Abstract
We show that Bacillus subtilis SMC (structural maintenance of chromosome protein) localizes to discrete foci in a cell cycle-dependent manner. Early in the cell cycle, SMC moves from the middle of the cell toward opposite cell poles in a rapid and dynamic manner and appears to interact with different regions on the chromosomes during the cell cycle. SMC colocalizes with its interacting partners, ScpA and ScpB, and the specific localization of SMC depends on both Scp proteins, showing that all three components of the SMC complex are required for proper localization. Cytological and biochemical experiments showed that dimeric ScpB stabilized the binding of ScpA to the SMC head domains. Purified SMC showed nonspecific binding to double-stranded DNA, independent of Scp proteins or ATP, and was retained on DNA after binding to closed DNA but not to linear DNA. The SMC head domains and hinge region did not show strong DNA binding activity, suggesting that the coiled-coil regions in SMC mediate an association with DNA and that SMC binds to DNA as a ring-like structure. The overproduction of SMC resulted in global chromosome compaction, while SMC was largely retained in bipolar foci, suggesting that the SMC complex forms condensation centers that actively affect global chromosome compaction from a defined position on the nucleoid.
Before cells can divide, they must duplicate and separate their chromosome(s) for faithful distribution to the daughter cells. Key players in many chromosome dynamics are SMC (structural maintenance of chromosome) proteins, which represent a ubiquitous family of proteins found in almost all organisms. SMCs are essential for chromosome condensation, sister chromatid cohesion, DNA recombination, and gene dosage compensation (15, 39). SMCs have a head-rod-tail conformation, each domain endowed with common structural motifs. The N-terminal region contains a nucleotide binding Walker A motif, two central coiled coils are separated by a hinge domain, and the C-terminal region contains the DA box, which resembles the Walker B motif. The functional ATPase pocket is jointly formed by the N- and C-terminal regions, as shown by the crystal structure (18, 27). SMCs generally form dimers (16, 29). Initially, it was thought that dimer formation is mediated by an antiparallel arrangement of the coiled-coil regions. However, recent studies have shown that the coiled-coil regions rather fold back onto each other in an intramolecular fashion and that dimer formation is instead mediated by the hinge domain (11). Interestingly, Rad50, an SMC family member that does not bear a hinge domain, uses a CXXC motif conserved in the middle section of the coiled-coil regions to form an interlocking Zn2+ binding hook (17), leading to dimerization. So, SMCs appear to generally form symmetrical dimers.
Eukaryotic SMCs function as heterodimers, while their bacterial counterparts are homodimers, with only one or no smc gene being present per genome (37). So far, in eukaryotes, six genes, encoding SMC1 to SMC6, have been identified. Each of the SMCs has a specific partner, resulting in three different complexes performing vital tasks in chromosome dynamics. The SMC1-SMC3 dimer in the cohesin complex functions in sister chromatid cohesion, while SMC2-SMC4 acts in chromosome condensation (condensin complex), and the latest known, SMC5-SMC6, functions in DNA repair and the checkpoint response (12). The condensin complex is active in DNA condensation only in the presence of SMC2-SMC4 and all non-SMC subunits (21), stressing the importance of the auxiliary proteins for SMC function.
In bacteria, SMCs are essential for chromosome condensation and segregation (4, 10, 30). Escherichia coli has a functional analog of SMC, called MukB, whose loss likewise leads to a profound defect in chromosome segregation (32). E. coli MukB forms a complex with two other proteins encoded by its operon, MukE and MukF (45). Recently, it was shown that Bacillus subtilis SMC interacts with two highly conserved prokaryotic proteins, ScpA and ScpB (28, 38). Despite all this information, the mechanism of SMC action remains poorly understood.
Unlike eukaryotes, bacteria lack a true mitotic apparatus, yet chromosomes are highly efficiently and dynamically separated during ongoing DNA replication (6, 7, 43). The large circular chromosome is compacted about 2,000-fold into a structure called the nucleoid and has a rather specific arrangement within the cell. Origins of replication separate rapidly toward opposite cell poles and remain at these locations throughout the rest of the cell cycle (42). Replication termini are located near the middle of cells, while positions between origins and termini are situated between these sites in cells (33, 41). Thus, the B. subtilis and E. coli chromosomes are folded approximately according to their physical structures. SMC and MukB are required to maintain the conserved arrangement of chromosomes and for the efficient separation of whole chromosomes but not for the bipolar movement of replication origins (8, 44). DNA polymerase is located at the middle of cells during most of the cell cycle (24), and the movement of chromosomes through the stationary polymerase has been proposed to serve as a motor for the separation of chromosomes (23).
In this article, we report the dynamic cell cycle-dependent localization of B. subtilis SMC and provide evidence that SMC forms a complex with ScpA and ScpB, which bind to the SMC head domains. SMC binds to double-stranded DNA in an unusual manner, probably by embracing DNA strands. Our data support the model that the SMC complex forms a subcellular structure that locally condenses DNA close to the cell poles, mediating chromosome organization and facilitating segregation.
MATERIALS AND METHODS
Construction of strains.
The strains used in this study are listed in Table 1. All clonings were done with E. coli strain XL1-Blue or GM48 (Stratagene). Cultures were grown in Luria-Bertani medium supplemented with 50 μg of ampicillin/ml. Bacillus strains were grown in Luria-Bertani medium. Promoters were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (for Pspac) or with 0.5% xylose (for Pxyl). The antibiotics chloramphenicol (5 μg/ml), spectinomycin (50 μg/ml), erythromycin (1 μg/ml), and lincomycin (25 μg/ml) were used when necessary. For microscopy, cells were grown in S750 medium (19). B. subtilis PY79 (42) was used as a wild-type strain.
TABLE 1.
