Abstract
Amplification, overexpression, or rearrangement of the c-rel gene, encoding the c-Rel NF-κB subunit, has been reported in solid and hematopoietic malignancies. For example, many primary human breast cancer tissue samples express high levels of nuclear c-Rel. While the Rev-T oncogene v-rel causes tumors in birds, the ability of c-Rel to transform in vivo has not been demonstrated. To directly test the role of c-Rel in breast tumorigenesis, mice were generated in which overexpression of mouse c-rel cDNA was driven by the hormone-responsive mouse mammary tumor virus long terminal repeat (MMTV-LTR) promoter, and four founder lines identified. In the first cycle of pregnancy, the expression of transgenic c-rel mRNA was observed, and levels of c-Rel protein were increased in the mammary gland. Importantly, 31.6% of mice developed one or more mammary tumors at an average age of 19.9 months. Mammary tumors were of diverse histology and expressed increased levels of nuclear NF-κB. Analysis of the composition of NF-κB complexes in the tumors revealed aberrant nuclear expression of multiple subunits, including c-Rel, p50, p52, RelA, RelB, and the Bcl-3 protein, as observed previously in human primary breast cancers. Expression of the cancer-related NF-κB target genes cyclin D1, c-myc, and bcl-xl was significantly increased in grossly normal transgenic mammary glands starting the first cycle of pregnancy and increased further in mammary carcinomas compared to mammary glands from wild-type mice or virgin transgenic mice. In transient transfection analysis in untransformed breast epithelial cells, c-Rel-p52 or -p50 heterodimers either potently or modestly induced cyclin D1 promoter activity, respectively. Lastly, stable overexpression of c-Rel resulted in increased cyclin D1 and NF-κB p52 and p50 subunit protein levels. These results indicate for the first time that dysregulated expression of c-Rel, as observed in breast cancers, is capable of contributing to mammary tumorigenesis.
Nuclear factor (NF)-κB/Rel is a structurally and evolutionary conserved family of transcription factors distinguished by the presence of a 300 amino acid region, termed the Rel homology domain (RHD), based on its homology with v-Rel, the transforming protein encoded by the RevT avian retrovirus. The RHD is responsible for DNA-binding, dimerization, nuclear translocation, and binding of Rel factors to the IκB inhibitory proteins (reviewed in reference 21). Mammals express five NF-κB members that belong to two classes. The first class includes c-Rel, RelB, and RelA (p65), which are synthesized as mature products and contain a C-terminal transactivation domain. The second class consists of NF-κB1 and NF-κB2, which are synthesized as longer precursors, p105 and p100. Those proteins require C-terminal proteolytic processing to produce the mature p50 and p52 subunits, respectively, which contain the RHD but lack a transactivation domain. Although, both p50 and p52 have been found to transactivate when in association with Bcl-3 protein (7, 20). In most untransformed cells, other than B lymphocytes, NF-κB complexes are sequestered in the cytoplasm bound to specific inhibitory proteins, of which IκB-α is the paradigm. Activation of NF-κB involves phosphorylation and rapid degradation of IκB, allowing for translocation of free NF-κB to the nucleus, where it controls genes involved in cell growth and survival, adhesion, and immune and inflammatory responses, including cyclin D1, c-myc, and bcl-xl (reviewed in references 47 and 56).
Evidence from several laboratories has suggested NF-κB is critically involved in regulation of tumorigenesis. We and others demonstrated aberrant constitutive activation of NF-κB factors in breast cancer (43, 59). High levels of nuclear NF-κB were found in human breast tumor cell lines, carcinogen-transformed mammary epithelial cells, and the majority of primary human and rodent breast tumor tissue samples. Accelerated degradation of the IκB-α inhibitory protein was observed (34), suggesting aberrant regulation of nuclear translocation in breast cancer cells. Inhibition of the constitutive NF-κB activity in human breast cancer cell lines induced apoptosis (59) or led to reduced tumorigenicity (50). Conversely, ectopic expression of c-Rel resulted in resistance to TGF-β-mediated inhibition of proliferation (58). Interestingly, we observed that 21 out of 25 primary human breast cancer tissues examined expressed high levels of nuclear c-Rel (59); similar observations were made by Cogswell and coworkers (15). The overexpression of c-Rel has been implicated in other hematopoietic and solid malignancies as well. For example, c-rel gene amplification was seen in ∼20% of non-Hodgkin's B-cell lymphomas, including diffuse large-B-cell lymphomas (DBCL) (reviewed in reference 47). In addition, the c-rel gene was also found rearranged or overexpressed in some follicular lymphomas and DBCL. The higher level of expression of c-Rel, plus RelA, in the activated B-cell (ABC) form of DBCL, was found associated with poorer prognosis (2). Furthermore, inhibition of c-Rel induced apoptosis in immature B-cell lymphomas (66).
The c-rel gene encodes a 68-kDa protein which is active mostly in lymphocytes and monocytic, granulocytic, and erythroid cells. Mice lacking c-rel are viable but show severely impaired lymphocyte proliferation and immune function, with impaired interleukin-2 expression (36). The X-ray crystal structure of the c-Rel homodimer bound to a DNA target site was resolved recently (28). It confirmed that c-Rel homodimers recognize a different set of κB element DNA sequences compared to c-Rel heterodimers or p50-containing dimers, suggesting that those complexes may have a different range of target genes. The v-rel gene, carried by the highly oncogenic avian reticuloendotheliosis virus strain T (Rev-T), is able to cause tumors in birds and transgenic animal models. The v-Rel oncogenic protein differs from its avian progenitor c-Rel by the presence of multiple mutations that increases its expression, nuclear localization, DNA binding, transactivation capability and neoplastic transformation potential (reviewed in reference 22). The direct role of c-Rel in tumorigenesis is still a matter of debate. Overexpression of either avian or human c-Rel was shown to transform primary chicken lymphoid cells (1, 23), although with lower efficiency than v-Rel. Retroviral delivery of avian c-rel in young chickens gave rise to lymphoid tumors, and cell transformation involved selection of a C-terminal deleted c-Rel protein with increased oncogenic activity (27). All these observations led us to test the oncogenic properties of c-Rel in vivo, in the mammary gland. Here, we have constructed a mouse model in which the mouse c-rel cDNA was driven by the hormone-dependent mouse mammary tumor virus (MMTV) promoter. MMTV-c-rel female mice developed late-onset mammary tumors, which were related to increased levels of expression of the NF-κB growth and survival target genes cyclin D1, c-myc, and bcl-xl. These results provide the first in vivo evidence for a causal role of c-Rel activation in the pathogenesis of breast cancer.
