Abstract
The DNA-dependent protein kinase (DNA-PK) plays an essential role in nonhomologous DNA end joining (NHEJ) by initially recognizing and binding to DNA breaks. We have shown that in vitro, purified DNA-PK undergoes autophosphorylation, resulting in loss of activity and disassembly of the kinase complex. Thus, we have suggested that autophosphorylation of the DNA-PK catalytic subunit (DNA-PKcs) may be critical for subsequent steps in DNA repair. Recently, we defined seven autophosphorylation sites within DNA-PKcs. Six of these are tightly clustered within 38 residues of the 4,127-residue protein. Here, we show that while phosphorylation at any single site within the major cluster is not critical for DNA-PK's function in vivo, mutation of several sites abolishes the ability of DNA-PK to function in NHEJ. This is not due to general defects in DNA-PK activity, as studies of the mutant protein indicate that its kinase activity and ability to form a complex with DNA-bound Ku remain largely unchanged. However, analysis of rare coding joints and ends demonstrates that nucleolytic end processing is dramatically reduced in joints mediated by the mutant DNA-PKcs. We therefore suggest that autophosphorylation within the major cluster mediates a conformational change in the DNA-PK complex that is critical for DNA end processing. However, autophosphorylation at these sites may not be sufficient for kinase disassembly.
Although DNA is the genetic blueprint for all living organisms, it is extremely sensitive to various forms of damage, including oxidation, hydrolysis, and methylation. Thus, efficient DNA repair systems are essential for the maintenance of chromosomal integrity. DNA double-strand breaks (DSBs) are perhaps the most lethal form of DNA damage. In eukaryotes, primarily two pathways repair DSBs: homologous recombination and nonhomologous DNA end joining (NHEJ). In higher eukaryotes, NHEJ is thought to be the major pathway that repairs these breaks (6, 29, 37, 38). Because NHEJ also functions in developing lymphocytes to repair the DSBs introduced during antigen receptor gene rearrangement, defects in this pathway result in a block in lymphocyte development and the disease known as severe combined immunodeficiency (SCID) (reviewed in references 14 and 29).
In the past decade, an intensive research effort has focused on NHEJ, resulting in a reasonable understanding of how DSBs are resolved. There are six known factors which unequivocally function in the NHEJ pathway. Three of these comprise the DNA-dependent protein kinase (DNA-PK) (reviewed in references 19, 20, and 25): the two subunits of the DNA end binding heterodimer Ku and the catalytic subunit of DNA-PK (DNA-PKcs) (18). Two other factors, XRCC4 and DNA ligase IV, form a stable DNA ligase complex (15, 28, 34). The sixth factor, Artemis, was described in 2001 (35); recent data indicate that it may play an important role in DNA end processing during NHEJ (31).
DNA-PK plays a central role in NHEJ because it initially recognizes and binds to damaged DNA and then targets other repair activities to the site of damage. The first step in NHEJ is binding of the Ku70/86 heterodimer to DNA ends at the site of DNA damage. When bound, Ku encircles the DNA. The Ku binding site covers approximately two turns of the DNA helix, but only the central 3 to 4 bp are completely encircled by the polypeptide chain. The DNA terminus is cupped in an accessible binding pocket (46), theoretically allowing DNA-PKcs to interact directly with the DNA end. Recruitment of DNA-PKcs results in translocation of Ku to internal sites of the linear DNA (51). When assembled onto DNA ends, DNA-PK is activated; it can then phosphorylate a variety of substrates including both Ku subunits, DNA-PKcs, XRCC4, and Artemis (4, 5, 8, 24, 31). Recent data from our laboratory and others demonstrate that the protein kinase activity of DNA-PK is essential during NHEJ (1, 21, 22), suggesting that phosphorylation by DNA-PK activates a downstream factor in the pathway.
In assay systems utilizing purified proteins, autophosphorylation of DNA-PK results in the release of DNA-PKcs from DNA-bound Ku and a decrease in the measurable protein kinase activity (4, 5, 8, 33). We suggest a model whereby autophosphorylation of DNA-PKcs and the resulting remodeling and/or disassembly of the DNA-PK complex could facilitate subsequent steps in the DNA repair process. To test this model, we have generated point mutations of the six clustered autophosphorylation sites previously mapped within DNA-PKcs (9). We find that phosphorylation of any single site is not critical to DNA-PK's function; however, mutation of several phosphorylation sites abolishes the ability of DNA-PK to function in NHEJ. Finally, both in vivo and in vitro studies of the mutant protein suggest that autophosphorylation at the major cluster specifically facilitates DNA end processing, whereas further autophosphorylation is required to promote kinase disassembly.
MATERIALS AND METHODS
Cell lines and culture conditions.
The DNA-PKcs-deficient DSB repair mutant CHO cell line V3 (47) was the generous gift of Martin Gellert. Cells were maintained in α-MEM with 10% fetal calf serum (Gibco BRL, Gaithersburg, Md.). Stable transfectants were maintained with 400 μg of G418/ml.
Oligonucleotides.
Oligonucleotides used in this study are as follows (5′ to 3′). The two oligonucleotides flanking the phosphorylation site cluster were KAM97 (GATGTGCTGATTCAAGGATTGA) and KAM98 (ATAAACCGTCTGCGCAGTCGT).
Mutagenic oligonucleotides (complements not shown) were KAM99 (mutant A) (TTTGTGGAGGCTCAGGCCTCC), KAM101 (mutant B) (GTGCTCAGGAAGGGGCTCTCTCA), KAM103 (mutant C) (ATAAGGGCCGCTCAGCAGCAG), KAM105 (mutant D) (TTCACACTGGCCCAGACTGCA), KAM132 (mutant CD) (CCGCTCAGCAGCAGCATGACTTCACACTGGCCCAGACT), KAM146 (mutant E) (CAGGCCGCTCAGGGCACTCTCCAGACC), and KAM165 (S→D, mutant ABCDE) (AGGATCAGGCCGATCAGGGCACTCTCCAGACCCGTGATCAGGAAGGGGATCTCTCAGCTCGCTGGCCAGTGGCAGGGCAGATAAGGGCCGATCAGCAGCAGCATGACTTCACACTGGATCAGACT). Positions at which serine or threonine was changed to alanine or aspartic acid are shown in boldface.
Oligonucleotides used in ligation-mediated PCR (LMPCR) were KAM150 (LMPCR ligation oligonucleotide, blunt ends, top strand) (GCTATGTACTACCCGGGAATTCGTG), KAM201 (LMPCR ligation oligonucleotide, 3′ overhang ends, top strand) (GCTATGTACTACCCGGGAATTCGTGCCGG), KAM200 (LMPCR ligation oligonucleotide, bottom strand) (CACGAATTCCC), KAM214 (LMPCR primer, linker outer) (GCTATGTACTACCCGGGAAT), KAM216 (LMPCR primer, linker inner signal ends) (GGGAATTCGTGCACAGTG), KAM206 (LMPCR primer, linker inner 3′ overhang coding ends) (GGGAATTCGTGCCGGATC), KAM218 (LMPCR primer, CAT, outer) (ATATCCAGCTGAACGGTCTG), KAM219 (LMPCR primer, CAT, inner) (GTTATAGGTACATTGAGCAAC), KAM217 (LMPCR primer, oop, outer) (GACCCCCCATTCAAGAACAGC), KAM215 (LMPCR primer, oop, inner) (GCAGCATTGAGAACTTTGG), and KAM88 (LMPCR hybridization probe) (CCAGTGATTTTTTTCTCCATTTTAG).
Oligonucleotides used in the electrophoretic mobility shift assay (EMSA) were DAR166 (CAGCTGGGAATTCCATATGAGTACTGCAGATGCACTTGCTCGATAGATCTAACATGAGCC) and DAR167 (GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGAG).
Construction and transfection of expression plasmids.
Construction of the wild-type human DNA-PKcs expression vector has been described previously (43) and was accomplished by assembling the following three fragments spanning the DNA-PKcs coding sequence from eight shorter reverse transcription-PCR fragments isolated from the Ramos human B-cell lymphoma cell line: 1-3860, 3559-8173, and 7911-12358. To generate expression plasmids encoding phosphorylation site mutants, duplex oligonucleotides encoding each mutation were utilized in PCR using single-strand overlap extension as described previously (21) in conjunction with oligonucleotides KAM97 and KAM98. The resulting PCR fragment was cloned by using the Topo TA cloning kit according to the manufacturer's protocol (Invitrogen, Carlsbad, Calif.). This PCR fragment spans unique AgeI and FseI sites at positions 7620 and 8166 in the open reading frame of the human DNA-PKcs cDNA. It was subsequently subcloned into the wild-type DNA-PKcs expression vector. There is an additional AgeI site within the vector sequence. To prepare the vector for ligations, partial AgeI digests were performed with a limiting enzyme, or in some experiments, partial digestion was achieved by protecting the vector site (from AgeI digestion) by treating the DNA with a complementary oligonucleotide and recombinant RecA protein. For certain multiple phosphorylation site mutants, the QuikChange mutagenesis kit (Stratagene, La Jolla, Calif.) was utilized to introduce mutations into the subcloned PCR fragment spanning the FseI and AgeI sites. Mutant expression constructs were confirmed by sequence analysis of the entire FseI-AgeI fragment after it was cloned back into the complete DNA-PKcs expression construct.