Strains used in this study
| B. subtilis strain | Genotype | Reference or source |
|---|---|---|
| PY79 | Wild type | 43 |
| PGΔ388 | smc::kan | 10 |
| JM8 | scpA-yfp | 28 |
| JM9 | scpB-yfp | 28 |
| JM10 | Pxyl-scpB-cfp::amy | 28 |
| JM11 | scpA::tet | 28 |
| PG31 | ypuI::tet | 28 |
| PG32 | scpB::tet | 28 |
| JM24 | smc-yfp | This work |
| JM25 | smc-yfp; Pspac for downstream gene | This work |
| JM26 | smc-cfp; Pspac for downstream gene | This work |
| JM27 | smc-yfp dnaX-cfp | This work |
| JM28 | smc-yfp spo0J::spec | This work |
| PG44 | Pxyl-smc-yfp; scpB-cfp at amy locus | This work |
| JM29 | smc-cfp scpA-yfp | This work |
| JM30 | smc-cfp scpB-yfp | This work |
| JM31 | smc-yfp (Pspac) scpA::tet | This work |
| JM32 | smc-yfp (Pspac) scpB::tet | This work |
| JM33 | smc-yfp (Pspac) ypuI::tet | This work |
| JM34 | smc-yfp (Pspac) scpAB::tet | This work |
| CAS4 | Phyperspank-smc at amy locus | This work |
| CAS5 | Phyperspac-smc | This work |
| JM35 | Phyperspac-smc-yfp | This work |
| JM36 | Phyperspac-smc scpB-yfp | This work |
| PG45 | Phyperspank-smc at amy locus; smc-yfp | This work |
To track SMC, a C-terminus-encoding fragment (539 bp) of smc was PCR amplified and cloned into ApaI and ClaI sites in plasmid pSG1187 (25). The resulting plasmid, pSy, was transformed into PY79 to yield JM24. A similar C-terminus-encoding fragment of smc was cloned into KpnI and ClaI sites in plasmids pMutin-YFP and pMutin-CFP (20). PY79 was transformed with the resulting plasmids, pMutinSy and pMutinSc, with selection for erythromycin-lincomycin resistance, to yield JM25 (smc-yfp) and JM26 (smc-cfp), respectively. To study the colocalization of SMC and Scp proteins, JM10 (Pxyl- scpB-cfp::amy) was transformed with chromosomal DNA from JM25 to yield PG44 (Pxyl-scpB-cfp::amy smc-yfp). JM8 and JM9 were transformed with chromosomal DNA from JM26 to generate JM29 (smc-cfp scpA-yfp) and JM30 (smc-cfp scpB-yfp), respectively. To simultaneously visualize DNA polymerase and SMC, chromosomal DNA from JM24 was used to transform PG28, resulting in JM27 (smc-yfp dnaX-cfp). scpA, scpB, and ypuI were deleted from smc-yfp cells by transformation of JM25 with chromosomal DNAs from JM11 (scpA::tet), PG31 (scpB::tet), and PG32 (ypuI::tet) to yield JM31, JM32, and JM33, respectively. To overproduce SMC, an N-terminus-encoding fragment of smc (510 bp) was amplified by PCR and cloned into HindIII and SphI sites in plasmid pJQ43 (35). PY79 was transformed with the resulting plasmid, pCAS4, to yield CAS4. The complete smc gene was PCR amplified (a six-His-encoding sequence was included in the downstream primer) and cloned into NheI and SphI sites in plasmid pdr111 (gift from D. Rudner, Harvard University). PY79 was transformed with the resulting plasmid, pCAS5, and double-crossover integration at the amyE locus was tested by a starch assay. CAS4 and CAS5 were transformed with chromosomal DNA from JM25 to yield JM36 and PG45, respectively.
Construction of six-His-tagged SMC, ScpA, ScpB, and hinge and head domains of SMC and purification of proteins.
DNA fragments comprising smc, scpA, scpB, and the SMC hinge domain (bp 1383 to 2028 in smc), a 711-bp fragment encoding the SMC N-terminal domain, and a 630-bp fragment encoding the C-terminal domain were amplified by PCR, digested with NcoI and BglII, and inserted into plasmid pQE60 (Qiagen). The six-His-tagged N-terminal domain, together with the promoter-operator element, was amplified from the corresponding plasmid and inserted into the XbaI site of the SMC C-terminal construct described above, resulting in the dimeric head domain construct. Plasmids were transformed into E. coli M42 (Qiagen), and the production of proteins was induced by the addition of 1 mM IPTG. Proteins were purified by using standard Ni-nitrilotriacetic acid chromatography (Qiagen) and dialyzed in HEPES buffer (50 mM HEPES, 300 mM NaCl [pH 8.0]) for further experiments.
Fluorescence microscopy.
Fluorescence microscopy was performed as described previously (8). Fluorescence measurements were obtained by using METAMORPH 4.0: maximum fluorescence was scored in circles of 0.3 by 0.3 μm containing SMC-green fluorescence protein (GFP) foci or cellular spaces devoid of foci. Background fluorescence in wild-type cells was subtracted, and the increase in fluorescence was calculated relative to the average fluorescence of SMC-GFP foci in JM24 cells. For DNA staining, 4′,6′-diamidino-2-phenylindole (DAPI) was used at 200 ng/ml, and FM4-64 vital membrane stain was used at 2 nM.
Native PAGE experiments.
For native polyacrylamide gel electrophoresis (PAGE), ScpA (2 μM), ScpB (10 μM), and SMC (2 μM) were incubated alone or in different combinations in binding buffer A (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM MgCl2) for 10 min at room temperature before being loaded onto 7.5% native polyacrylamide gels.
Analytical gel filtration and sucrose gradient centrifugation.
A Pharmacia Superdex 75 column was used for gel filtration studies; proteins were rebuffered in 50 mM NaH2PO4- 100 mM KCl [pH 7.5]. For gradient centrifugation, a 5 to 20% sucrose gradient was spun at 165,000 × g for 15 h. Fractions were removed manually and subjected to sodium dodecyl sulfate-PAGE. Standard proteins used were bovine serum albumin, ovalbumin, chymotrypsinogen, cytochrome c, and aprotinin (66, 45, 25, 14, and 6.5 kDa, respectively).
Mass spectroscopy.
ScpB was dialyzed in 10 mM ammonium acetate buffer (pH 8.0), and different concentrations (60 to 600 μM) were subjected to electrospray ionization-time-of-flight analysis in an API QSTAR apparatus.
DNA binding assays.
Gel shift assays were done with 7.5% native polyacrylamide gels in Tris-borate-EDTA buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.0]). Twenty picomoles of SMC (2.7 μg), ScpA (0.6 μg), ScpB (0.44 μg), head domain, and hinge was incubated with 1.5 pmol of DNA (500 ng; 500 bp) in binding buffer B (20 mM Tris-HCl [pH 8.6], 50 mM NaCl, 5 mM MgCl2) at room temperature for 30 min before being applied to the gels.
Surface plasmon resonance experiments.
Protein-protein and protein-DNA interactions were analyzed by surface plasmon resonance with a Biacore X instrument. Proteins were dialyzed in HEPES buffer. CM5 chips were derivatized with proteins by amine coupling according to the manufacturer's recommendations, and SA chips were used for biotin coupling. The signal in the reference cell was subtracted online during all measurements. The soluble binding partner (analyte) was injected at a range of concentrations at a flow rate of 10 μl/min. For the removal of unbound analyte, 50 mM NaOH was used; for the complete removal of DNA from SA chips, 250 mM NaOH was used. Control proteins were 2% bovine serum albumin, 10 μM peptidyl carrier protein (kind gift from M. Mofid, Marburg, Germany), AbrB (kind gift from G. Schimpf-Weihland, Marburg, Germany), and Fis (kind gift from G. Muskhelishvili, Marburg, Germany).
RESULTS
Cell cycle-dependent localization of SMC in live cells.
We wished to investigate the localization of SMC in live cells. Initial attempts to create a functional SMC-GFP fusion were unsuccessful, so we used a technique devised by Ohsumi and coworkers (34), introducing four additional glycine residues into the SMC-YFP linker region (YFP is a color variant of GFP). Integration of this construct into the smc locus resulted in wild-type-like colonies at 25°C but not at 37°C. Nucleoids appeared normal in the SMC-YFP-expressing strain (JM24; data not shown), suggesting that SMC-YFP is functional at room temperature. To further improve the fusion, we used a vector system established by Kaltwasser et al. (20), which includes a promoter to drive the expression of genes downstream of the YFP fusion (the gene downstream of smc, ftsY, confers an important function for viability at 37°C). The resultant strain, JM25, grew normally even at 37°C in the presence but not in the absence of IPTG, indicating that continued transcription of ftsY is critical in the smc-yfp fusion strain at 37°C. Both strains, JM24 and JM25, showed indistinguishable fluorescent foci at 25°C in minimal medium, which was used throughout this study for the investigation of a simple cell cycle devoid of overlapping rounds of DNA replication and thus chromosome segregation. Like its interacting proteins, ScpA and ScpB, SMC localized in a cell cycle-dependent manner; one or two fluorescent foci were present in the middle of small (and thus young) cells, while in larger cells (and thus later in the cell cycle), one or two fluorescent foci were present close to each cell pole (Fig. 1A). These results were supported by statistical analyses of the position of SMC in a growing cell population (data not shown; see reference 28). The fluorescence of cells outside SMC-YFP foci was similar to that of cells that did not carry the fusion, indicating that most SMC molecules are present within the foci.