MATERIALS AND METHODS
Isolation of founder MMTV-c-rel transgenic mice.
The 2.5-kb EcoRI/HindIII fragment from the pSVSport-c-Rel vector, containing the full-length murine c-rel cDNA (kindly provided by T. Gilmore, Boston University, Boston, Mass.), was blunt end ligated into the MMTV-LTR plasmid, containing the MMTV long terminal repeat (MMTV-LTR), which directs expression chiefly to the mammary epithelium, with ras 5′ untranslated sequences provided upstream of the cDNA and a simian virus 40 (SV40) intron and polyadenylation signal downstream (38, 55), yielding the pMMTV-c-rel plasmid. The direction of the insert was confirmed by restriction mapping and DNA sequencing at the Molecular Biology Core at Boston University Medical School. Plasmid sequences were removed by restriction digestion at the SalI and SpeI sites, leaving the MMTV-LTR sequence, ras 5′ untranslated sequences, c-rel cDNA, and the SV40 intron and poly(A) addition signal sequence. The excised transgene construct was gel purified and microinjected into pronuclei of fertilized one-cell zygotes from FVB/N mice. These zygotes were reimplanted into pseudopregnant foster mothers, and the offspring were screened for presence of the transgene by Southern blotting (see below). Carriers were bred to establish four independent transgenic lines. Female transgenic mice were continuously bred to induce transgene expression though activation of the hormone-dependent MMTV-LTR promoter. Mice were monitored biweekly for the appearance of tumors. Mice were housed in a two-way barrier at the Boston University School of Medicine Transgenic mouse facility in accordance with the regulations of the American Association for the Accreditation of Laboratory Animal Care.
DNA analysis.
Genomic tail DNA was isolated as described previously (46), samples (10 μg) were digested with PstI, and the resulting fragments were separated on a 0.8% agarose gel and transferred to GeneScreenPlus (DuPont NEN) nylon membranes. DNA was cross-linked to the membrane by UV irradiation (Stratalinker; Stratagene, La Jolla, Calif.) at 0.12 J/cm2 for 30 s. Hybridization was performed using the 2.2-kb DNA fragment, encompassing the MMTV-LTR promoter and an ∼1-kb fragment of the mouse c-rel cDNA, released from the pMMTV-c-rel plasmid by digestion with PstI as a probe. The DNA was radiolabeled by random priming and used as described previously (35), except that 2.0 × 106 cpm/ml 32P-labeled DNA was employed.
RNA analyses.
Frozen breast tissue was pulverized in liquid nitrogen with a mortar and pestle, and total RNA extracted with the Ultraspec-II RNA Isolation System (Biotecx Laboratories Inc., Houston, Tex.). To remove contaminating DNA, RNA samples were digested for 30 min at 37°C with RQ1 RNase free DNase (Promega Corporation, Madison, Wis.), according to the manufacturer's directions. RNA was reextracted and ethanol precipitated. For reverse transcriptase PCR (RT-PCR), 5 μg RNA samples were reverse transcribed with SUPERSCRIPT RNase H-RT in the presence of 200 ng of random primers (all reagents from Invitrogen Life Technologies, Carlsbad, Calif.). For PCR, a 236-bp fragment of the transgene was amplified with a sense oligonucleotide primer from the mouse c-rel cDNA coding sequence (5′-GTGACCCCTAAGGGTTTTCTG-3′) and an antisense oligonucleotide from the SV40 poly(A) tail of the vector construct (5′-CCCATTCATAAGTTCCATAG-3′). PCR were performed in a thermal cycler (MJ Research, Watertown, Mass.) by denaturing at 95°C for 45 s, annealing at 46°C for 90 s, and extending at 72°C for 90 s for 35 cycles. Primer pairs specific for the mouse cyclin D1 gene were 5′-CACAACGCACTTTCTTTCCA-3′ and 5′-GACCAGCCTCTTCCTCCAC-3′, and amplified a 164-bp fragment with an annealing temperature of 55°C. The primer pairs specific for mouse bcl-xl were as described previously (52). As a control for RNA quality, a 750-bp fragment of β-actin mRNA was amplified by 25 or 30 PCR cycles with the following primers: 5′-ACCAGTTCGCCATGGATGACGATA-3′ and 5′-AGCTCATAGCTCTTCTCCAGGGAG-3′, which were used with an annealing temperature of 55°C. For radiolabeled PCR, 0.85 μCi of [α-32P]dCTP and [α-32P]dGTP were added in the PCR. For RNase protection assays (RPAs), multiprobe RPA kits (Pharmingen, San Diego, Calif.) were used according to the manufacturer's directions. The identity of the RNase protected bands in the gel were established using the undigested probes as markers and a control RNA for mouse cyclin or apoptosis gene mRNA expression (Pharmingen). For Northern blot analysis, RNA samples (5 to 15 μg) were denatured and separated by electrophoresis in 1.0% formaldehyde agarose gels (16). Probes included a 2.4-kb pM-c-myc54 mouse c-myc cDNA (60), the full-length human cyclin D1 cDNA subcloned into the pBPST-R1 vector, kindly provided by R. G. Pestell (Albert Einstein College of Medicine, New York, N.Y.), and a 750-bp fragment of the mouse bcl-x cDNA amplified by PCR using the previously described primers (52). Quantitation by scanning densitometry was performed with a KDS1D device (version 2.0; Kodak, New Haven, Conn.).
EMSA.
