang, F. Karimi-Busheri, D. D. Lasko, M. Weinfeld, and K. W. Caldecott. 2001. XRCC1 stimulates human polynucleotide kinase at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104:107-117. [DOI] [PubMed] [Google Scholar]
- 34.Wu, Y., R. Hickey, K. Lawlor, P. Wills, F. Yu, H. Ozer, R. Starr, J. Y. Quan, M. Lee, and M. Malkas. 1994. A 17S multiprotein form of murine DNA polymerase mediates polyoma virus DNA synthesis. J. Cell. Biochem. 54:32-46. [DOI] [PubMed] [Google Scholar]
- 35.Zahradka, P., and K. Ebisuzaki. 1982. A shuttle mechanism for DNA-protein interactions. The regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127:579-585. [PubMed] [Google Scholar]
- 36.Zdzienicka, M. Z., G. P. Vanderschans, A. T. Natarajan, L. H. Thompson, I. Neuteboom, and J. W. I. M. Simmons. 1992. A Chinese hamster ovary cell mutant (EMC-11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis 7:265-269. [DOI] [PubMed] [Google Scholar]