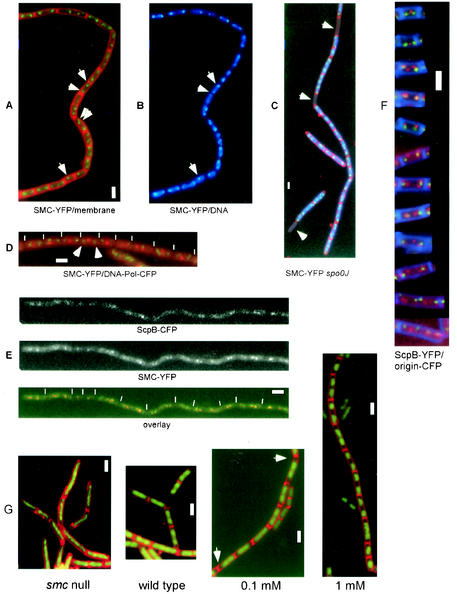
FIG. 1.
Fluorescence microscopy of B. subtilis cells growing at mid-exponential phase. (A to D) SMC-YFP (green), membranes (red; A and C), DNA (blue; B and C), and tau subunit of DNA polymerase (red; D). (A and B) Strain JM24 (smc-yfp); arrowheads indicate SMC at midcell or in a bipolar location. (C) Strain JM28 (smc-yfp spo0J::spec); arrowheads indicate the absence of SMC fluorescence in anucleate cells, which arise at a frequency of about 1% in spo0J mutant cells. (D) Strain JM27 (smc-yfp dnaX-cfp); arrowheads indicate SMC foci flanking a central DNA polymerase focus. (E) Strain PG44 (scpB-cfp smc-yfp); upper panel shows CFP fluorescence, middle panel shows YFP fluorescence, and lower panel shows an overlay. (F) Representative cells showing the localization of ScpB-YFP (red), origin regions (green), and membranes (blue). (G) DNA (green) and membranes (red); first panel shows strain PGΔ388 (smc::kan), second panel shows strain PY79 (wild type), third panel shows strain CAS5 (Phyperspac-smc) with 0.1 mM IPTG, and fourth panel shows strain CAS5 (Phyperspac-smc) with 1 mM IPTG. Arrowheads in the third panel indicate increased DNA-free spaces in cells. Ends of cells are indicated by thin white lines. Thick white bars, 2 μm.
In general, SMC was present at the outer edge of nucleoids (Fig. 1B) and, like ScpA and ScpB, SMC was absent from anucleate (DNA-less) cells in an spo0J mutant (Fig. 1C), showing that SMC is tightly associated with the chromosome. SMC was well separated from the centrally located DNA polymerase complex (Fig. 1D). Even in small cells with a central SMC focus, SMC did not colocalize with DNA polymerase (data not shown). These data show that while DNA polymerase remains stationary at midcell throughout most of the cell cycle (24), SMC moves toward the cell poles early in the cell cycle. Our findings corroborate and extend earlier findings obtained with strains expressing SMC-YFP and MukB-GFP (4, 5, 34) and suggest that immunolabeling of wild-type cells with SMC antibodies (10) exaggerated the bipolar localization of SMC.
SMC colocalizes with ScpA and ScpB.
For dual labeling, we constructed several strains in which SMC and the Scp proteins were fused to CFP or YFP variants of GFP. It was difficult to localize a SMC-CFP fusion in ScpA-YFP- and ScpB-YFP-expressing cells (see below). In the rare instances when clear CFP and YFP foci were visible, these were coincident. Conclusive results were obtained with a strain (PG44) in which a functional ScpB-CFP fusion at an ectopic site in the chromosome was combined with SMC-YFP. In all samples showing clear signals in both channels (>60 cells), ScpB and SMC foci were coincident (Fig. 1E). Because ScpA and ScpB colocalize in B. subtilis cells (28), these results show that most if not all SMC molecules are present within the subcellular regions containing both Scp proteins. Therefore, investigation of any of the three complex partners provides information about the positions of all three proteins of the complex.
Dynamic localization of the SMC complex.
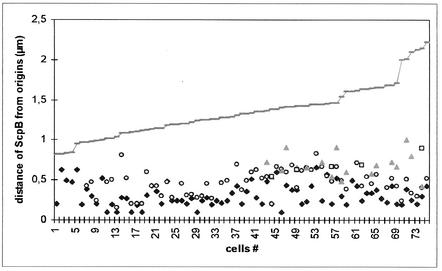
To further characterize the positions of the SMC condensation centers during the cell cycle, we monitored the location of ScpB relative to the origins of replication during the cell cycle. Initial studies showed that ScpB mostly localizes close to the origin regions, often with two foci flanking each origin (28). Figure 2 shows the distance measured between ScpB foci and origin regions relative to cell size (the size of half of a cell because one origin is generally present in each cell half under the conditions used), and Fig. 1F shows representative images of cells at different times in the cell cycle (at least 10 cells similar to each panel were found from among about 600 cells monitored). In small cells (<1 μm), one or two central SMC/Scp foci were flanked by two well-separated origin signals, so at this early stage, Scp foci were well separated from origin regions (Fig. 2). Later, SMC/Scp foci moved toward the cell poles, close to or coincident with origin regions (Fig. 2, cells between 1 and ∼1.25 μm). It should be noted that even when Scp foci and origins were very close, the measurements were never below 0.2 μm because of the resolution limit of light microscopy. Occasionally, one ScpB focus was found very close to the origin, while the other was still well separated from the other origin, being close to midcell (Fig. 1F, third panel). These findings agree well with the asymmetrical movement of SMC/Scp foci found in time-lapse experiments (see below). After this initial period, the distance between SMC or Scp foci and origin regions increased (Fig. 2, cells >1.25 μm), approximately depending on cell size, although ScpB foci could be found very close to origin regions even in large cells. In general, Scp foci moved away from the origins toward the middle of cells (Fig. 1F). Sometimes, the largest cells contained four bipolar origins, while SMC/Scp foci were located near quarter sites corresponding to the future middle of newborn cells after cell division, indicating that a new round of segregation had occurred before cell division (Fig. 1F, last panel). Thus, condensation centers are not static but appear to move away from the origins of replication toward midcell, where newly replicated DNA is expected to leave DNA polymerase (23).
FIG. 2.
Statistical analysis of the distance of ScpB foci from origins of replication, depending on cell size. Symbols: ♦, ScpB focus closest to origin; ○, focus in other cell half; □ and ▴, third focus and fourth focus, respectively, in cells with more than two foci. The grey line indicates the size of half of a cell.