V3 transfectants expressing human DNA-PKcs were derived by stably transfecting cells with 40 μg of a PvuI-linearized expression plasmid and 1 μg of a NotI-linearized pcDNAI/Neo plasmid (Invitrogen). Transfections were performed in 60-mm-diameter dishes by using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were placed under selection conditions (400 μg of G418/ml) and isolated clones were obtained. Individual colonies were screened for DNA-PKcs expression by immunoblot analysis and were further cultured.
Immunoblot analysis.
The indicated amounts of whole-cell extracts were analyzed after electrophoresis on a sodium dodecyl sulfate (SDS)-5% polyacrylamide gel and transfer to a polyvinylidene difluoride membrane. A monoclonal antibody raised against DNA-PKcs (42-27; a generous gift of Tim Carter, St. John's University, New York, N.Y.) was used as the primary antibody (dilution, 1:300). A goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase was used as the secondary antibody. The membrane was then incubated with a chemiluminescent substrate (ECL; DuPont) according to the manufacturer's recommendations.
Assessment of radiation sensitivity.
Cells (3 × 103) were exposed to various amounts of ionizing radiation using a 60Co source and immediately seeded in complete medium containing 10% fetal bovine serum. After 7 days, cell colonies were fixed with ethanol and stained with crystal violet, and colony numbers were assessed.
V(D)J recombination assay.
Extrachromosomal recombination assays were performed essentially as described previously (21). To assess V(D)J recombination in V3 cells, RAG-1 and RAG-2 expression constructs (3 μg each), expression plasmids encoding DNA-PKcs, or the pCMV6 vector control (6 μg) and a recombination substrate (pJH201 or pJH290; 1 μg) were transfected by liposome using the FuGENE 6 transfection reagent (Roche Molecular Biochemicals). In some experiments, 6 μg of a terminal deoxynucleotidyl transferase (Tdt) expression plasmid was included in the transfections. Forty-eight hours later, plasmid substrates were rescued from the cells by alkaline lysis. Alkaline lysates were restricted with DpnI and then used to transform chemically competent Escherichia coli. Transformed bacteria were spread onto two Luria-Bertani agar plates, one containing 100 μg of ampicillin/ml and the other containing 100 μg of ampicillin/ml plus 22 μg of chloramphenicol/ml. For sequence analyses, plasmid DNA was prepared from individual chloramphenicol-resistant colonies. Sequencing was performed by the Michigan State University sequencing facility.
DNA cellulose pulldown of DNA-PK and measurement of protein kinase activity.
Whole-cell extracts were prepared by a modification of the method of Finnie et al. (11). Briefly, 20 × 106 cells were harvested and washed three times in phosphate-buffered saline, and cell pellets were frozen at −80°C. Frozen cell pellets were resuspended in 20 μl of extraction buffer (20 mM HEPES [pH 7.8], 450 mM NaCl, 50 mM NaF, 25% [vol/vol] glycerol, 0.2 mM EDTA) with Complete protease inhibitor (EDTA free; Roche Molecular Biochemicals). The resuspended pellets were then subjected to three freeze-thaw cycles (liquid nitrogen; 37°C) and centrifuged at 8,160 × g for 7 min at 4°C. Supernatants were stored at −80°C prior to use, and concentrations were determined by Bradford analysis using bovine serum albumin as a standard.
The SignaTECT DNA-PK assay system (Promega Corp., Madison, Wis.) was used to assay DNA-PK activity with the following modifications. Whole-cell extracts (amounts indicated in each figure legend) were incubated with 20 μl of preswollen double-stranded DNA-cellulose beads (Amersham Pharmacia Biotech) for 30 min at 4°C. The double-stranded DNA-cellulose was then washed three times with 1 ml of buffer A (25 mM HEPES [pH 7.9], 50 mM KCl, 10 mM MgCl2, 10% [vol/vol] glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT]). To measure protein kinase activity, the DNA-cellulose was resuspended in 20 μl of a DNA-PK reaction buffer containing 100 μg of bovine serum albumin/ml. Protein kinase reactions were conducted with 10-μl aliquots of the resuspended cellulose beads and were performed both in the presence and in the absence of a biotinylated p53-derived substrate peptide and [γ-32P]ATP. Reactions were terminated and analyzed after spotting onto a SAM2 membrane, washing, and counting in a scintillation counter according to the manufacturer's instructions (Promega Corp., Madison, Wis.). All assays were performed in duplicate with at least three different extract preparations. In some assays, recombinant XRCC4 or Artemis was utilized as a substrate instead of the p53 peptide. Ni+-agarose purification of these proteins was performed as described previously (24). In these experiments, phosphorylation was assessed by autoradiography of SDS-polyacrylamide gel electrophoresis (PAGE) gels.
Assessment of autophosphorylation.
To assess autophosphorylation, 1 mg of whole-cell extracts was absorbed onto 20 μl of DNA-cellulose for 30 min on ice and then washed three times in buffer A. DNA-cellulose fractions were incubated in buffer A with [γ-32P]ATP for 15 min at room temperature and then analyzed by SDS-PAGE and autoradiography.
Purification of recombinant DNA-PKcs.
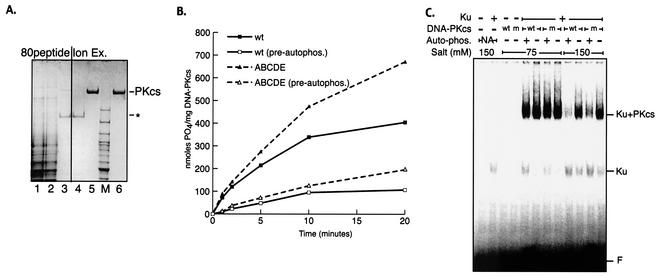
The purified recombinant human Ku heterodimer was generated as previously described (36). Purified human DNA-PKcs was obtained from 5 liters of V3 cells stably transformed with either wild-type or ABCDE mutant DNA-PKcs. The washed cell pellet was extracted by homogenization in 50 ml of a buffer containing 50 mM Tris-HCl (pH 7.5), 600 mM KCl, 10% glycerol, 1 mM DTT, 5 mM EDTA, and 0.5 ml of a protease inhibitor cocktail (P8340; Sigma) and was clarified by centrifugation at 60,000 × g for 1 h. Nucleic acids were depleted by stirring the extract for 15 min with 5 ml of a 70% slurry of polyethyleneimine-cellulose (Sigma). Polyethyleneimine-cellulose with absorbed nucleic acid was removed by filtration. The filtrate was than dialyzed to buffer A (25 mM HEPES-KOH [pH 7.5], 10% glycerol, 1 mM DTT, 0.1 mM EDTA) plus 50 mM KCl (see Fig. 3A, lane 1) and loaded onto an affinity column (80peptide) generated by coupling 5 mg of a peptide from the C terminus of Ku80 (KGSGEEGGDVDDLLDMI; derived from the work of Gell and Jackson [13]) to a 1-ml, N-hydroxysuccinimide-activated HiTrap column (Amersham Biosciences). Bound proteins were eluted by a gradient to buffer A plus 1 M KCl. DNA-PKcs-containing fractions were identified by SDS-PAGE analysis, pooled (Fig. 3A, lane 3), and dialyzed against buffer A plus 100 mM KCl. The dialysate was then loaded onto a 100-μl Mono S ion-exchange column (Amersham Biosciences) and eluted with a 2-ml gradient to buffer A plus 350 mM KCl (Fig. 3A, lane 5). Ion-exchange chromatography both removed the remaining major contaminant (∼180-kDa band in Fig. 3A, lanes 3 and 4) and concentrated the protein ∼25-fold. Fractions were flash-frozen in liquid nitrogen in small aliquots.
FIG. 3.