Frozen tissue powders were resuspended in homogenization buffer (1 g/ml) in 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 50 mM sucrose, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), leupeptin (5 μg/ml), and aprotinin (5 μg/ml) (Sigma Chemical Co., St. Louis, Mo.). Samples were Dounce homogenized for 20 strokes with a loose pestle and then 20 strokes with a tight pestle. The KCl concentration was then adjusted to 100 mM and the nuclei were washed twice with the homogenization buffer with 100 mM KCl. Nuclear proteins were extracted on ice for 30 min in 2 packed nuclear volumes containing 10 mM HEPES (pH 7.9), 400 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF, leupeptin (0.5 μg/ml), and aprotinin (5 μg/ml). Protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). The sequence of the URE NF-κB-containing oligonucleotide from the c-myc gene is as follows: 5′-GATCCAAGTCCGGGTTTTCCCCAACC-3′ (17). The core element is underlined. The Octomer-1 (Oct-1) oligonucleotide has the following sequence: 5′-TGTCGAATGCAAATCACTAGAA-3′. Nuclear extracts samples (5 μg) were subjected to electrophoretic mobility shift analysis (EMSA) as described previously (59). For antibody supershift analysis, the binding reaction was performed in the absence of the probe, the appropriate antibody was added and the mixture incubated for 16 h at 4°C. The probe was then added and the reaction incubated an additional 30 min at 25°C and the complexes resolved by gel electrophoresis, as above. Antibodies used included anti-RelA (C-20), sc-372; anti-c-Rel (N), sc-70; and anti-p50 (NLS), sc-114 (all from Santa Cruz Biotechnology, Santa Cruz, Calif.). In addition, rabbit polysera 1266, 1050, and 1051 specific for c-Rel were kindly provided by N. Rice and M. Ernst (National Cancer Institute, Frederick Md.). Data were quantified by densitometry using a Molecular Dynamics densitometer.
Cell lines and transfection conditions.
NMuMG, which is an untransformed, immortalized mouse mammary epithelial cell line, was cultured as described previously (57). MCF-10F is a human mammary epithelial cell line established from a patient with fibrocystic disease, which does not display malignant characteristics (9). The RelA and p50 expression vectors have been described elsewhere (39). The p52 and Bcl-3 expression vectors were kindly provided by U. Siebenlist (National Institutes of Health, Bethesda, Md.). The cyclin D1 promoter constructs CD1 −66 WT-Luc and CD1 −66 Mut-Luc, with wild-type (WT) and mutant NF-κB elements, respectively, were a kind gift of R. G. Pestell (25). For luciferase assays, NMuMG cells were transfected in six-well plates using the Fugene reagent (Roche Diagnostics Corporation, Indianapolis, Ind.), and the SV40 promoter-β-galactosidase (pSV40-β-gal) reporter vector was used to normalize transfection efficiency, as previously described (3). For stable transfectants, MCF-10F cells were transfected in P100 dishes with 10 μg of pBluescript or pSVSport-c-Rel vector along with 1 μg of pGKpuro selection plasmid, selected with puromycin (4 μg/ml; Sigma) for 4 days, and then grown in the presence of puromycin (1 μg/ml).
Immunoblot analysis.
Whole-cell extracts (WCEs) were prepared in RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.1% SDS, 1% sodium sarcosyl, 0.2 mM PMSF, leupeptin [10 μg/ml], 1 mM DTT). Nuclear extracts were prepared as described above. Samples (40 μg) were separated by electrophoresis in 8% polyacrylamide-SDS gels, transferred to a 0.45-μm-pore-size polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) and subjected to immunoblotting, as described (59). Antibodies included anti-RelB (C-19) sc-226, anti-p52 (K-27) sc-298; Bcl-3 (C-14) sc-185, and Sp1 (PEP 2) sc-59 (all from Santa Cruz Biotechnology) and anti-cyclin D1 monoclonal Ab-3 (Oncogene, Boston, Mass.). Antibodies specific for other NF-κB subunits were as described above.
Histology.
Upon necropsy, tumors and other mammary glands, heart, lung, liver, kidney, spleen, and the adrenal gland were removed and immediately fixed in Optimal Fix (American Histology Reagent Co., Lodi, Calif.) and shipped in alcohol. The tissues were processed, embedded in paraffin, and sectioned at a thickness of 7 μm. The sections were mounted on glass slides and stained with hematoxylin and eosin using routine laboratory procedures in the Transgenic Core Pathology Laboratory at the University of California, Davis. Sections were compared with other specimens in the extensive mouse mammary tumor database (http://www-mp.ucdavis.edu/tgmice/firststop.html).
RESULTS
Generation and characterization of MMTV-c-rel transgenic mice.
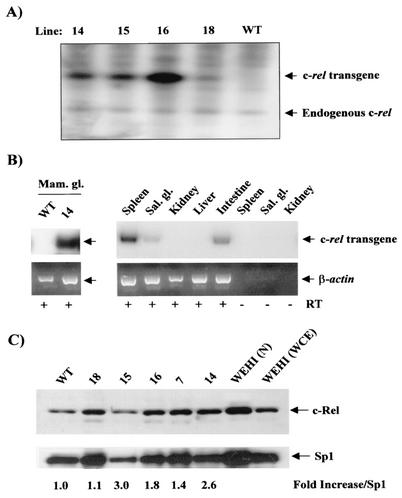
To determine the role of c-Rel in mammary tumorigenesis, we generated a mouse model where c-rel cDNA was expressed under the control of the MMTV-LTR promoter, an effective vector for expressing oncogenes and for transforming the mammary epithelium (11, 61). A construct containing the full-length mouse c-rel cDNA was inserted in a plasmid containing the MMTV-LTR promoter and an SV40 T antigen intron-poly(A) cassette to ensure efficient expression in vivo. The insert c-rel DNA was then utilized to generate transgenic mice in the FVB/N mouse strain. Integration of the construct into the genome of potential founders was assessed by Southern blot analysis of tail DNA with c-rel cDNA as a probe (Fig. 1A and data not shown). Five founders successfully passed the transgene through the germ line. Founder line 18 had approximately two to three copies of the transgene, while lines 7, 14, and 15 had four to five copies and line 16 had approximately nine copies, as estimated by comparison with bands resulting from hybridization with the endogenous gene on the Southern blots. MMTV-c-rel transgenic mice of all founders developed and bred normally. Transgenic females were able to nurse their pups.
FIG. 1.