Specific localization of SMC depends on ScpA and ScpB.
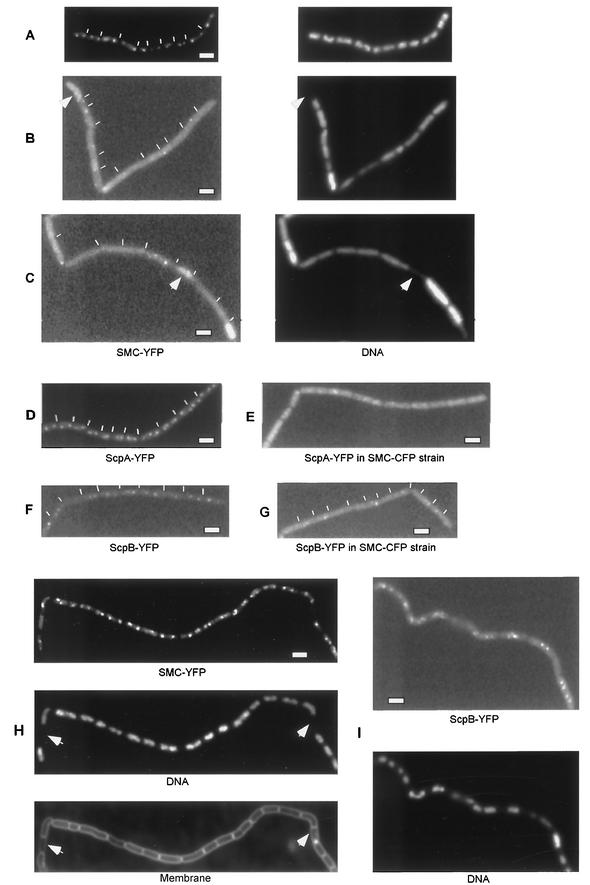
Mascarenhas et al. (28) previously showed that the formation of foci for both Scp proteins is dependent on the presence of SMC. To investigate the important question as to whether SMC can form foci in the absence of Scp proteins, we constructed several strains in which scpA, scpB, or both genes were deleted from strain JM25 (smc-yfp). We also constructed a control strain in which the gene downstream of scpA and scpB, ypuI, was deleted (strain JM33). As shown in Fig. 3, JM33 grew like the wild type and had SMC as bipolar foci (Fig. 3A). The formation of bipolar SMC foci was lost in the absence of ScpA as well as of ScpB (Fig. 3B and C) or of both genes (data not shown). In most samples, fluorescence was seen distributed throughout the cell; however, some cells had foci that were located on nucleoids, but in an aberrant fashion. In a few samples, fluorescence was seen to accumulate near the membrane. Interestingly, SMC-YFP was also present in anucleate cells in this experiment (Fig. 3B and C, arrowheads), in contrast to anucleate cells in the presence of ScpA and ScpB (Fig. 1C). These results show that while SMC can still form foci on DNA, both Scp proteins are required for proper localization on nucleoids. From these data, we infer that Scp proteins play an essential role in the formation of condensation centers, supporting the notion that the proteins form a ternary complex in vivo.
FIG. 3.
Fluorescence microscopy of B. subtilis mutant cells. (A to C) (Left panels) Fluorescence of SMC-YFP. (Right panels) DNA stained by DAPI. (A) Strain JM33 (smc-yfp ypuI::tet). (B) Strain JM31 (smc-yfp scpA::tet). (C) Strain JM32 (smc-yfp scpB::tet). Arrowheads in panels A to C indicate anucleate cells containing SMC-YFP. (D) Strain JM8 (scpA-yfp). (E) Strain JM29 (scpA-yfp smc-cfp). (F) Strain JM9 (scpB-yfp). (G) Strain JM30 (scpB-yfp smc-cfp). (H) Strain JM35 (phyperspac-smc-yfp) with 1 mM IPTG; arrowheads indicate anucleate cells. (I) Strain JM36 (phyperspac-smc scpB-yfp) with 1 mM IPTG. Thin white lines indicate septa between cells. Thick white bars, 2 μm.
SMC can condense whole nucleoids largely from a single position on the nucleoid.
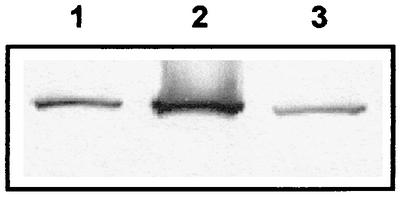
To test whether SMC can work as an active condensation factor, we sought to overproduce the protein in vivo. To do this, we used a new version of the Pspac promoter, hyperspac, in which transcription can no longer be repressed but can be strongly induced upon the addition of an inducer (IPTG). We also used a set of plasmids in which the hyperspac promoter is converted into a completely repressible version called hyperspank. Both promoters allowed for a gradual two- to fourfold increase in the production of SMC compared to the wild-type situation. Figure 1G shows the effects of different levels of SMC in B. subtilis cells. In the absence of SMC (and after a decrease in SMC production; data not shown), chromosomes were highly decondensed, giving rise to slow temperature-sensitive growth and the production of about 15% anucleate cells (first panel). Wild-type cells contained amorphous condensed nucleoids, and longer cells contained two visible nucleoids (Fig. 1G, second panel). A slight overproduction of SMC (about twofold; 0.1 mM IPTG) (Fig. 4, lane 1) already led to a visible overcompaction of DNA (Fig. 1G, third panel), while full induction of the hyperspac promoter (1 mM IPTG) caused severe chromosome compaction (Fig. 1G, fourth panel) accompanied by the formation of about 5% anucleate cells (Fig. 3H). Under these conditions, SMC levels were about fourfold higher than in wild-type cells (Fig. 4, compare lanes 2 and 3). Thus, SMC can actively induce chromosome compaction, even when only slightly overproduced.
FIG. 4.
Western blot with SMC antiserum. Lane 1, induction of SMC from phyperspac by 0.1 mM IPTG; lane 2, full induction of SMC from phyperspac by 1 mM IPTG, lane 3, wild-type cells.
It was possible that chromosome compaction was caused by uncontrolled binding of SMC throughout the nucleoids. To visualize SMC, we introduced the functional C-terminal YFP fusion of SMC into the overproduction strain. At 0.025 mM IPTG, the morphology of nucleoids was indistinguishable from that of wild-type cells, and SMC-YFP localized in a bipolar manner similar to that in the parent strain (Fig. 3A and data not shown). Strikingly, at 1 mM IPTG, SMC-YFP was still largely retained in foci, with very little fluorescence outside these subcellular regions (Fig. 3H), while chromosomes were highly compacted. To support these results, we measured fluorescence intensities of GFP foci and of (comparably sized, ∼0.3 by 0.3 μm) cellular spaces devoid of foci in more than 200 cells. While the intensity of the SMC-GFP foci increased by 80% after phyperspac induction (from 172 to 206 U, on average), background fluorescence inside cells increased only about 5% (from 138 to 140 U, compared with 132 U in wild-type cells). Likewise, ScpB-YFP was still present in the condensation centers at 1 mM IPTG (Fig. 3I), indicating that the whole SMC complex was retained at its specific location. To rule out the possibility that the YFP fusion causes any artifact, we overproduced SMC at an ectopic location on the chromosome, while the YFP fusion was driven by the original promoter at the smc locus. Full induction of this construct resulted in a similar level of SMC produced (data not shown) accompanied by a similar degree of chromosome compaction, while the specific localization of SMC-YFP was retained (data not shown). These results confirm that the SMC complex can induce global chromosome compaction largely from a defined structure located on the nucleoid.