Autophosphorylation within the 2609-to-2647 cluster is not required for autophosphorylation-induced kinase dissociation. (A) Wild-type and ABCDE mutant DNA-PKcs's were purified as described in Materials and Methods. Coomassie-stained SDS-PAGE gel shows analysis of fractions from a purification of the recombinant ABCDE mutant of DNA-PKcs. Lane 1, 10 μg of clarified extract from V3 cells expressing the ABCDE mutant; lane 2, 10 μg of flowthrough after the extract was loaded onto an affinity column coupled with a peptide from the C terminus of Ku80 (80peptide); lane 3, 2 μg of pooled fractions containing DNA-PKcs from 80peptide eluate; lane 4, 2 μg of flowthrough of Mono S column; lane 5, 2 μg of pooled DNA-PKcs-containing fractions from Mono S; lane 6, 2 μg of purified DNA-PKcs from placenta. Asterisk indicates location of major contaminant in eluate from 80peptide column. Similar results were obtained by purifying recombinant wild-type DNA-PKcs. (B) Kinase activities of purified wild-type and ABCDE mutant DNA-PKcs's were assessed as described in Materials and Methods. DNA-protein complexes were preformed by incubation (at 30°C for 10 min) of 1 nM purified recombinant Ku, 1 nM either purified wild-type (wt) or purified ABCDE mutant DNA-PKcs, and 10 μg of sheared calf thymus DNA/ml. Reactions were started by transfer to 37°C and addition of 400 μM peptide substrate, 200 μM [γ-32P]ATP (0.5 Ci/mmol), and 5 mM MgCl2. The impact of prior autophosphorylation (pre-autophos.) was assessed in parallel reactions by including the same concentrations of ATP and MgCl2 in the preincubation step as well. (C) The stabilities of DNA-PK complexes containing wild-type or mutant DNA-PKcs were assessed by EMSA as described in Materials and Methods. DNA-protein complexes were formed with a 60-bp radiolabeled duplex, Ku, and wt or ABCDE mutant (m) DNA-PKcs as indicated. Complexes were preincubated at 75 mM salt by using either mock-treated DNA-PKcs, active kinase (+ Auto-phos.), or wortmannin-inactivated kinase (− Auto-phos.). Autophosphorylation was arrested by addition of 5 mM EDTA. Reactions were then adjusted to the indicated salt concentrations and incubated for a further 10 min at room temperature before surviving DNA-protein complexes were fixed by cross-linking with 0.25% glutaraldehyde. DNA-protein complexes were resolved by electrophoresis on a 3.5% native polyacrylamide gel.
Assays using purified proteins.
The kinase activity of purified DNA-PKcs was assessed in a buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 0.1 mM EDTA, and 0.2 mM EGTA. The peptide substrate used was biotin-EPPLSQEAFADLWKK. Phosphorylation of the peptide was assessed as described previously (7). For EMSA analysis, we used a 60-bp double-stranded DNA substrate made by annealing the 5′ 32P-labeled oligonucleotide DAR166 (defined above) to DAR167. One hundred nanomolar DNA-PKcs stocks were inactivated by treatment with 10 μM wortmannin or mock treated with an equivalent amount of a solvent (dimethyl sulfoxide) for 10 min on ice prior to dilution into the reaction mixture. Samples were prepared for EMSA analysis in two steps. In the first step (complex assembly and autophosphorylation), DNA-protein complexes were formed with 10 nM each protein and 100 nM duplex DNA by using a buffer containing 25 mM Tris-HCl (pH 8.0) supplemented with 5 mM MgCl2 and 200 μM ATP. Reaction mixtures were incubated for 10 min at 30°C before autophosphorylation was arrested by addition of 5 mM EDTA. In the second step (probing the stability of complexes), reaction products were adjusted with added NaCl to the indicated salt concentrations and were incubated for a further 10 min at room temperature. Stable DNA-protein complexes were then fixed by cross-linking using 0.25% glutaraldehyde and were resolved by electrophoresis on a 3.5% native PAGE gel in 1× Tris-borate-EDTA at 18 V/cm. The composition of each indicated species was confirmed by antibody supershift analysis (data not shown).
Construction of an Artemis-expressing baculovirus and coimmunoprecipitation.
A cDNA encoding full-length human Artemis was assembled from overlapping reverse transcription-PCR fragments. Although numerous splice variants were isolated, we assembled a full-length coding sequence analogous to that shown previously to complement the V(D)J recombination deficit in Artemis-deficient cell lines (35). The termination codon was not included, and an in-frame 3′ V5-His tag was added at the C terminus. The assembled fusion protein construct was subcloned into the pAcHCT-C transfer vector (Pharmingen, San Diego, Calif.). A recombinant baculovirus encoding human Artemis was obtained by using Baculogold-linearized viral DNA (Pharmingen) and standard techniques. For the pulldown experiments for which results are shown in Fig. 5, whole-cell extracts of V3 transfectants (1 mg) and whole-cell extracts from either Artemis-infected or control Sf9 cells were coincubated for 30 min. Subsequently, 50 μl of Ni+ agarose was added, and the extracts were absorbed in buffer A with 25 mM imidazole. After 1 h, the Ni+ agarose was washed three times with buffer A containing 50 mM imidazole. Proteins were eluted with SDS-PAGE buffer and analyzed by immunoblotting.
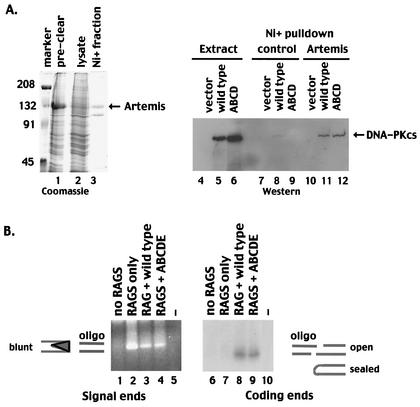
FIG. 5.
Autophosphorylation within the 2609-to-2647 cluster is not required for the interaction of DNA-PKcs with Artemis or to facilitate opening of coding end hairpins. (A) (Left) SDS-PAGE analyses of the following fractions of Sf9 cells infected with a baculovirus encoding an Artemis-V5 His-tagged fusion protein: precleared whole-cell lysate (lane 1) and cleared lysate (lane 2). Marker, marker proteins, with molecular weights (in thousands) given on the left. (Right) Western blot of whole-cell extracts or Ni+-agarose fractions. Lanes 4 to 6, whole-cell extracts of V3 cells transfected with vector alone (lane 4), wild-type DNA-PKcs (lane 5), or mutant ABCD (lane 6); lanes 7 to 9, Ni+-agarose fractions of control Sf9 extracts coincubated with extracts from V3 cells transfected with vector alone (lane 7), wild-type DNA-PKcs (lane 8), or mutant ABCD (lane 9); lanes 10 to 12, Ni+-agarose fractions of extracts from Sf9 cells expressing Artemis coincubated with extracts from V3 cells transfected with vector alone (lane 10), wild-type DNA-PKcs (lane 11), or mutant ABCD (lane 12). (B) LMPCR was performed on Hirt supernatants prepared from V3 cells transiently transfected with substrate alone (lanes 1 and 6), substrate and RAG expression vectors (lanes 2 and 7), substrate plus RAG and wild-type DNA-PKcs expression vectors (lanes 3 and 8), or substrate plus RAG and mutant ABCDE expression vectors (lanes 4 and 9). Lanes 5 and 10 included no input DNA in the LMPCRs. In lanes 1 to 5, a blunt oligonucleotide was ligated to Hirt fractions, and PCR amplifications to detect signal ends were performed. In lanes 6 to 10, an oligonucleotide with a 4-bp overhang complementary to a potential opened coding end in substrate pJH290 was ligated to Hirt fractions, and PCR amplifications to detect coding ends were performed.
Analyses of V(D)J recombination intermediates.
V3 cells were transfected either with the pJH290 recombination substrate alone, with the substrate and the RAG expression plasmids, or with the substrate and the RAG and DNA-PKcs expression plasmids as described above. Hirt supernatants were prepared from transfected cells as follows. Forty-eight hours after transfection, cells were harvested, resuspended in 400 μl of Hirt buffer 1 (10 mM Tris [pH 8.0], 1 mM EDTA, 0.6% SDS), and incubated for 15 min at room temperature. One hundred microliters of Hirt buffer 2 (10 mM Tris [pH 8.0], 1 mM EDTA, 5 M NaCl) was added, and the samples were incubated overnight at 4°C. After being spun for 10 min at 10,000 × g at 4°C, supernatants were extracted with phenol-chloroform, ethanol precipitated, and then resuspended in 10 μl of double-distilled water. Five microliters of each Hirt supernatant was ligated to 500 pmol of annealed oligonucleotides KAM150-KAM200 or KAM201-KAM200 at 16°C overnight. Ligated Hirt supernatants were ethanol precipitated and used in nested PCR amplifications. Signal ends were amplified first with the primer combination KAM214-KAM217 and then with KAM216-KAM215. Coding ends were amplified first with the primer combination KAM214-KAM218 and then with KAM206-KAM219. Forty cycles of amplification were performed using the following conditions: for primary PCRs, 94°C for 30 s, 58°C for 1 min, and 68°C for 1 min; for secondary PCRs, 94°C for 30 s, 56°C for 1 min, and 68°C for 1 min). Amplification products were analyzed by Southern hybridization using oligonucleotide KAM88 as a hybridization probe.
RESULTS
Six of seven autophosphorylation sites are located within a 38-amino-acid cluster within human DNA-PKcs.
Recently, the locations of seven in vitro autophosphorylation sites within DNA-PKcs were reported (9). As diagrammed in Fig. 1A, six of the seven phosphorylation sites are tightly clustered in a short region of primary sequence in the central region of the molecule (amino acids 2609 to 2647). Four of the six clustered autophosphorylation sites (T2609, S2612, T2638, and T2647) are also phosphorylated in vivo in okadaic acid-treated cells. All four of these in vivo sites are completely conserved among the six sequenced vertebrate DNA-PKcs genes (human, horse, dog, mouse, chicken, and frog).