MMTV-LTR-driven c-rel transgene expression in FVB/N mice. (A) Identification of founder lines. Genomic tail DNA was prepared from the indicated potential founders MMTV-c-rel transgenic mice, and samples (10 μg) digested with PstI and subjected to Southern blot analysis for c-rel using the 2.2-kb fragment, encompassing the MMTV-LTR promoter and an ∼1-kb fragment of mouse c-rel cDNA, released from the MMTV-c-rel plasmid digested with PstI, as a probe. The positions of the bands derived from the c-rel transgene and the endogenous c-rel gene are as indicated. (B) Transgenic c-rel expression. Total RNA was isolated from the indicated organs of WT FVB/N or line14 MMTV-c-rel mice at day 18.5 of the first pregnancy, and subjected to DNase treatment. Samples (5 μg) were subjected to RT-PCR analysis, in the presence (+) or absence (−) of RT to control for DNA contamination, using c-rel transgene-specific oligonucleotides, amplifying a 236-bp fragment. Similar analysis of β-actin RNA levels confirmed the integrity of the reverse transcription reaction. (C) Total c-Rel expression. Mammary glands were removed from WT FVB/N or the indicated transgenic line mice at day 18.5 of the first pregnancy. Nuclear extract were prepared, and samples (20 μg) subjected to immunoblot analysis of c-Rel, and Sp1, as control for loading. As additional controls, nuclear extracts and WCEs from the WEHI 231 immature B-lymphoma cells, which express high constitutive levels of c-Rel (40), were similarly analyzed. The values of c-Rel normalized to Sp1 level relative to the WT sample are displayed below.
To characterize the expression pattern of the c-rel transgene, line 14 MMTV-c-rel mice or WT FVB/N mice, as control, were bred to activate the MMTV-LTR promoter, which contains hormonally responsive elements activated by progestins and corticosteroids. At day 18.5 of the first pregnancy, total RNA was isolated from the mammary glands and various other organs. RNA samples were subjected to a radiolabeled transgene-specific RT-PCR assay, performed with a 5′ mouse c-rel cDNA sense primer and a 3′ SV40 poly(A) antisense primer (Fig. 1B). Expression of c-rel transgene mRNA was observed in the mammary gland of line 14, but not the WT mouse, as expected (Fig. 1B, left panel). RNA quality and equal loading was confirmed by analysis of β-actin mRNA expression profiles by RT-PCR (Fig. 1B, bottom panel), as well as in ethidium bromide-stained gels (data not shown). At day 18.5 of pregnancy, expression of transgene c-rel mRNA was also observed in the spleen, salivary gland, and intestine of mice of line 14 (Fig. 1B) and line 16 (data not shown), while undetectable or low levels were observed in the kidney and liver (Fig. 1B) and heart and lung (data not shown). Where indicated, reactions were performed in the absence of RT, which confirmed the absence of DNA contamination. Thus, the c-rel transgene mRNA is expressed mostly in glandular organs and lymphoid tissues, which is consistent with previous studies with the MMTV-LTR promoter (38, 55).
We next sought to determine total levels of c-Rel protein expression, which includes both endogenous and transgenic c-Rel. At day 18.5 of the first pregnancy, nuclear extracts were prepared from mammary glands of transgenic lines 7, 14, 15, 16, and 18 mice, and from a WT FVB/N mouse as control. Samples were subjected to immunoblot analysis for c-Rel and Sp1 to normalize for loading (Fig. 1C). Nuclei from the WT mammary gland contained basal levels of c-Rel, as has been reported recently (10). All of the transgenic lines displayed higher normalized levels of nuclear c-Rel. The lowest level was seen in line 18, consistent with its low transgene copy number, as seen above; therefore, the other four lines (i.e., lines 7, 14, 15, and 16) were chosen for further study.
MMTV-c-rel transgenic mice develop late-onset mammary carcinomas.
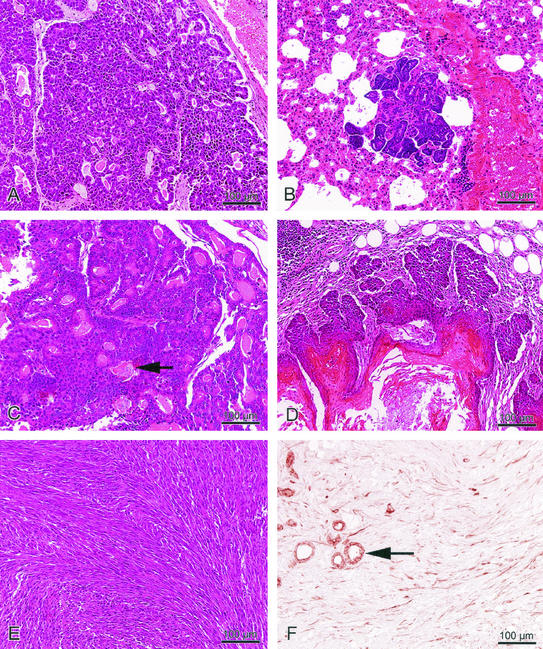
To promote c-rel transgene overexpression, MMTV-c-rel female mice were continuously bred to induce the MMTV-LTR promoter. A cohort of 38 multiparous female mice from the 4 expressing lines was monitored for tumor incidence over 2 years. Mice were subjected to biweekly palpable examination, and when the presence of a tumor was detected, the mammary glands and other organs were subjected to histopathological analysis. Thirty-one percent of the mice developed mammary carcinomas at an average age of 19.9 months (Table 1). In contrast, mammary tumors develop with a very low incidence (<1%) in WT female FVB/N mice that have been similarly bred (38). An identification number, with the origin of the line was attributed to each tumor, and characteristics of the different breast tumors are described in the Table 1. Mice from each of the four transgenic lines developed mammary carcinomas. The tumor incidence was 33.3, 41.7, 20, and 33.3% in MMTV-c-rel transgenic lines 7, 14, 15, and 16, respectively (data not shown), suggesting that tumor development is related to c-rel transgene expression rather than random insertional events. In all but one case, the tumors arose as solitary masses in a single mammary gland. Of the 12 mammary tumors, 3 were pure adenocarcinomas (Fig. 2A), 3 were adenosquamous carcinomas (Fig. 2C), 4 were squamous cell carcinomas (Fig. 2D), 1 was classified as a papillary adenocarcinoma, and 1 was a spindle cell carcinoma (Fig. 2E). Spindle cell carcinomas are often related to an epithelial to mesenchymal cell transition (EMT), which is the transformation of epithelial cells into cells with features of mesenchymal cells, favoring the progression of a carcinoma towards a dedifferentiated and more malignant state (reviewed in reference 63). To test for cells of epithelial origin, immunohistological staining was carried out using antibodies specific for the epithelial cell marker cytokeratin 8 (Fig. 2F). The spindle cell carcinoma stained positively for cytokeratin 8, consistent with an EMT tumor. One of the adenocarcinomas was metastatic to the lung (Fig. 2B).