ScpA, ScpB, and the hinge domain form dimers in solution.
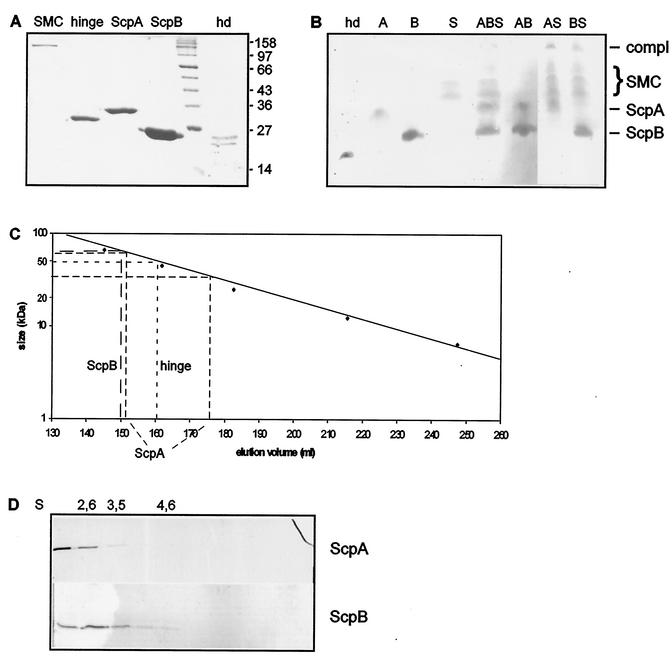
To investigate the biochemical properties of ScpA, ScpB, and SMC, we purified all three proteins and the SMC hinge domain by Ni-nitrilotriacetic acid affinity chromatography to apparent homogeneity (Fig. 5A). To ensure the proper function of SMC-six-His, we constructed a strain which carries an smc-his6 fusion at the amylase (amy) locus on the chromosome. This fusion was able to complement the function of SMC at 25°C but not at 37°C. Therefore, all biochemical experiments were performed at 25°C. In addition, we cloned the N-terminal domain and the C-terminal domain into a six-His vector, such that both fusions were simultaneously expressed. X-ray crystallography has shown that both domains together form the SMC head domain (27), so we refer to this dimeric construct as the SMC head domain. As expected, both domains coeluted after affinity chromatography (Fig. 5A, lane hd) and migrated as a single band on native PAGE (Fig. 5B, lane hd).
FIG. 5.
Protein analyses. (A) Coomassie blue-stained sodium dodecyl sulfate-7.5% polyacrylamide gel of purified proteins. Lane hd, head domain; note that this lane was taken from another gel at its appropriate position relative to the marker. Selected sizes of marker proteins are indicated on the right in kilodaltons. (B) Coomassie blue-stained 7.5% native polyacrylamide gel. Lanes: A, ScpA (2 μM); B, ScpB (10 μM); S, SMC (2 μM); hd, head domain (1.8 μM; this lane was taken from another gel at its appropriate position relative to ScpB and ScpA); other lanes, combinations of A, B, and/or S. Lines at right indicate migration positions for ScpA, ScpB, and the complex (compl); the brace indicates the migration position for SMC. (C) Gel filtration analysis of ScpA, ScpB, and the SMC hinge domain. Standard proteins are indicated by diamonds. (D) Five to 20% sucrose gradient centrifugation of ScpA and ScpB. Migration positions for marker proteins are indicated above the gels.
ScpA, ScpB, and the hinge domain were analyzed by size exclusion chromatography and by analytical centrifugation. ScpA eluted in two peaks, corresponding to 35 kDa (monomer) and 60 kDa (dimer) (Fig. 5C); ScpB eluted exclusively at 65 kDa, corresponding to a trimer (Fig. 5C). The hinge domain eluted exclusively at 50 kDa, in agreement with the dimer formation found for the hinge domain in Thermotoga maritima SMC by X-ray crystallography (11). To calculate the native masses of the Scp proteins, sucrose gradient centrifugation was performed; this analysis showed that ScpA has a major sedimentation value of about 2.0S (similar to the value of 1.8S for cytochrome c, which was exclusively found in fraction 1), while ScpB has a value of about 2.8S (Fig. 5D). With a Stokes radius of 2.8, as determined from gel filtration, ScpA is clearly mostly a monomer in solution (native mass, 30 kDa) but can also exist as a dimer. With a calculated Stokes radius of 3.4, ScpB has a native mass of 39 kDa, suggesting that ScpB forms a dimer. To verify this notion, mass spectroscopy was performed with different concentrations of ScpB. Even at very dilute concentrations, ScpB showed two mass peaks, at 22 and 44 kDa (data not shown), proving that ScpB forms a dimer in solution. The high Stokes radii of ScpA and ScpB suggest that the monomer or the dimer has an elongated structure, consistent with the prediction that ScpA and ScpB contain coiled-coil elements and a high degree of α helices (28), which might mediate dimer formation.
ScpA and ScpB bind to the SMC head domain.
Three lines of evidence suggest that SMC, ScpA, and ScpB form a ternary complex. First, we constructed strains in which SMC is tagged with CFP at its C terminus (and thus at each head domain) and in which ScpA and ScpB are tagged with YFP (strains JM29 and JM30, respectively). In contrast to the rates of growth of strains carrying any single GFP-tagged construct, which were indistinguishable from the rate of growth of wild-type cells, the rate of growth of the ScpB- and SMC-tagged strain was somewhat reduced, while that of the ScpA- and SMC-tagged strain was strongly compromised. Fluorescence microscopy showed that ScpB-YFP was still properly localized in strain JM30 (Fig. 3G), although not as regularly as in the ScpB-YFP-expressing strain (Fig. 3F). In contrast, ScpA-YFP was almost completely delocalized in strain JM29 (compare Fig. 3E with Fig. 3D). These results show that simultaneous tagging of SMC and ScpA strongly interferes with the correct localization of the complex, while the tagging of SMC and ScpB has a weaker, yet detectable, effect on the correct localization of the complex.
Second, to support these interactions in vitro, we assayed purified proteins by native PAGE. The incubation of SMC with ScpA (Fig. 5B, lane AS) or with ScpA and ScpB (lane ABS) resulted in an additional, slowly migrating band (but not with other proteins assayed; data not shown), indicating complex formation among these proteins. Incubation of SMC with ScpB (Fig. 5B, lane BS) resulted in a diffuse, slowly migrating band only at high concentrations of ScpB. Note that SMC runs as three visible bands, probably due to different conformations in solution.