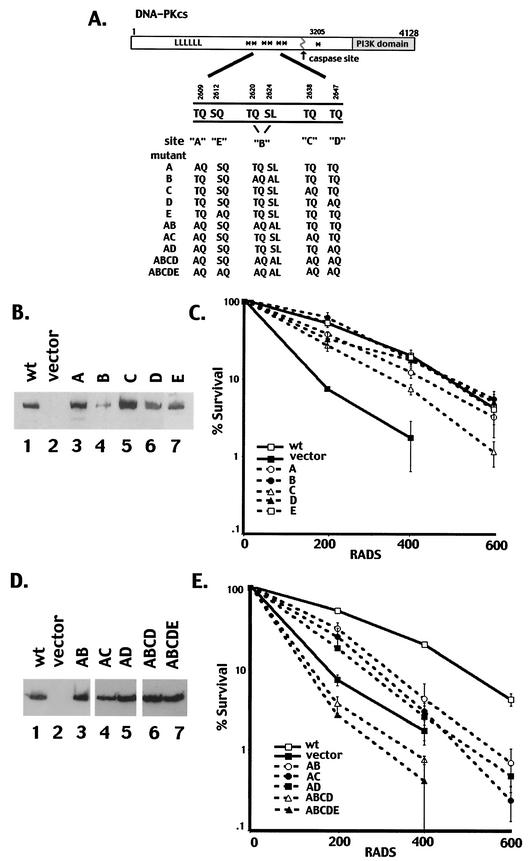
FIG. 1.
Single phosphorylation site mutants complement the radiosensitivity of V3 cells, whereas multiple phosphorylation site mutants do not. (A) Diagrammatic representation of seven autophosphorylation sites within DNA-PKcs (asterisks). Mutations are as follows: A, T2609A; B, T2620A and S2624A; C, T2638A; D, T2647A; E, S2612A. The A, B, C, D, and E mutations were introduced either alone or in combinations as indicated. (B) Immunoblot analyses of whole-cell extracts from V3 transfectants expressing either full-length DNA-PKcs (lane 1), vector alone (lane 2), mutant A (lane 3), mutant B (lane 4), mutant C (lane 5), mutant D (lane 6), or mutant E (lane 7). (C) The radiation resistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone, or each of the single phosphorylation site mutants was assessed as described in Materials and Methods. Data are presented as percent survival relative to that of nonirradiated controls (set at 100%). Error bars, standard errors of the means of three separate experiments. (D) Immunoblot analyses of whole-cell extracts from V3 transfectants expressing either full-length DNA-PKcs (lane 1), vector alone (lane 2), mutant AB (lane 3), mutant AC (lane 4), mutant AD (lane 5), mutant ABCD (lane 6), or mutant ABCDE (lane 7). (E) The radiation resistance of V3 transfectants expressing either wild-type DNA-PKcs, vector alone, or mutant AB, AC, AD, ABCD, or ABCDE was assessed. Data are presented as percent survival relative to that of nonirradiated controls (set at 100%). Error bars, standard errors of the means of three separate experiments.
To begin to assess the functional relevance of autophosphorylation of DNA-PKcs, several point mutations of the DNA-PKcs cDNA were generated. Initially, five mutant constructs, designated A, B, C, D, and E, were generated, as indicated in Fig. 1A. Constructs A, C, D, and E each contain a single mutation of serine or threonine to alanine, as indicated. Mutation B includes two mutations of serine or threonine to alanine (T2620 and S2624); to date, phosphorylation of these two sites has been observed in vitro but not in vivo.
Single phosphorylation site mutant constructs complement V3's radiosensitivity similarly to constructs encoding wild-type DNA-PKcs.
To assess the function of each phosphorylation site mutant, expression vectors carrying each coding sequence were transfected into V3 cells (Fig. 1B). The V3 cell line is DNA-PKcs deficient and thus is defective in DNA DSB repair (47). Clones with similar levels of DNA-PKcs expression were selected for each mutant. We next performed cell irradiation assays for this panel of DNA-PKcs mutants and compared the radioresistance to that of clones expressing wild-type human DNA-PKcs as well as to that of an empty vector control clone. As seen in Fig. 1C, each of the single phosphorylation site mutant constructs complemented the radiosensitivity of V3 cells similarly to the construct encoding wild-type DNA-PKcs. In some experiments (four of six) (data not shown), the transfectant expressing mutant C was slightly less radioresistant than the wild-type transfectant, suggesting that phosphorylation of Thr2638 is functionally relevant. Indeed, it was found previously that Thr2638 was a major in vitro phosphorylation site in DNA-PKcs (9). Recently, Chan et al. (3) reported that an alanine mutant of Thr2609 (analogous to mutant A) could only partially complement the radioresistance of V3 cells. In our studies, the V3 T2609A clone generated displayed a radiosensitivity similar to that of transfectants expressing wild-type DNA-PKcs. Two additional clones expressing mutant A were also studied, and these clones also displayed radiosensitivity similar to that of clones expressing wild-type DNA-PKcs (data not shown). In the report of Chan et al., the single T2609A mutant V3 transfectant studied showed 10% survival at a dose of only 2.4 Gy, whereas the wild-type-expressing V3 transfectant showed 10% survival at a dose of 5 Gy. In our studies, the wild-type transfectant showed 10% survival at an average dose of 5.28 Gy (range, 4.28 to 6.24), similar to the dose for the wild-type transfectant studied by Chan et al. The average dose for 10% survival for the mutant A V3 transfectant was 4.47 Gy (range, 3.28 to 4.67). The discrepancy in these results is most likely due to individual clonal variation. In any case, our data demonstrate that the radioresistance of transfectants expressing single phosphorylation site substitutions is not dramatically different from that of wild-type transfectants. Thus, we conclude that phosphorylation of any single site within the cluster is not essential to DNA-PK's function in NHEJ.
V3 transfectants expressing DNA-PKcs with multiple phosphorylation site mutations are extremely radiosensitive.
The fact that six of seven autophosphorylation sites are tightly clustered within approximately 1% of the total amino acid sequence of DNA-PKcs suggested that concurrent phosphorylation of several sites within the cluster might be functionally important. Thus, we next generated expression constructs with two, three, five, or six mutations of serine or threonine to alanine. These are designated mutants AB, AC, AD, ABCD, and ABCDE, and they combine the mutations described previously. Stable cell clones with similar DNA-PKcs expression levels were selected for each mutant (Fig. 1D), and their radiosensitivities were assessed (Fig. 1E). Mutants AB, AC, and AD only partially complemented the radiosensitive phenotype of the V3 cells. Significantly, clones expressing DNA-PKcs with five or six phosphorylation site substitutions (mutants ABCD and ABCDE) displayed radiosensitivities analogous to (or more severe than) those of the vector-only control clones. These data demonstrate that concurrent autophosphorylation of DNA-PKcs on at least two of these clustered sites is a critical step in DNA damage repair, and they define DNA-PKcs as a physiologically relevant target of its own protein kinase activity.
Transiently expressed DNA-PKcs with single but not multiple phosphorylation site mutations supports wild-type levels of coding end joining in V3 cells.
We next assessed the ability of the mutant DNA-PKcs to support V(D)J recombination (Table 1). Wild-type DNA-PKcs substantially complemented the coding end joining deficit of V3 cells (assessed with substrate pJH290). Similarly, all the mutants, with the exception of AC, ABCD, and ABCDE, supported near-wild-type levels of coding joint formation. Mutant AC supported reduced levels of coding end joining, whereas the abilities of the multiple phosphorylation mutants ABCD and ABCDE to support V(D)J recombination were severely impaired. These data are consistent with reports that minimal function of the NHEJ pathway can suffice to support V(D)J recombination (21, 39, 49). Thus, only transfectants that are severely impaired in NHEJ show reduced levels of V(D)J recombination.
TABLE 1.