TABLE 1.
Tumor incidence and histopathology in female MMTV-c-rel transgenic mice after multiple cycles of pregnancy and regressiona
| Mam. tumor diagnosis (other tumors)b | Agec (mo) | No. of:
|
Line | Mouse no.d | |
|---|---|---|---|---|---|
| Mam. tumors | Other tumors | ||||
| Mam. squamous cell carcinoma and hyperplasia (bronchial adenocarcinoma) | 22 | 1 | 1 | 7 | 4026 |
| Mam. adenosquamous carcinoma | 22 | 1 | 0 | 7 | 4042 |
| Mam. adenosquamous carcinoma | 18 | 1 | 0 | 14 | 3814 |
| Mam. adenosquamous carcinoma (papillary bronchial adenomas) | 20 | 1 | 1 | 14 | 3983 |
| Mam. adenocarcinomas with pulmonary metastases | 23 | 4 | 1 | 14 | 3996 |
| Mam. papillary carcinoma, lobular hyperplasia, squamous nodules | 23 | 1 | 0 | 14 | 4936 |
| Mam. squamous cell carcinoma and hyperplasia | 15 | 1 | 0 | 14 | 4556 |
| Mam. adenocarcinoma (papillary bronchial adenocarcinomas) | 19 | 1 | 1 | 15 | 127 |
| Mam. squamous cell carcinoma | 22 | 1 | 0 | 16 | 3872 |
| Mam. adenocarcinoma with poorly differentiated large cells | 18 | 1 | 0 | 16 | 3875 |
| Mam. squamous cell carcinoma and hyperplasia (centrocytic lymphoma) | 18 | 1 | 1 | 16 | 4521 |
| Spindle cell tumor, originating from a mam. adenocarcinoma | 19 | 1 | 0 | 16 | 4528 |
Thirty-eight multiparous female mice from the four transgenic lines (7, 14, 15, and 16) were monitored for tumor incidence by biweekly palpable examination.
Histopathological analysis of mammary (mam.) glands and other organs (heart, lung, liver, kidney, spleen, and adrenal gland) from the same animal was performed when a tumor was detected.
Age when tumor was detected.
An identification number was given to the individual mice of the cohort for further analysis of the tumors.
FIG. 2.
Representative histopathologies of mammary tumors that developed in MMTV-c-rel transgenic mice after multiple cycles of pregnancy and regression. (A) Adenocarcinoma; (B) pulmonary metastasis in a mouse with mammary adenocarcinomas; (C) adenosquamous carcinoma showing areas with extracellular squamous differentiation (arrow); (D) squamous cell carcinoma; (E) spindle cell carcinoma; (F) immunohistochemistry for cytokeratin 8 expression in the spindle cell carcinoma shown in 2E. Note the staining of many of the spindle cells and staining of luminal epithelium in the glands (arrow).
Lastly, mammary glands of three other multiparous 2-year-old transgenic mice, which were not included in the cohort, were subjected to histopathological analysis even without the presence of a palpable tumor. This examination revealed the presence of an adenosquamous carcinoma and a mammary squamous cell carcinoma in two of the mice (data not shown). In addition to the tumors, poor regression of the alveolar tree of the mammary gland after pregnancy was another histological abnormality that was frequently seen (data not shown). Therefore, the histology of mammary tumors in MMTV-c-rel mice appears variable, suggesting changes in mammary epithelial cells during c-Rel-induced tumorigenesis.
c-Rel expression is elevated in mammary glands and tumors of MMTV-c-rel transgenic mice.
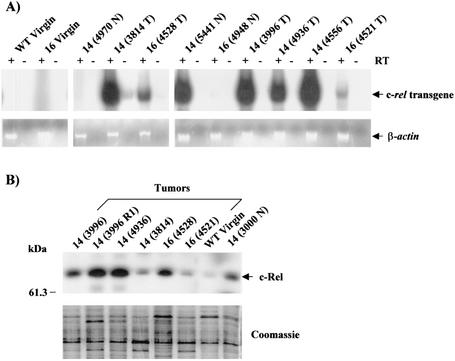
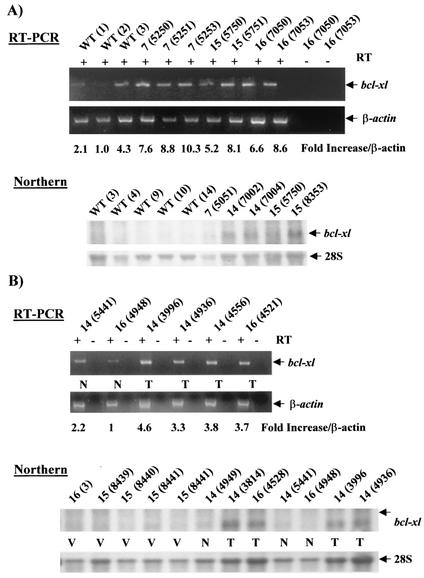
Previous studies showed that the MMTV-LTR promoter is still active in regressing mammary glands, leading to sustained transgene expression over the animal's lifetime (38, 55). We therefore investigated transgenic c-rel expression in mammary glands and tumors from multiparous transgenic mice. RNA was isolated from mammary tumors of mice from transgenic lines 14 and 16 and from grossly normal mammary glands from age-matched transgenic mice (16 to 24 months) that had bred at least three times. In addition, RNA was isolated from mammary glands of a nulliparous (virgin) WT FVB/N mouse and a virgin transgenic line 16 female as negative controls. Samples were subjected to a radiolabeled transgene-specific RT-PCR assay (Fig. 3A). As a control for DNA contamination, reactions were performed in the presence or absence of RT. RNA quality and essentially equal loading was confirmed by analysis of β-actin mRNA expression profiles by RT-PCR (Fig. 3A, bottom panel), and by ethidium bromide staining of gels (data not shown). All tumors and most normal mammary glands of multiparous MMTV-c-rel transgenic mice exhibited expression of transgene mRNA, although at variable levels (Fig. 3A, top panel). In tumors, the lowest expression level of transgene mRNA was observed in the mammary tumor developed in mouse 4521 (line 16), whose histologic analysis showed a complex cell pattern, consisting of mammary squamous cell carcinoma and hyperplasic epithelial cells (Table 1). In contrast, virgin WT and transgenic mammary glands did not express detectable levels of transgene mRNA, as expected. Thus, ectopic c-rel transgene expression is induced in the mammary gland by pregnancy, and is expressed in the tumors and grossly normal mammary glands of multiparous MMTV-c-rel mice.