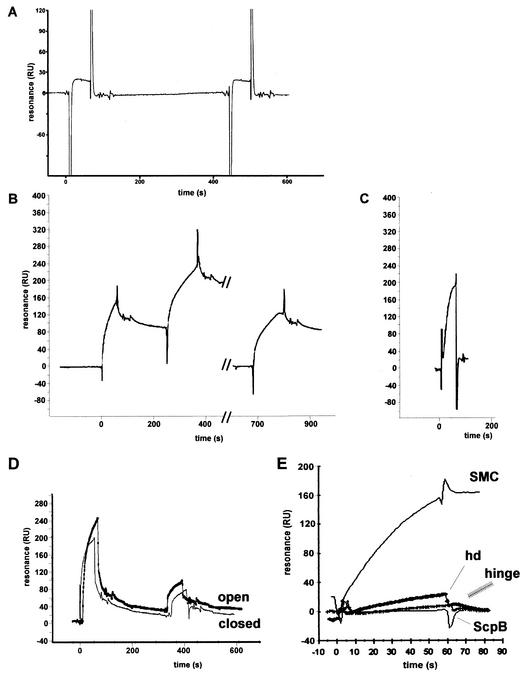
Third, we used surface plasmon resonance to detect protein-protein interactions. Direct tests of full-length SMC and ScpA were inconclusive, because of technical difficulties. However, when ScpA was covalently immobilized on the Biacore chip, a weak interaction was detectable with the SMC head domain but not with ScpB or with the hinge domain (data not shown). The interaction between ScpA and the head domain became robust when soluble ScpB was simultaneously injected with the head domain (Fig. 6A). To test whether DNA had an influence on the protein interactions, the head domain and ScpB were coinjected with a 500-bp DNA fragment that showed binding to SMC (see below). However, the presence of free DNA had no significant effect on the interaction of the head domain and ScpB with ScpA (Fig. 6A).
FIG. 6.
Surface plasmon resonance experiments. (A) ScpA (180 resonance units [RU]) was covalently immobilized on a Biacore chip. An equimolar mixture of the head domain and ScpB (2 μM each) was injected, followed by injection (at 450 s) of the head domain and ScpB (2 μM each) and a 500-bp linear DNA fragment. (B) A Streptavidin chip was coated with 350 RU of a 500-bp linear DNA fragment carrying biotin labels at both ends (closed). SMC (2 μM) was injected, followed by a second injection (2 μM, double amount at 250 s). The chip was washed with 50 mM NaOH, and SMC (2 μM, double amount at 680 s) was injected. (C) Same DNA as in panel B, except that the DNA was biotinylated only at the 3′ end (open). SMC (2 μM) was injected. Peaks flanking the binding curves were due to buffer fluctuations between the reference and the assay chamber at the beginning and end of each injection. (D) AbrB binding open or closed DNA. First injection, 12 μM AbrB; second injection, 6 μM AbrB. (E) Binding of different proteins to closed DNA: SMC (2 μM), head domain (hd; 2 μM), hinge (2 μM), or ScpB (20 μM).
Full-length SMC, but neither the head domain nor the hinge domain, binds nonspecifically to double-stranded DNA.
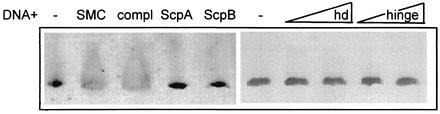
To test for DNA binding of SMC, ScpA, and ScpB, we performed gel shift experiments with double-stranded DNA fragments of different lengths. For all fragments tested, neither ScpA, ScpB, the hinge domain, nor the head domain showed any DNA binding, while SMC induced a hypershift of the DNA in the form of a smear (Fig. 7). Since this assay was not very satisfactory, we turned to surface plasmon resonance with 100- and 500-bp DNA fragments that were biotinylated at one or both ends and contained chromosomal DNA from an rRNA operon close to the replication origin. Figure 6B shows that SMC bound strongly to the DNA in which both ends were attached to the chip surface and was largely retained on the DNA after flushing of the chamber. The mode of binding strongly suggests a sequence-independent interaction of SMC with DNA, because more and more SMC bound during the injection period. In contrast, specific binding would result in a rapid occupation of binding sites followed by a state of equilibrium, because dissociating molecules would be replaced by others passing over the chip surface. A second injection of a twofold-larger amount of SMC (at 220 s) resulted in further, similar, but longer binding of SMC. After mild washing of the chamber, SMC bound again with similar kinetics, suggesting reversible binding to DNA. To further test whether SMC interacts with different DNA sequences, we also loaded onto the chip a 500-bp region close to the replication terminus (180°). SMC bound to this DNA with similar kinetics, although slightly less strongly (data not shown). We also immobilized SMC on a chip surface and tested it for DNA binding. The association of DNA with SMC was detectable, but at a much lower level than in the reverse experiment. Either immobilization compromises DNA binding in SMC or SMC molecules need to interact with each other to achieve strong binding to DNA. Interestingly, when the DNA was attached to the surface at only one end, SMC bound to the DNA with kinetics similar to those for its binding to the closed DNA (compare Fig. 6C with Fig. 6B) but was largely removed from the substrate during flushing of the chamber (Fig. 6C). These results suggest that SMC binds to DNA in a ring-like structure with the heads closed and DNA trapped between the arms (see Fig. 8), because such a structure would slip off the ends of an open DNA helix and would dissociate very slowly from closed DNA.
FIG. 7.
Ethidium-bromide stained 7% native polyacrylamide mobility shift assay. Linear DNA (1.5 pmol; 500 ng; 500 bp) was run in the absence (minus) or presence of SMC (20 pmol); SMC, ScpA, and ScpB (20 pmol each; compl); ScpA (20 pmol); ScpB (20 pmol); head domain (hd; 20 and 40 pmol, from left to right); or hinge (20 and 40 pmol).
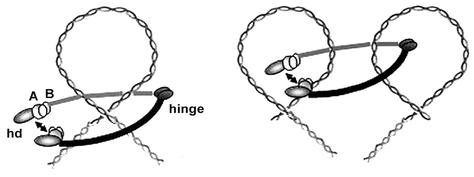
FIG. 8.
Model for architecture and DNA binding of the bacterial SMC complex. hd, SMC head domain; A, ScpA; B, ScpB. The SMC complex could condense DNA by introducing loops or by interlocking different DNA loops.
We also tested by surface plasmon resonance two known DNA binding proteins, B. subtilis AbrB and E. coli Fis (both global transcription factors [22, 31]), for their interactions with DNA. Figure 6D shows that AbrB bound to DNA in a nonspecific manner (there is no known specific binding site for AbrB in the ribosomal DNA used) and was released from both open and closed DNAs with similar kinetics. Likewise, Fis was released from both kinds of DNA, but at an even lower rate than AbrB (data not shown). Because a 3- to 6-fold-higher concentration of AbrB than of SMC was used (Fig. 6D), it is clear that SMC had a higher affinity for the DNA tested, whereas the affinity for the DNA was about 10-fold higher with Fis (data not shown) than with SMC. While AbrB showed a slightly higher affinity for open DNA (Fig. 6D, thick line versus thin line), Fis did not show a clear preference for either type of DNA (data not shown). In contrast to AbrB and Fis, SMC was immediately released from open DNA after flushing of the chamber (Fig. 6C), as opposed to a defined rate of release for high-affinity DNA binding.
Because SMC has weak DNA-stimulated ATPase activity (14), we tested the effect of ATP or its analogs on the DNA binding of SMC; unfortunately, however, we were not able to obtain conclusive results.