DNA-PKcs with single phosphorylation site mutations, but not with multiple phosphorylation site mutations, supports normal levels of V(D)J recombinationa
| Transfected plasmidb | No. of Ampr Cmr colonies/ no. of Ampr coloniesc | Recombina- tion (%)d |
|---|---|---|
| pJH290 only | 0/46,060 | 0 |
| 0/99,568 | 0 | |
| 4/81,634 | 0.005 | |
| pJH290 + RAGS | 1/30,968 | 0.003 |
| 0/60,270 | 0 | |
| 2/72,716 | 0.003 | |
| pJH290 + RAGS + wild type | 444/35,868 | 1.24 |
| 257/25,520 | 1.01 | |
| 203/57,918 | 0.35 | |
| pJH290 + RAGS + mut A | 283/31,130 | 0.91 |
| 99/38,906 | 0.25 | |
| 100/32,928 | 0.30 | |
| pJH290 + RAGS + mut B | 141/22,540 | 0.63 |
| 120/29,792 | 0.40 | |
| 436/42,924 | 1.02 | |
| pJH290 + RAGS + mut C | 209/22,990 | 0.91 |
| 201/46,354 | 0.43 | |
| 53/23,912 | 0.22 | |
| pJH290 + RAGS + mut D | 107/45,080 | 0.24 |
| 221/40,572 | 0.54 | |
| 235/34,888 | 0.67 | |
| pJH290 + RAGS + mut E | 20/3,300 | 0.61 |
| 136/9,020 | 1.51 | |
| 18/5,060 | 0.36 | |
| pJH290 + RAGS + mut AB | 81/20,384 | 0.40 |
| 138/32,340 | 0.43 | |
| 210/23,520 | 0.89 | |
| pJH290 + RAGS + mut AC | 47/23,520 | 0.20 |
| 60/37,828 | 0.16 | |
| 29/32,732 | 0.09 | |
| pJH290 + RAGS + mut AD | 141/25,450 | 0.55 |
| 58/39,298 | 0.15 | |
| 129/41,160 | 0.31 | |
| pJH290 + RAGS + mut ABCD | 2/5,940 | 0.03 |
| 0/2,900 | 0 | |
| 0/2,888 | 0 | |
| pJH290 + RAGS + mut ABCDE | 5/28,900 | 0.017 |
| 0/21,000 | 0 | |
| 1/7,600 | 0.013 | |
| pJH290 + RAGS + S/T→D | 38/61,800 | 0.061 |
| 22/23,900 | 0.092 | |
| 6/14,900 | 0.040 | |
| PJH201 only | 0/5,100 | 0 |
| 0/6,100 | 0 | |
| 0/6,996 | 0 | |
| 0/1,350 | 0 | |
| PJH201 + RAGS | 1/2,500 | 0.04 |
| 15/15,800 | 0.095 | |
| 7/4,950 | 0.14 | |
| 5/1,900 | 0.26 | |
| PJH201 + RAGS + wild type | 32/600 | 5.3 |
| 49/1,700 | 2.88 | |
| 57/1,848 | 3.08 | |
| 98/1,800 | 5.44 | |
| PJH201 + RAGS + mut ABCDE | 6/1,200 | 0.5 |
| 9/2,300 | 0.39 |
Transient V(D)J recombination assays were performed as described in Materials and Methods. RAG expression from plasmid vectors initiates recombination in V3 cells, as assessed by the plasmid substrate pJH290, which detects coding joints, or the pJH201 substrate, which detects signal joints, as indicated.
mut, mutant.
Numbers from at least three separate experiments are presented.
Calculated as (number of chloramphenicol-resistant colonies/number of ampicillin-resistant colonies) × 100.
Signal end joining is variably depressed in different DNA-PKcs-deficient cell lines and in different animal models of DNA-PKcs deficiency (10, 32, 43, 52). The V3 cell line displays a fairly significant defect in signal end joining (22). This is evidenced by the fact that wild-type DNA-PKcs increased the level of signal joints retrieved in transient assays approximately 30-fold (Table 1, pJH201 transfections). In contrast, signal joining was still impaired in cells transfected with the multisite mutant ABCDE. We conclude that the multiple phosphorylation site mutant cannot support normal levels of either signal or coding end joining during V(D)J recombination.
V3 transfectants expressing DNA-PKcs with multiple phosphorylation site mutations display wild-type levels of DNA-PK activity.
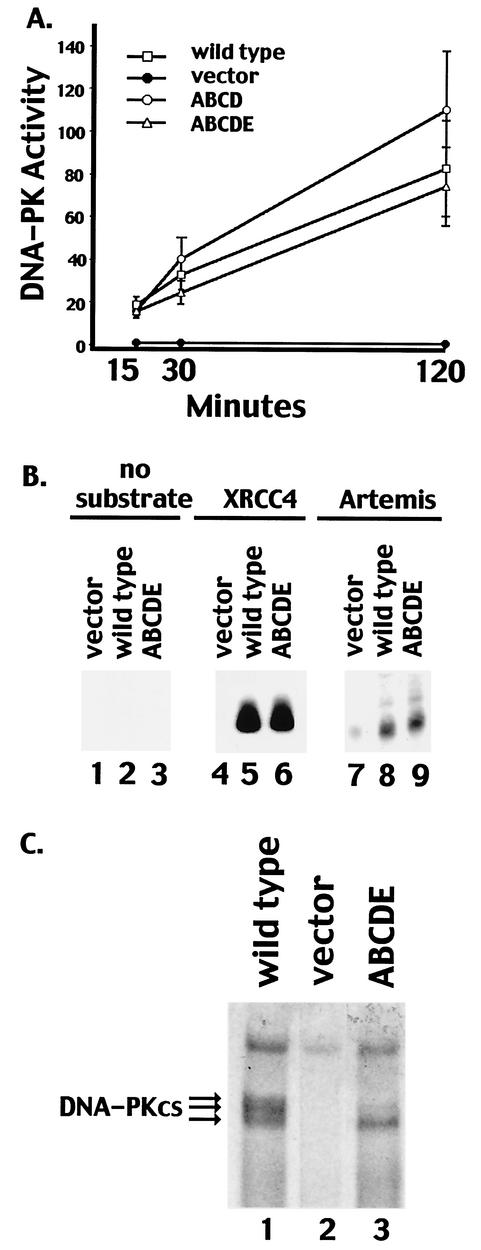
DNA-PK activity in extracts derived from V3 transfectants was next assessed by using a previously described DNA-cellulose pulldown assay (11) and a peptide substrate (derived from the N terminus of p53). Transfectants expressing mutant ABCD or ABCDE displayed DNA-PK levels similar to those of transfectants expressing wild-type DNA-PKcs. Though DNA-PK activity was consistently somewhat higher in the transfectant expressing mutant ABCD, this increase might be explained by the slightly higher DNA-PKcs expression in the clone analyzed (Fig. 1D). We also utilized recombinant XRCC4 and Artemis as substrates in pulldown assays; the ABCDE mutant phosphorylated XRCC4 and Artemis similarly to wild-type DNA-PKcs (Fig. 2B). Finally, by using SDS-PAGE with a low percentage of acrylamide, a marked reduction in mobility was observed for highly phosphorylated wild-type DNA-PKcs. (The indicated phosphoproteins were confirmed to be DNA-PKcs by immunoblotting [data not shown].) As can be seen (Fig. 2C), although the ABCDE mutant protein can still be phosphorylated (possibly at position 3205 outside the major phosphorylation site cluster or perhaps at another, as yet unidentified site) hyperphosphorylation of the mutant protein (as evidenced by a gel shift) was not observed. In summary, DNA-PKcs containing multiple autophosphorylation site mutations is active toward exogenous substrates such as a synthetic peptide, XRCC4, and Artemis; therefore, the radiosensitive phenotype of cells expressing mutant ABCDE is not due to a lack of protein kinase activity per se.
FIG. 2.
Transfectants expressing multiple phosphorylation site substitutions express normal levels of DNA-PK activity. (A) Whole-cell extracts (500 μg) prepared from V3 cells transfected with vector alone, wild-type DNA-PKcs, mutant ABCD, or mutant ABCDE were assayed for DNA-PK activity as described in Materials and Methods. Phosphorylation of the p53 substrate was assessed after 15, 30, and 120 min as indicated. Each cell extract was tested in duplicate, and three independent extracts were tested for each cell line. Error bars, standard deviations. (B) Whole-cell extracts (250 μg) from V3 cells transfected with vector alone, wild-type DNA-PKcs, or mutant ABCDE were assayed for the capacity to phosphorylate no substrate (lanes 1 to 3), recombinant XRCC4 (lanes 4 to 6), or recombinant Artemis (lanes 7 to 9). (C) DNA-cellulose fractions from 500 μg of whole-cell extracts of V3 cells expressing wild-type DNA-PKcs (lane 1), vector alone (lane 2), or mutant ABCDE (lane 3) were incubated in protein kinase buffer with [γ-32P]ATP. Subsequently, bound proteins were analyzed by SDS-4% PAGE and autoradiography.
DNA-PK complexes containing wild-type or ABCDE mutant DNA-PKcs display similar stabilities.
To begin to assess the biochemical basis of defective NHEJ in cells expressing the ABCDE mutant, we purified the wild-type and ABCDE mutant proteins from the V3 transfectants (see Materials and Methods) (Fig. 3A). We have shown previously using purified proteins that autophosphorylation of DNA-PKcs results in dissociation of DNA-PKcs from Ku bound to DNA and loss of kinase activity. Thus, we tested whether preautophosphorylation of the mutant protein could affect kinase activity. Although the mutant protein is slightly more resistant to preautophosphorylation, preautophosphorylated DNA-PK complexes containing either wild-type or mutant DNA-PKcs have dramatically lower protein kinase activity than nonphosphorylated DNA-PK (Fig. 3B). The modest difference in resistance to autophosphorylation likely explains the variance in protein kinase activity observed over time with the mutant versus wild-type proteins without preautophosphorylation. Still, these data suggest that phosphorylation at additional sites facilitates kinase disassembly.