FIG. 3.
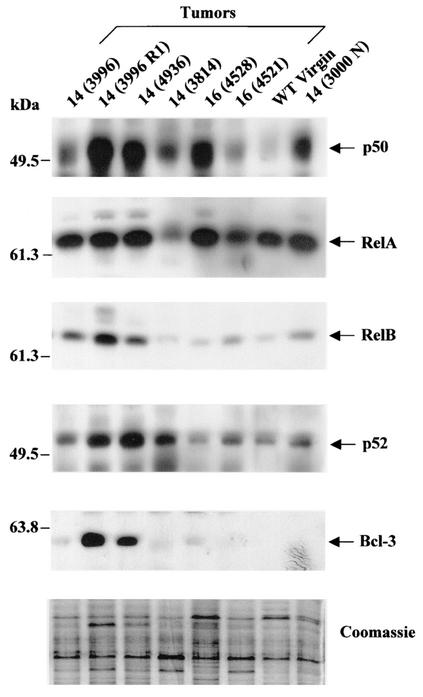
Expression of c-Rel in transgenic female MMTV-c-rel breast tumors. Mammary glands were removed from virgin WT FVB/N (WT Virgin) or transgenic line 16 (16 Virgin) mice and from tumor (T) and grossly normal (N) tissues of multiparous line 14 and 16 transgenic mice. The identification number given to the individual mice is indicated in parentheses. Characteristics of the tumor samples are given in the Table 1. (A) Transgenic c-rel expression. RNA was prepared from mammary gland and tumor tissues, and samples subjected to RT-PCR with primers specific for transgenic c-rel, as in the legend to Fig. 1. (B) Total protein c-Rel expression. Nuclear extracts were prepared, and samples (40 μg) subjected to immunoblot analysis of c-Rel levels. Coomassie blue staining of SDS-PAGE gels was used as control for equal loading (bottom panel). While the overall levels of staining were essentially equivalent, the analysis revealed that the patterns of protein expression are different between tumors, WT and normal transgenic mammary gland samples. This variability likely results from differences in cell type composition in these tissues.
We next assessed total c-Rel protein expression in mammary glands and tumors of transgenic mice by immunoblot analysis. Nuclear extracts were prepared from the indicated tumors (Fig. 3B). As a control, nuclear extracts were isolated from mammary glands of a WT virgin FVB/N mouse and from nonmalignant mammary glands of an age-related line 14 mouse (3000 N) that had undergone three cycles of pregnancy and regression. The mammary gland of the line 14 mouse (3000 N) and all tumors displayed elevated levels of c-Rel expression compared to the WT mammary gland, which showed only a low basal expression (Fig. 3B and data not shown). This is consistent with the pattern of transgenic c-rel expression obtained above. In addition, about half of the tumors showed higher expression levels of c-Rel than grossly normal transgenic mammary glands (Fig. 3B and data not shown). When the results were scanned, a 3- to 150-fold (average, [68.0 ± 60.1]-fold) increase in nuclear c-Rel expression was observed in tumor and normal transgenic mammary glands compared to the WT sample. Equal loading was confirmed by Coomassie blue staining of gels (Fig. 3B, bottom panel), although the patterns of total protein expression appeared different between tumor samples and WT or normal transgenic mammary gland samples. This variability likely results from differences in cell type composition between the various mammary tumors and the normal tissue. Overall, these results indicated that c-rel transgenic mRNA and total c-Rel protein expression are upregulated in MMTV-c-rel mammary glands and tumors compared to WT mice.
Expression of NF-κB family members in mammary glands and carcinomas of MMTV-c-rel transgenic mice.
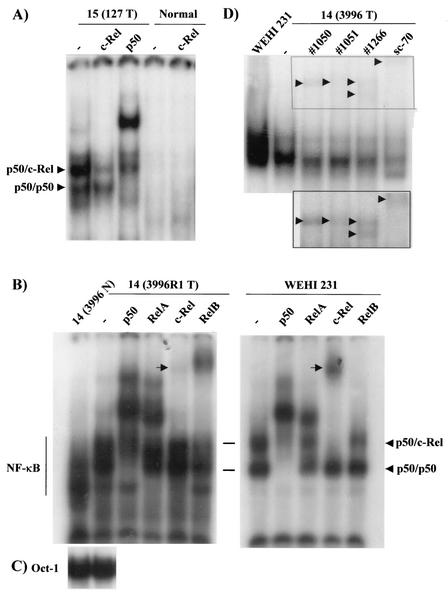
To test for constitutive nuclear c-Rel binding activity, nuclear proteins were isolated from a mammary tumor (adenocarcinoma) and grossly normal mammary glands of line 15 mouse 127 and were subjected to EMSA analysis with an oligonucleotide probe containing the NF-κB element upstream of the c-myc promoter, which binds all Rel family members (39) (Fig. 4A). The normal mammary gland displayed a low level of NF-κB binding (better seen on a darker exposure), consistent with our previous findings with nuclear extracts from normal rat mammary glands and histologically normal mammary tissue from MMTV-Her-2/neu transgenic mice (34, 45, 59). The extracts from the c-Rel-induced tumor displayed two major complexes of NF-κB binding. To identify the nature of the subunit components, we used antibodies against either c-Rel or p50 in EMSA. Addition of an antibody that preferentially recognizes p50 in a homodimer complex, shifted the bottom band completely, and reduced the more slowly migrating complex. Addition of an antibody against c-Rel selectively reduced the top band. These results suggest that the upper complex consisted of p50/c-Rel heterodimers, while the bottom complex was a homodimer of p50.