In contrast to SMC, neither ScpA nor ScpB showed any interaction with DNA in surface plasmon resistance analysis (Fig. 6E). The results showed that SMC has considerable affinity for double-stranded DNA in vitro, that it binds to DNA in a nonspecific and unusual manner, and that it is not released from closed DNA after binding. Moreover, the data suggested that SMC is the sole DNA binding component within the SMC/Scp complex.
Previous reports suggested that the C-terminal domain of SMC has DNA binding activity (1). However, the three-dimensional structure of SMC has shown that N- and C-terminal domains come together to form a single domain (27), so it has become clear that the isolated C-terminal domains are not useful for DNA binding assays. To test whether the complete head domain in SMC is the DNA binding site, we performed gel shift and surface plasmon resonance experiments with the SMC head domain and the hinge domain. Neither construct showed a pronounced affinity for DNA (Fig. 7, lanes hd and hinge; and Fig. 6E), suggesting that either the coiled-coil domains alone or in concert with the head and/or hinge domains mediate nonspecific DNA binding in SMC.
DISCUSSION
This report establishes that SMC, ScpA, and ScpB constitute a dynamic chromosome condensation-segregation complex in vivo and in vitro. SMC localizes to discrete regions on the nucleoids, in a cell cycle-dependent manner similar to that of ScpA and ScpB (28), and indeed colocalizes with both Scp proteins. These findings shows that most if not all SMC molecules are present in the observed condensation centers. Time-lapse microscopy showed that early in the cell cycle, the SMC complex moves from one or two positions close to the middle of the cell toward opposite cell poles in a rapid and dynamic manner (data not shown; see www.chemie.uni-marburg.de/∼graumann/SMCmovie.htm), reminiscent of the bipolar movement of origin regions on chromosomes (42). However, while replication origins move rather synchronously toward both poles (like several other specific regions on chromosome that have been assayed [41]) and before the bipolar separation of SMC and Scp foci, the latter moved in an asymmetrical manner. That is, while one focus moved toward and very close to one polar replication origin, the other remained close to midcell and moved to the opposite pole considerably later in the cell cycle. These findings suggest that the SMC complex does not strictly comigrate with a specific region on the chromosome. Indeed, the condensation centers do not appear to be associated with a single, specific region on the chromosome, because the SMC and Scp foci moved away from origin regions toward midcell during the cell cycle. Thus, the SMC complex could associate with different sites on chromosomes as they move from the central DNA polymerase toward the cell poles (for a model, see reference 9). Further cytological data suggest that the SMC complex is closely associated with the chromosome, because the SMC complex is not detectable in DNA-less cells.
While this work was under review, a somewhat different pattern of localization of SMC was reported. Lindow et al. (26) showed that SMC localized mostly close to midcell and thus close to DNA polymerase, while it moved to a bipolar arrangement (as was predominantly found in this work) later during the cell cycle. This apparent discrepancy can be explained by the fact that the SMC-GFP fusion used by Lindow et al. caused growth slower than that of wild-type cells (and compared to that of SMC-YFP-expressing strain JM24). When we reduced the rate of growth of JM24 cells (by using succinate instead of glucose defined medium), we indeed found that a higher proportion of cells contained a central SMC focus (data not shown). Under slow growth conditions, SMC might be present at midcell for a longer time because the initiation of replication occurs at a later point in the cell cycle, compared to a shorter cell cycle with higher growth rates, during which SMC moves toward cell poles early in the cell cycle.
The specific localization of SMC depends on both ScpA and ScpB (Fig. 3) and vice versa (28). Thus, all three proteins are required for proper subcellular localization. It is possible that one of the proteins is anchored to a specialized structure within the cells and needs the other proteins for efficient anchoring. Alternatively, the proteins could form a ternary complex that binds to and translocates on the DNA. The second scenario is supported by our in vivo findings and by in vitro data. Purified ScpA can bind to SMC head domains only in the presence of ScpB, supporting the formation of a ternary complex. Direct binding of ScpA to SMC head domains in vivo is supported by our finding that GFP tagging of both SMC and ScpA interferes with the specific localization of ScpA, accompanied by a reduction in the growth rate, although both GFP fusions are fully functional per se. Simultaneous GFP tagging of SMC and ScpB also has an effect on growth and on the localization of ScpB, but it is much weaker than the effect on ScpA, supporting the notion that ScpB might bind only indirectly to SMC via ScpA. Thus, like the non-SMC subunits of eukaryotic condensin and cohesin (2, 11, 46), the prokaryotic counterparts also bind to SMC head domains. Interestingly, ScpB forms strong dimers in solution, while ScpA exists in monomeric and dimeric forms. It is possible that an ScpA dimer binds to both SMC head domains, which would be bridged in a manner analogous to the bridging of cohesin head domains by Scc1 (11). This notion is supported by the finding that the N and C termini of ScpA bear significant sequence similarity to the equivalent regions in Scc1 (data not shown) (36). Dimeric ScpB could mediate the tight binding of ScpA to both head domains. Dimer formation for both Scp proteins is supported by computer sequence analysis indicating coiled-coil regions in both proteins (28). The SMC hinge domain eluted as a dimer in gel filtration, indicating that the hinge-mediated dimerization that has been established for eukaryotic cohesin (11) is also true for prokaryotic SMC.
Because the SMC complex is closely associated with DNA in vivo, we investigated the DNA binding properties of the individual components. In contrast to ScpA and ScpB (alone and in combination), SMC showed strong binding to double-stranded DNA, both in gel shift experiments and on DNA-coated chips in surface plasmon resonance experiments. These findings show that SMC not only has an affinity for single-stranded DNA (14) but also can efficiently and directly bind to chromosomal DNA and establish that SMC is the DNA binding component in the SMC/Scp complex in B. subtilis. To identify the DNA binding region on SMC, we tested the SMC head domain and the hinge domain for DNA binding. In contrast to reports suggesting DNA binding properties for the C-terminal domain (which is part of the head domain) in SMC (1), we found that neither the hinge domain nor the complete head domain showed significant DNA binding activity. Therefore, SMC does not bind to DNA through its head domain alone (nor through the hinge domain), but the coiled-coil regions mediate DNA binding, either alone or in concert with the head and/or hinge domains. It is possible that the long extended coils present several low-affinity binding sites for DNA that might wrap around both coils. Alternatively, SMC could bind to DNA by forming a ring-like structure around its substrate (Fig. 8), a notion which is supported by our results. The association of SMC with DNA was unusual, because SMC was loaded onto DNA in a dose-dependent and sequence-independent manner and was released from linear (open) DNA but not from DNA in which both ends were attached to the surface (closed DNA). Thus, SMC binds to DNA in a rather nonspecific manner, possibly by embracing the DNA with the long coiled-coil arms (Fig. 8). This notion is feasible because the SMC arms can open and close (13, 29) and because the head domain can dimerize, as was found for the Rad50 crystal structure containing ATP (18) to close the ring. Due to their binding to the head domain, ScpA and ScpB could stabilize ring closure in vivo.
Our proposed mode of DNA binding is similar to that suggested for cohesin (11). Moreover, ScpA has significant similarity with Scc1 (data not shown) (36) and, like cohesin, the SMC complex appears to consist of only three proteins (unpublished observation), in contrast to the four subunits found in condensin. Thus, the prokaryotic SMC complex could be a cohesin ancestor. However, the SMC complex clearly condenses DNA and does not localize to the sites in B. subtilis where chromosome cohesion, such as that seen in E. coli (40), would occur (at midcell, where both sister chromosomes leave DNA polymerase). Therefore, B. subtilis SMC could condense DNA through cohesin-like DNA binding, by interlocking DNA strands from different DNA loops or by introducing DNA loops (Fig. 8).