To more formally assess complex stability after autophosphorylation, a modified EMSA was also performed. Samples were prepared for analysis in two steps. First, complexes were allowed to assemble onto the DNA probe by using either wortmannin-inactivated DNA-PKcs or non-wortmannin-treated DNA-PKcs in the presence of ATP to allow autophosphorylation. After kinase activity was stopped (by the addition of EDTA), the relative stabilities of the complexes were probed by a second incubation in increasing NaCl concentrations. As can be seen (Fig. 3C), under more stringent conditions, neither autophosphorylated wild-type nor autophosphorylated ABCDE mutant DNA-PKcs forms stable complexes with Ku-bound DNA, although complexes containing the mutant protein are consistently slightly more stable than wild-type complexes. These data suggest that further autophosphorylation is required for kinase disassembly.
An ABCDE mutant with aspartic acid substitutions is partially competent in NHEJ.
The data presented above suggest that autophosphorylation outside of the major cluster might be important to signal kinase inactivation, at least in vitro. In an attempt to mimic constitutively phosphorylated DNA-PKcs in living cells, we next constructed an expression vector with aspartic acid substitutions for each of these six phosphorylation sites within the major cluster. We considered two possibilities. If phosphorylation within the major cluster induces release of DNA-PKcs from Ku bound to DNA, then the S/T→D-substituted DNA-PKcs should not be able to associate at all with Ku bound to DNA ends. In this case the mutant protein should not complement the NHEJ defect in V3 cells, and we would expect to observe reduced kinase activity in extracts from these cells. Another possibility is that autophosphorylation within the major cluster is important for another reason, perhaps by inducing some conformational change in the repair complex that facilitates subsequent steps in the repair process. In this case, the mutant protein should at least partially complement the NHEJ deficit in V3 cells, and we would expect the mutant protein to be an active protein kinase.
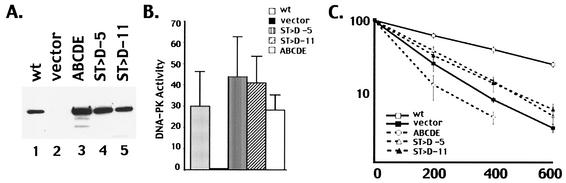
The expression vector with aspartic acid substitutions at ABCDE (designated S/T→D) was transfected into the V3 cell line, and two cell clones with similar DNA-PKcs expression levels were studied (Fig. 4A). DNA-PK activity was assessed using the DNA-cellulose pulldown assay and the p53 peptide substrate. Transfectants expressing the S/T→A mutant displayed DNA-PK levels similar to those of transfectants expressing wild-type DNA-PKcs or the ABCDE mutant (Fig. 4B). As can be seen in Fig. 4C, although cells expressing the S/T→D mutant are still very radiosensitive compared to cells expressing wild-type DNA-PKcs, these cells are slightly more radioresistant than the vector-only control and significantly more radioresistant than the ABCDE mutant. Similarly, coding end resolution was slightly better in transient transfections comparing the S/T→D mutant to the ABCDE mutant (Table 1). We attribute this weak partial phenotype to poor mimicking of phosphoserine or phosphothreonine by the aspartic acid substitutions, although other explanations are also possible. Still, these data are consistent with a model whereby phosphorylation within the major cluster induces a conformational change that is requisite for subsequent steps in DNA end joining.
FIG. 4.
An ABCDE mutant with aspartic acid substitutions is partially competent in NHEJ. (A) Immunoblot analyses of whole-cell extracts from V3 transfectants expressing wild-type (wt) DNA-PKcs (lane 1), vector alone (lane 2), mutant ABCDE (lane 3), the ST→D mutant (clone 5), (lane 4), and the ST→D mutant (clone 11) (lane 5). (B) Whole-cell extracts (500 μg) prepared from V3 cells transfected with either vector alone, wt DNA-PKcs, mutant ABCDE, or one of two independent clones (clones 5 and 11) expressing the ST→D mutation were assayed for DNA-PK activity. Phosphorylation of the p53 substrate was assessed after 60 min. Each cell extract was tested in duplicate, and three independent extracts were tested for each cell line. Error bars, standard deviations. (C) Radiation resistance of V3 transfectants expressing wt DNA-PKcs, vector alone, mutant ABCDE, or the ST→D mutant. Data are presented as percent survival relative to that of nonirradiated controls (set at 100%). Error bars, standard errors of the means of three separate experiments.
Autophosphorylation within the 2609-to-2647 cluster is not required for the interaction of DNA-PKcs with Artemis or to facilitate opening of coding end hairpins.
The recent elegant studies of Ma et al. demonstrated that DNA-PKcs interacts with Artemis to generate an active DNA end-processing complex (31). To assess whether autophosphorylation within the 2609-to-2647 cluster is required for the interaction of Artemis with DNA-PKcs, a baculovirus vector expressing V5-His-tagged Artemis was constructed. Although only a fraction of the baculovirus-expressed, V5-His-tagged Artemis was soluble (Fig. 5A), a significant portion of the soluble protein fractionated onto Ni+ agarose beads. Extracts from V3 transfectants expressing either wild-type DNA-PKcs or the DNA-PKcs mutant ABCD were incubated with extracts from Artemis-infected Sf9 cells or extracts from control Sf9 cells and then absorbed onto Ni+ agarose. Wild-type DNA-PKcs and the multiple phosphorylation site mutant copurified equivalently with Ni+ agarose-immobilized Artemis (Fig. 5A, lanes 11 and 12), suggesting that autophosphorylation within the major phosphorylation site cluster is not required for the interaction of DNA-PKcs with Artemis.
Ma et al. also demonstrated that the DNA-PKcs-Artemis complex opens hairpinned DNA ends (like the ones that result from RAG-mediated cleavage) and that DNA-PK's kinase activity is requisite for this activity. However, it was not determined whether DNA-PK's phosphorylation of Artemis or phosphorylation of DNA-PKcs itself is important for activating the hairpin nuclease. Thus, we next assessed broken DNA intermediates generated during transient V(D)J assays by using an LMPCR assay (Fig. 5B). Signal ends were detected by using a blunt-ended oligonucleotide in the ligation step. As expected, blunt signal ends are readily detected from Hirt supernatants prepared from cells transfected with RAG expression constructs in the presence or absence of DNA-PKcs expression. Previous studies have suggested that hairpin opening likely occurs 2 bp from the hairpinned termini, leaving a 4-bp 3′ overhang (31). Thus, to detect opened hairpin coding termini, an oligonucleotide with a 4-bp 3′ overhang, complementary to a hairpin opened 2 bp from the tip of the pJH290 substrate, was utilized in the ligation step. No opened coding end intermediates were detected in transfections with substrate alone or from transfections including substrate plus the RAG proteins (Fig. 5B, lanes 6 and 7). This is consistent with previous studies demonstrating that hairpinned termini in DNA-PKcs-deficient cells remain sealed (41, 53). In contrast, opened coding ends were detected in transfectants including either wild-type DNA-PKcs or the ABCDE mutant (Fig. 5B, lanes 8 and 9). Thus, we conclude that the ABCDE mutant is fully capable of activating the hairpin-opening activity of Artemis.
Coding joints mediated by mutant ABCDE have minimal nucleotide loss from joined coding ends.
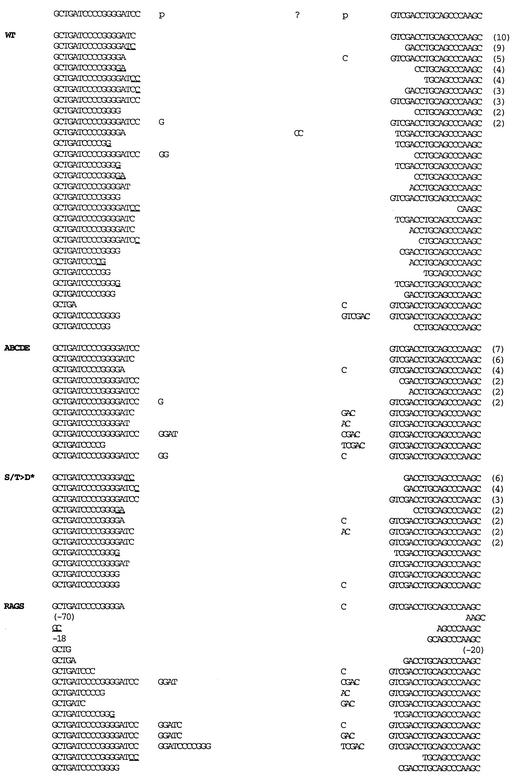
Structural analyses of signal and coding joints as well as recombination intermediates has provided significant insight into the mechanistic details of both V(D)J recombination and NHEJ. Thus, we next sequenced rare coding joints mediated by the ABCDE mutant isolated from transient V(D)J recombination assays and compared them to sequences obtained from transfections including wild-type DNA-PKcs. As expected, coding joints obtained from recombination assays including wild-type DNA-PKcs yielded a diverse collection of sequences (Fig. 6; Table 2). Nucleotide loss from each joint ranged from 0 to 14 nucleotides, with an average of 4.61 bp lost per joint. Of the joints with at least one complete coding end (37 of 122 [30%]), P segments were apparent in 10 of 37 (27%). Short sequence homologies of 1 to 2 bp could be observed in 44% of the joints. Though only a few rare coding joints were sequenced from transfections with no DNA-PKcs, structurally these were consistent with classic SCID joints. These joints display excessive nucleotide loss and a high incidence of long P segments when a coding end is complete (12 of 13 [92%]).