FIG. 4.
MMTV-c-rel tumors display elevated NF-κB binding. (A) Nuclear extracts were prepared from MMTV-c-rel line 15 (127) mouse mammary tumor and grossly normal mammary glands, and samples (5 μg) subjected to EMSA for NF-κB binding. To identify subunit composition, the indicated samples were incubated overnight at 4°C in the absence (−) or the presence of a supershifting antibody specific for p50, or a blocking antibody specific for c-Rel. The positions of the identified p50/c-Rel and p50 homodimer complexes are as indicated. (B and C) Nuclear extracts were prepared from line 14 3996 mammary tumor (3996R1 T) and grossly normal mammary glands (N), and samples (5 μg) were subjected to EMSA for NF-κB (B) and, as a loading control, Oct-1 (C). For supershift analysis, samples were incubated overnight at 4°C in the absence (−) or the presence of supershifting antibodies specific for p50, RelA, c-Rel (sc-70), and RelB. The arrow shows the position of the c-Rel supershifted complex. Where indicated, nuclear extracts (5 μg) of WEHI 231 B cells, which express high levels of c-Rel/p50 complexes (40), were analyzed as a positive control in the same experiment, however a lighter exposure is shown for the WEHI 231 samples. (D) A nuclear extract was prepared from line 14 3996 mammary tumor (3996 T), and samples (5 μg) were subjected to EMSA for NF-κB, with a WEHI 231 nuclear extract, in the absence or presence of the indicated c-Rel-specific antibodies, as above. Inset represents a darker exposure of the region of the supershifted bands indicated in the figure.
We next performed EMSA on mammary tumor (adenocarcinoma) samples from a line 14 (3996) mouse. This mouse had developed four mammary adenocarcinomas, as well as pulmonary metastases (Table 1). Nuclear extracts isolated from one of the mammary tumors (3996R1) displayed multiple NF-κB complexes (Fig. 4B). The level of NF-κB binding was more intense than seen with the nuclear extract from a nonmalignant mammary gland from the same animal (line14 3996 N), while levels of control Oct-1 binding were similar in these samples (Fig. 4C). Of note, an NF-κB complex of higher mobility was present in the normal sample. We previously observed the presence of such a complex in nuclear extracts isolated from normal human and mouse breast tissue, and supershift analysis showed that these complexes contained predominantly p50 homodimers (reference 50 and data not shown). Addition of a c-Rel-specific antibody resulted in diminished binding and the formation of one slowly migrating supershifted complex (Fig. 4B, indicated by an arrowhead). Addition of an antibody against c-Rel reduced the binding and yielded a similar supershifted complex with control extracts from WEHI 231 immature B-lymphoma cells, which express high levels of activated c-Rel and p50, and lower levels of RelA (40, 48), indicating that the line14 3996R1 tumor sample contains p50/c-Rel complexes. Addition of supershifting antibodies against p50, RelA, or RelB to the line 14 3996R1 extracts resulted in shifted complexes. In particular, addition of a p50 antibody greatly reduced binding and yielded two major and a few minor supershifted bands. Interestingly, addition of a RelA antibody yielded three supershifted RelA-containing complexes, whereas,the expected one RelA-containing complex (e.g., p50/RelA) was seen with the WEHI 231 extract (40). These results suggest that RelA is present in multiple complexes in the mammary gland tumor sample. Lastly, a RelB antibody also reduced binding with the line 14 tumor extract, yielding one supershifted complex (Fig. 4B). Addition of the RelB antibody had no detectable effect on the WEHI 231 cells, consistent with the lack of nuclear RelB in these cells (48). To further characterize the c-Rel binding, we compared the effects of the anti-c-Rel sc-70 from Santa Cruz with three different c-Rel antibody rabbit polysera obtained from N. Rice and M. Ernst (sera 1050, 1051, and 1266), using nuclear extracts from a second tumor from line 14 3996. All of the antibodies reduced the upper complex which comigrated with the WEHI 231 c-Rel/p50 band to approximately the same extent (Fig. 4D). The positions of the supershifted complexes varied for all of the antibodies (Fig. 4D, inset). These finding confirm the presence of c-Rel in NF-κB DNA binding complexes, although, it was not the primary component in the nuclear extracts. Together, these results indicate that multiple NF-κB subunits are binding in the tumor samples, including c-Rel, p50, RelA, and RelB.
Mammary tumors contain multiple NF-κB subunits.
The finding that nuclear extracts of mammary tumors from the MMTV-c-rel mice contain multiple NF-κB complexes led us to more fully assess the nature of the NF-κB subunit expression in mammary glands and tumors. Immunoblot analysis was performed for the p50, RelA, RelB, and p52 NF-κB subunits, and for Bcl-3 protein in nuclear extracts from mammary tumors developed in MMTV-c-rel mice, and from uninvolved mammary glands from a multiparous age-related line 14 transgenic mouse (3000 N), and from a WT virgin FVB/N mouse (Fig. 5). Interestingly, expression of the subunits encoded by genes that are regulated by NF-κB, e.g., p50, p52, and RelB (reviewed in reference 44) appeared substantially increased in many of the samples from the transgenic mice. A dramatic increase in expression of p50 was seen in essentially all of the tumor specimens, as well as grossly normal transgenic mammary gland samples compared to the WT sample. Densitometry indicated a 2.1- to 618-fold induction (average, [330 ± 325]-fold) induction in p50 nuclear levels. Expression of RelB displayed a large increase in three of the specimens compared to the WT mammary gland sample, and a moderate increase in the other 4 samples. In contrast, RelA appeared only moderately increased in most transgenic mammary glands compared to WT mammary glands. Densitometry showed a 1.0- to 1.6-fold increase (average, [1.3 ± 0.4]-fold) in RelA nuclear expression. Interestingly, this contrasted with the relatively high level of RelA binding previously seen in the EMSA of line 14 3996R1 (Fig. 3B), and of other 2 tumor samples that were similarly analyzed (data not shown). Bcl-3 expression was detectable only in tumor samples, and two of them also displayed extremely high levels of p50 and p52 expression. No detectable IκB-α was seen in the nuclear extracts, and no differences were observed in IκB-β levels (data not shown). Overall, these findings indicate that in addition to the c-Rel subunit itself, the MMTV-c-rel mammary gland tumors display a wide range of constitutively active nuclear NF-κB subunits, including p50, p52, and RelB, and to a lesser extent RelA, as well as Bcl-3. Similar complex patterns were seen previously in primary human breast cancer specimens (15, 59).