Recently, condensin was shown to introduce supercoils into a circular (plasmid) DNA from a single point on the DNA (3). Intriguingly, we have found that the SMC complex can compact whole nucleoids when SMC is overproduced in vivo, while the complex largely remains localized within the specific bipolar condensation centers. This finding does not necessarily mean that the complex binds to a single region on the chromosome but strongly suggests that the SMC complex forms active condensation factories that have an impact on global chromosome compaction and arrangement. Our in vitro data suggest that SMC binds to DNA by closing of the SMC arms through dimerization of the head domain, possibly stabilized by ScpA and ScpB; this process could be a mechanism for active DNA condensation. Future experiments will address the role of SMC ATPase function and the effects of ScpA and ScpB on this activity and on the bridging of the SMC head domains.
Acknowledgments
We thank the Grossman Laboratory (Massachusetts Institute of Technology) for the hyperspac plasmid and David Rudner and members of the Losick Laboratory (Harvard University) for the kind gift of the hyperspank vector. We are grateful to Uwe Linne, Gabi Schimpff-Weihland, and Mohamed Marahiel for performance of mass spectroscopy and for the kind gift of AbrB protein; to G. Muskhelishvili for the gift of Fis protein; and to Martin Neeb and Norbert Hamp for expert help in sucrose gradient centrifugation.
This work was supported by the Deutsche Forschungsgemeinschaft (Emmy Noether Programm) and the Fonds der Chemischen Industrie.
A. Volkov and J. Mascarenhas contributed equally to this work.
Footnotes
This report is dedicated to Richard Losick in celebration of his 60th birthday.
REFERENCES
- 1.Akhmedov, A. T., C. Frei, M. Tsai-Pflugfelder, B. Kemper, S. M. Gasser, and R. Jessberger. 1998. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem. 273:24088-24094. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. E., A. Losada, H. P. Erickson, and T. Hirano. 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156:419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazett-Jones, D. P., K. Kimura, and T. Hirano. 2002. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9:1183-1190. [DOI] [PubMed] [Google Scholar]
- 4.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Blaauwen, T., A. Lindqvist, J. Lowe, and N. Nanninga. 2001. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol. Microbiol. 42:1179-1188. [DOI] [PubMed] [Google Scholar]
- 6.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 7.Gordon, S., D. Sitnikov, C. D. Webb, A. Teleman, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 8.Graumann, P. L. 2000. Bacillus subtilis SMC is required for proper arrangement of the chromosome and for efficient segregation of replication termini but not for bipolar movement of newly duplicated origin regions. J. Bacteriol. 182:6463-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graumann, P. L. 2001. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 83:53-60. [DOI] [PubMed] [Google Scholar]
- 10.Graumann, P. L., R. Losick, and A. V. Strunnikov. 1998. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol. 180:5749-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 12.Harvey, S. H., M. J. Krien, and M. J. O'Connell. 2002. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases Genome Biol. 3:REVIEWS3003. [DOI] [PMC free article] [PubMed]
- 13.Hirano, M., D. E. Anderson, H. P. Erickson, and T. Hirano. 2001. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 20:3238-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano, M., and T. Hirano. 1998. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 17:7139-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, T. 1999. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 13:11-19. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 17.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418:562-566. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner, K. P., A. Karcher, D. S. Shin, L. Craig, L. M. Arthur, J. P. Carney, and J. A. Tainer. 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101:789-800. [DOI] [PubMed] [Google Scholar]
- 19.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaltwasser, M., T. Wiegert, and W. Schumann. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 68:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, K., and T. Hirano. 2000. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc. Natl. Acad. Sci. USA 97:11972-11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, W., and M. A. Marahiel. 2002. Structure-function relationship and regulation of two Bacillus subtilis DNA-binding proteins, HBsu and AbrB. J. Mol. Microbiol. Biotechnol. 4:323-329. [PubMed] [Google Scholar]
- 23.Lemon, K. P., and A. D. Grossman. 2001. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 15:2031-2041. [DOI] [PubMed] [Google Scholar]
- 24.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-110. [DOI] [PubMed] [Google Scholar]
- 26.Lindow, J. C., M. Kuwano, S. Moriya, and A. D. Grossman. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46:997-1009. [DOI] [PubMed] [Google Scholar]
- 27.Lowe, J., S. C. Cordell, and F. van den Ent. 2001. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 306:25-35. [DOI] [PubMed] [Google Scholar]
- 28.Mascarenhas, J., J. Soppa, A. Strunnikov, and P. L. Grauman. 2002. Cell cycle dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 21:3108-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melby, T. E., C. N. Ciampaglio, G. Briscoe, and H. P. Erickson. 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29:179-187. [DOI] [PubMed] [Google Scholar]
- 31.Muskhelishvili, G., and A. Travers. 2003. Transcription factor as a topological homeostat. Front. Biosci. 8:D279-D285. [DOI] [PubMed]
- 32.Niki, H., A. Jaffe, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14:212-223. [PMC free article] [PubMed] [Google Scholar]
- 34.Ohsumi, K., M. Yamazoe, and S. Hiraga. 2001. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40:835-845. [DOI] [PubMed] [Google Scholar]
- 35.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schleiffer, A., S. Kaitna, S. Maurer-Stroh, M. Glotzer, K. Nasmyth, and F. Eisenhaber. 2003. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell 11:571-575. [DOI] [PubMed] [Google Scholar]
- 37.Soppa, J. 2001. Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene 278:253-264. [DOI] [PubMed] [Google Scholar]
- 38.Soppa, J., K. Kobayashi, M. F. Noirot-Gros, D. Oesterhelt, S. D. Ehrlich, E. Dervyn, N. Ogasawara, and S. Moriya. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45:59-71. [DOI] [PubMed] [Google Scholar]
- 39.Strunnikov, A. V., and R. Jessberger. 1999. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur. J. Biochem. 263:6-13. [DOI] [PubMed] [Google Scholar]
- 40.Sunako, Y., T. Onogi, and S. Hiraga. 2001. Sister chromosome cohesion of Escherichia coli. Mol. Microbiol. 42:1233-1242. [DOI] [PubMed] [Google Scholar]
- 41.Teleman, A. A., P. L. Graumann, D. C. H. Lin, A. D. Grossman, and R. Losick. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102-1109. [DOI] [PubMed] [Google Scholar]
- 42.Webb, C. D., P. L. Graumann, J. Kahana, A. A. Teleman, P. Silver, and R. Losick. 1998. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 28:883-892. [DOI] [PubMed] [Google Scholar]
- 43.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C.-H. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 44.Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol. 38:392-400. [DOI] [PubMed] [Google Scholar]
- 45.Yamazoe, M., T. Onogi, Y. Sunako, H. Niki, K. Yamanaka, T. Ichimura, and S. Hiraga. 1999. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18:5873-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimura, S. H., K. Hizume, A. Murakami, T. Sutani, K. Takeyasu, and M. Yanagida. 2002. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 12:508-513. [DOI] [PubMed] [Google Scholar]