FIG. 6.
Coding joints mediated by mutant ABCDE have minimal nucleotide loss from joined ends. The sequences of coding ends as they occur in the pJH290 substrate are shown above the sequences of the recombinant junctions. The number of observations of each sequence is given in parentheses to the right of the sequence. All duplicate sequences were derived from separate transfections. Nucleotides in columns headed by “p” are palindromic nucleotides added to each junction. Nucleotides that cannot be unequivocally assigned to a particular coding end are underlined and listed in the 5′-most location. Portions of the wild-type (WT) and RAG-only coding joints are from references 21 and 49.
TABLE 2.
Coding joints mediated by mutant ABCDE have minimal nucleotide loss from joined coding endsa
| DNA-PKcs | No. of sequences | No. of bases lost/joint | % Complete ends (no. complete/ total no.) | % SSHb (no. with SSH/ total sequences) | % P segments (no. of P segments/ no. complete ends) |
|---|---|---|---|---|---|
| Wild type | 61 | 4.61 | 30 (37/122) | 44 (27/61) | 27 (10/37) |
| ABCDE | 28 | 1.43 | 70 (39/56) | 0 (0/27) | 25 (10/40) |
| S/T→D | 25 | 3.08 | 38 (19/50) | 52 (13/25) | 26 (5/19) |
| RAGS only | 16 | 14.69 | 41 (13/32) | 31 (5/16) | 92 (12/13) |
Average nucleotide loss per joint, percent intact coding ends, percent sequences with short sequence homologies, and percent P segments per full-length end were calculated from sequences presented in Fig. 6.
SSH, short sequence homologies.
The most obvious difference between joints isolated from transfections with wild-type DNA-PKcs versus the ABCDE mutant is that there is remarkably less nucleotide loss at the site of joining in joints mediated by the mutant protein. Nucleotide loss from each joint ranged from 0 to 7 nucleotides, with an average of only 1.43 bp lost per joint. Furthermore, 70% of the coding ends (39 of 56) had no nucleotide loss versus only 30% (37 of 122) for joints mediated by wild-type DNA-PKcs. Of the joints with at least one complete coding end, P segments were apparent in 10 of 40 (25%). This is completely analogous to P segment incidence in joints mediated by wild-type DNA-PKcs and substantiates our conclusion that hairpin opening is normal in joints mediated by mutant ABCDE. Although four P segments were slightly longer than the normal 2 bp, slightly longer P segments were also occasionally observed in wild-type joints. Remarkably, short sequence homology at the junctures could not be observed in any of the 28 coding sequences, suggesting that this mechanism to facilitate end joining might be blocked in cells expressing the ABCDE mutant (although this may be partially attributed to the lower extent of base loss in these joints).
We also sequenced 25 coding joints mediated by the S/T→D mutant protein. These joints more closely resembled those mediated by wild-type DNA-PKcs: they averaged 3.08 bp lost per joint, and 19 of 50 coding ends (38%) were complete. Of the joints with at least one complete coding end, 5 of 19 (26%) had a P element, again analogous to P segment incidence in joints mediated by either wild-type DNA-PKcs or mutant ABCDE. Finally, short sequence homologies of 1 to 2 bp were apparent in 52% of the joints, also similar to the percentage in joints mediated by wild-type DNA-PKcs. In summary, these data suggest that end processing during V(D)J recombination is in some way impeded in the presence of the ABCDE mutant. We suggest that phosphorylation within the major cluster induces a conformational change that facilitates DNA end alignment and/or end processing prior to end joining.
DISCUSSION
DNA-PK has been vigorously studied for more than a decade, and numerous potential substrates have been reported. In fact, more than 40 different proteins have been shown to be efficient in vitro targets of DNA-PK. To date, DNA-PKcs itself is the only unequivocally defined, functionally relevant target of its own kinase activity (3, 44).
Numerous spontaneous and targeted mutations of DNA-PKcs have been studied previously. These include three spontaneous germ line mutations (30, 32, 48), five spontaneous mutations in cultured cell lines (26, 52), four targeted germline mutations (2, 12, 23, 45), and three reports of specific targeted mutations of DNA-PKcs expressed ectopically in cultured cells (21, 22, 49). In each of these, loss of DNA-PKcs function in vivo was associated with loss of DNA-PK activity. The multiphosphorylation site DNA-PKcs mutants are the first DNA-PKcs mutants that retain protein kinase activity but are incapable of supporting NHEJ. Thus, these mutants may be helpful in discerning other postulated functions (i.e., apart from NHEJ) of DNA-PKcs (16, 42).
We have explored several possible molecular mechanisms that could account for the radiosensitive phenotype of cells expressing the multisite autophosphorylation mutants of DNA-PKcs. Mutations in the autophosphorylation sites do not affect the in vitro protein kinase activity of DNA-PK, as judged by the ability of extracts from V3 cells containing DNA-PKcs with multiple autophosphorylation site mutations to phosphorylate synthetic peptide substrates, recombinant XRCC4, or recombinant Artemis. This suggests that the radiosensitive phenotype is due not to a lack of DNA-PK activity but rather is a consequence of the inability of DNA-PKcs to undergo autophosphorylation.
Similarly, mutations in the autophosphorylation sites of DNA-PKcs do not affect the ability of DNA-PKcs to interact with or activate Artemis. In fact, if lack of Artemis activation were the only important deficit in cells expressing mutant ABCDE, one might expect complementation of V3's signal joint deficit by mutant ABCDE, since Artemis is not required for signal end joining (35, 40). This is not the case. In sum, these data demonstrate that the function of DNA-PKcs in NHEJ goes beyond its role in activating the Artemis endonuclease. This is consistent with conclusions of a recent report directly comparing mice with targeted deletions of Artemis and DNA-PKcs (40).
The most revealing data as to the functional block of the multi-phosphorylation site mutants are the sequence analyses of coding joints mediated by the mutants. These joints have minimal nucleotide loss from rarely joined coding ends. Furthermore, there is no evidence of joining via short sequence homologies, a mechanism commonly observed in joints mediated by wild-type DNA-PKcs. These data imply that autophosphorylation of DNA-PKcs facilitates alignment via short sequence homologies and/or facilitates access of the DNA ends to DNA end-processing factors, perhaps including the endonuclease activity of Artemis and a DNA polymerase (31). We have preliminary data suggesting that the 3′ hydroxyls of the DNA break are not completely occluded by nonphosphorylated DNA-PKcs. Briefly, these data show that in transient recombination assays including an expression vector encoding Tdt, Tdt can add N segments to rare coding ends in the presence of the ABCDE mutant (data not shown).
Several studies have shown that DNA-PKcs facilitates alignment or synapses of DNA-PK-bound DNA ends (7, 33, 50). Furthermore, a recent report suggests that synapses of the two end binding complexes are required for kinase activation (7). Based on this report, it seems likely that mutant ABCDE forms synapses normally, since the kinase activity of the mutant protein is indistinguishable from that of wild-type DNA-PKcs. Chu and colleagues have proposed a model whereby DNA-PKcs may promote melting and alignment of DNA ends in a synaptic complex of two DNA-PK-bound DNA ends (17, 27). One attractive possibility is that autophosphorylation is required for this potential function of DNA-PKcs.
In summary, our results showing that autophosphorylation of DNA-PKcs between residues 2609 and 2647 is required during NHEJ lead us to favor a model whereby autophosphorylation induces a conformational change that leads to remodeling of the DNA-PK complex. Furthermore, this conformational change in DNA-PKcs is requisite for efficient end processing and DNA repair.
Acknowledgments
This work was supported by Public Health Service grants AI32600, AI42938 (to K.M.), and CA 84442-01 (to D.A.R.) from the National Institutes of Health. Work in the laboratory of S.P.L.-M. is supported by the Alberta Heritage Foundation for Medical Research and the Canadian Institutes for Health Research.
REFERENCES
- 1.Baumann, P., and S. C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95:14066-14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogue, M. A., C. Jhappan, and D. B. Roth. 1998. Analysis of variable (diversity) joining recombination in DNA-dependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc. Natl. Acad. Sci. USA 95:15559-15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, D. W., B. P. Chen, S. Prithivirajsingh, A. Kurimasa, M. D. Story, J. Qin, and D. J. Chen. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16:2333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, D. W., and S. P. Lees-Miller. 1996. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 271:8936-8941. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. W., R. Ye, C. J. Veillette, and S. P. Lees-Miller. 1999. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry 38:1819-1828. [DOI] [PubMed] [Google Scholar]
- 6.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 7.DeFazio, L. G., R. M. Stansel, J. D. Griffith, and G. Chu. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas, P., G. B. Moorhead, R. Ye, and S. P. Lees-Miller. 2001. Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 276:18992-18998. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, P., G. P. Sapkota, N. Morrice, Y. Yu, A. A. Goodarzi, D. Merkle, K. Meek, D. R. Alessi, and S. P. Lees-Miller. 2002. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 368:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Errami, A., D. M. He, A. A. Friedl, W. J. Overkamp, B. Morolli, E. A. Hendrickson, F. Eckardt-Schupp, M. Oshimura, P. H. Lohman, S. P. Jackson, and M. Z. Zdzienicka. 1998. XR-C1, a new CHO cell mutant which is defective in DNA-PKcs, is impaired in both V(D)J coding and signal joint formation. Nucleic Acids Res. 26:3146-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finnie, N. J., T. M. Gottlieb, T. Blunt, P. A. Jeggo, and S. P. Jackson. 1995. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 92:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D. T. Weaver, and F. W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 9:367-376. [DOI] [PubMed] [Google Scholar]
- 13.Gell, D., and S. P. Jackson. 1999. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27:3494-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellert, M. 2002. V(D)J recombination: rag proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 15.Grawunder, U., M. Wilm, X. Wu, P. Kulesza, T. E. Wilson, M. Mann, and M. R. Lieber. 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388:492-495. [DOI] [PubMed] [Google Scholar]
- 16.Gurley, K. E., and C. J. Kemp. 2001. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr. Biol. 11:191-194. [DOI] [PubMed] [Google Scholar]
- 17.Hammarsten, O., L. G. DeFazio, and G. Chu. 2000. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J. Biol. Chem. 275:1541-1550. [DOI] [PubMed] [Google Scholar]
- 18.Hartley, K. O., D. Gell, G. C. Smith, H. Zhang, N. Divecha, M. A. Connelly, A. Admon, S. P. Lees-Miller, C. W. Anderson, and S. P. Jackson. 1995. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82:849-856. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, S. P. 1997. DNA-dependent protein kinase. Int. J. Biochem. Cell Biol. 29:935-938. [DOI] [PubMed] [Google Scholar]
- 20.Jeggo, P. A. 1998. DNA breakage and repair. Adv. Genet. 38:185-218. [DOI] [PubMed] [Google Scholar]
- 21.Kienker, L. J., E. K. Shin, and K. Meek. 2000. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 28:2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurimasa, A., S. Kumano, N. V. Boubnov, M. D. Story, C. S. Tung, S. R. Peterson, and D. J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurimasa, A., H. Ouyang, L. Dong, S. Wang, X. Li, C. Cordon-Cardo, D. J. Chen, and G. C. Li. 1999. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc. Natl. Acad. Sci. USA 96:1403-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leber, R., T. W. Wise, R. Mizuta, and K. Meek. 1998. The XRCC4 gene product is a target for, and interacts with the DNA dependent protein kinase. J. Biol. Chem. 273:1794-1801. [DOI] [PubMed] [Google Scholar]
- 25.Lees-Miller, S. P. 1996. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell. Biol. 74:503-512. [DOI] [PubMed] [Google Scholar]
- 26.Lees-Miller, S. P., R. Godbout, D. W. Chan, M. Weinfeld, R. S. Day, G. M. Barron, and J. Allalunis-Turner. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 267:1183-1185. [DOI] [PubMed] [Google Scholar]
- 27.Leuther, K. K., O. Hammarsten, R. D. Kornberg, and G. Chu. 1999. Structure of DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J. 18:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Z., T. Otevrel, Y. Gao, H. L. Cheng, B. Seed, T. D. Stamato, G. E. Taccioli, and F. W. Alt. 1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J. recombination. Cell 83:1079-1089. [DOI] [PubMed] [Google Scholar]
- 29.Lieber, M. R. 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4:77-85. [DOI] [PubMed] [Google Scholar]
- 30.Lieber, M. R., J. E. Hesse, S. Lewis, G. C. Bosma, N. Rosenberg, K. Mizuuchi, M. J. Bosma, and M. Gellert. 1989. Abnormal V(D)J recombination in murine severe combined immune deficiency: absence of coding joints and formation of alternative products. Curr. Top. Microbiol. Immunol. 152:69-75. [DOI] [PubMed] [Google Scholar]
- 31.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 32.Meek, K., L. Kienker, C. Dallas, W. Wang, M. J. Dark, P. J. Venta, M. L. Huie, R. Hirschhorn, and T. Bell. 2001. SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. J. Immunol. 167:2142-2150. [DOI] [PubMed] [Google Scholar]
- 33.Merkle, D., P. Douglas, G. B. Moorhead, Z. Leonenko, Y. Yu, D. Cramb, D. P. Bazett-Jones, and S. P. Lees-Miller. 2002. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41:12706-12714. [DOI] [PubMed] [Google Scholar]
- 34.Modesti, M., J. E. Hesse, and M. Gellert. 1999. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 18:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 36.Nick McElhinny, S. A., C. M. Snowden, J. McCarville, and D. A. Ramsden. 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastink, A., J. C. Eeken, and P. H. Lohman. 2001. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 480-481:37-50. [DOI] [PubMed] [Google Scholar]
- 38.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell. Biol. 11:52-59. [DOI] [PubMed] [Google Scholar]
- 39.Riballo, E., S. E. Critchlow, S. H. Teo, A. J. Doherty, A. Priestley, B. Broughton, B. Kysela, H. Beamish, N. Plowman, C. F. Arlett, A. R. Lehmann, S. P. Jackson, and P. A. Jeggo. 1999. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 9:699-702. [DOI] [PubMed] [Google Scholar]
- 40.Rooney, S., J. Sekiguchi, C. Zhu, H. L. Cheng, J. Manis, S. Whitlow, J. DeVido, D. Foy, J. Chaudhuri, D. Lombard, and F. W. Alt. 2002. Leaky scid phenotype associated with defective V(D)J coding end processing in artemis-deficient mice. Mol. Cell 10:1379-1390. [DOI] [PubMed] [Google Scholar]
- 41.Roth, D. B., J. P. Menetski, P. B. Nakajima, M. J. Bosma, and M. Gellert. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983-991. [DOI] [PubMed] [Google Scholar]
- 42.Sekiguchi, J., D. O. Ferguson, H. T. Chen, E. M. Yang, J. Earle, K. Frank, S. Whitlow, Y. Gu, Y. Xu, A. Nussenzweig, and F. W. Alt. 2001. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. USA 98:3243-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin, E. K., T. Rijkers, A. Pastink, and K. Meek. 2000. Analyses of TCRB rearrangements substantiate a profound deficit in recombination signal sequence joining in SCID foals: implications for the role of DNA-dependent protein kinase in V(D)J recombination. J. Immunol. 164:1416-1424. [DOI] [PubMed] [Google Scholar]
- 44.Soubeyrand, S., L. Pope, B. Pakuts, and R. J. Hache. 2003. Threonines 2638/2647 in DNA-PK are essential for cellular resistance to ionizing radiation. Cancer Res. 63:1198-1201. [PubMed] [Google Scholar]
- 45.Taccioli, G. E., A. G. Amatucci, H. J. Beamish, D. Gell, X. H. Xiang, M. I. T. Arzayus, A. Priestley, S. P. Jackson, A. M. Rothstein, P. A. Jeggo, and V. L. M. Herrera. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9:355-366. [DOI] [PubMed] [Google Scholar]
- 46.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607-614. [DOI] [PubMed] [Google Scholar]
- 47.Whitmore, G. F., A. J. Varghese, and S. Gulyas. 1989. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int. J. Radiat. Biol. 56:657-665. [DOI] [PubMed] [Google Scholar]
- 48.Wiler, R., R. Leber, B. B. Moore, L. F. VanDyk, L. E. Perryman, and K. Meek. 1995. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. USA 92:11485-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woods, T., W. Wang, E. Convery, A. Errami, M. Z. Zdzienicka, and K. Meek. 2002. A single amino acid substitution in DNA-PKcs explains the novel phenotype of the CHO mutant, XR-C2. Nucleic Acids Res. 30:5120-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaneva, M., T. Kowalewski, and M. R. Lieber. 1997. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 16:5098-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo, S., and W. S. Dynan. 1999. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 27:4679-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zdzienicka, M. 1999. Mammalian X-ray-sensitive mutants which are defective in non-homologous (illegitimate) DNA double-strand break repair. Biochimie 81:107-116. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, C., and D. B. Roth. 1995. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination. Immunity 2:101-112. [DOI] [PubMed] [Google Scholar]