FIG. 5.
MMTV-c-rel tumors express multiple NF-κB subunits and the Bcl-3 protein. Nuclear extracts were prepared from the indicated mammary tumors, grossly normal mammary glands (N) of multiparous line 14 MMTV-c-rel mice, and from mammary glands of a WT nulliparous FVB/N mouse (WT Virgin). Samples (40 μg), subjected to immunoblot analysis as in Fig. 3B, were reprobed for expression of the p50, RelA, RelB, and p52 NF-κB subunits and the Bcl-3 protein. The positions of molecular mass markers are indicated.
MMTV-c-rel mammary glands and carcinomas display elevated expression of downstream target gene cyclin D1.
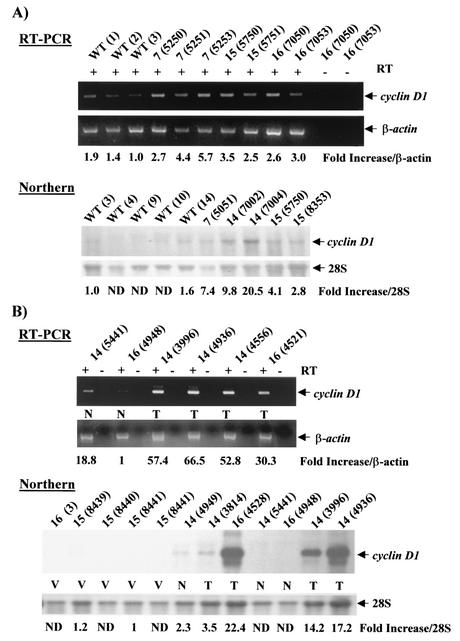
The NF-κB target gene cyclin D1 (25, 26) has been implicated in breast cancer (reviewed in reference 32). Its expression, which is upregulated in ∼50% of human breast tumors, is required for proliferation of breast cancer cells in culture, and MMTV-cyclin D1 mice develop mammary adenocarcinomas (64). Therefore, we sought to test whether cyclin D1 mRNA levels were increased during pregnancy in the mammary glands of MMTV-c-rel versus WT mice. Lines 7, 14, 15, and 16 MMTV-c-rel mice and age-matched WT FVB/N mice (3 to 6 months old) were bred to activate the MMTV-LTR promoter in transgenic mammary glands. At day 18.5 of the first pregnancy, total RNA was isolated from the mammary glands and samples subjected to analysis of cyclin D1 mRNA levels using either a semiquantitative RT-PCR assay specific for cyclin D1 in the presence or absence of RT, as control, or Northern blot analysis (Fig. 6A). For the RT-PCR assay, RNA quality and loading were normalized by evaluation of β-actin levels using 25 cycles of PCR, which is within the linear phase of amplification. The level of cyclin D1 mRNA was higher in all of the transgenic mouse mammary glands compared with the three WT mouse samples. When results of this and a duplicate experiment were scanned and normalized to β-actin mRNA levels, a (2.5 ± 0.8)-fold increase (P < 0.003) in cyclin D1 mRNA levels was observed in transgenic compared to WT samples. Similarly, higher levels of cyclin D1 mRNA were observed in the transgenic mouse mammary glands compared with WT mouse samples in the Northern blot analysis. RNA loading was normalized to the levels of 28S rRNA. Expression of cyclin D1 mRNA could be detected in two of the five WT mice tested (WT 3 and WT 14). All transgenic mice displayed detectable expression levels of cyclin D1 mRNA, and these levels were higher than those of the WT 3 and WT 14 samples. When results were scanned and normalized to 28S rRNA levels, a (6.8 ± 5.4)-fold increase in cyclin D1 mRNA levels were observed in the transgenic samples compared to WT 3 and WT 14 samples. Thus, overexpression of transgenic c-Rel in the mammary gland during the first pregnancy is sufficient to induce a substantial increase in cyclin D1 mRNA expression.
FIG. 6.
MMTV-c-rel mammary glands and carcinomas overexpress cyclin D1 mRNA. (A) Mammary glands. Total RNA was prepared from mammary glands of 3- to 6-month-old mice or WT FVB/N mice at day 18.5 of the first pregnancy. RT-PCR: RNA was subjected to DNase treatment, and analysis on ethidium bromide stained gels verified quality and essentially equal loading (data not shown). Samples (5 μg) were subjected to RT-PCR analysis of cyclin D1 and β-actin mRNA levels in the presence (+) or absence (−) of RT to control for DNA contamination. For the β-actin mRNA analysis, levels appeared saturated after 30 cycles of PCR and identical in all of the samples tested, while levels were undetectable below 20 PCR cycles (data not shown), so 25 PCR cycles was selected for normalization. The values of cyclin D1 signal intensity normalized to β-actin mRNA levels are presented relative to the WT (3) sample. RNA samples (5 μg) were subjected to Northern blot analysis of cyclin D1 gene levels using a radiolabeled human full-length cyclin D1 cDNA as probe. Ethidium bromide staining of the 28S rRNA was used as a control for loading. The values of cyclin D1 signal normalized to 28S rRNA relative to WT (3) are given below. ND, not detectable by scanning. (B) Mammary carcinomas. Total RNA was prepared from the indicated mammary tumors (T) and grossly normal mammary glands (N) of age-related multiparous line 14 and 16 MMTV-c-rel mice, as well as from virgin (V) transgenic mouse mammary glands. RNA samples (5 μg) were subjected to semiquantitative RT-PCR analysis to assess cyclin D1 mRNA levels, as described above. The values of cyclin D1 signal intensity normalized to β-actin RNA levels are presented relative to the line 16 (4948 N) normal sample (which was better seen on a darker exposure). RNA samples (5 μg) were subjected to Northern blot analysis to assess cyclin D1 mRNA levels, as described above. The values of cyclin D1 signal normalized to 28S rRNA relative to 15 (8441) are given below. ND, not detectable by scanning.
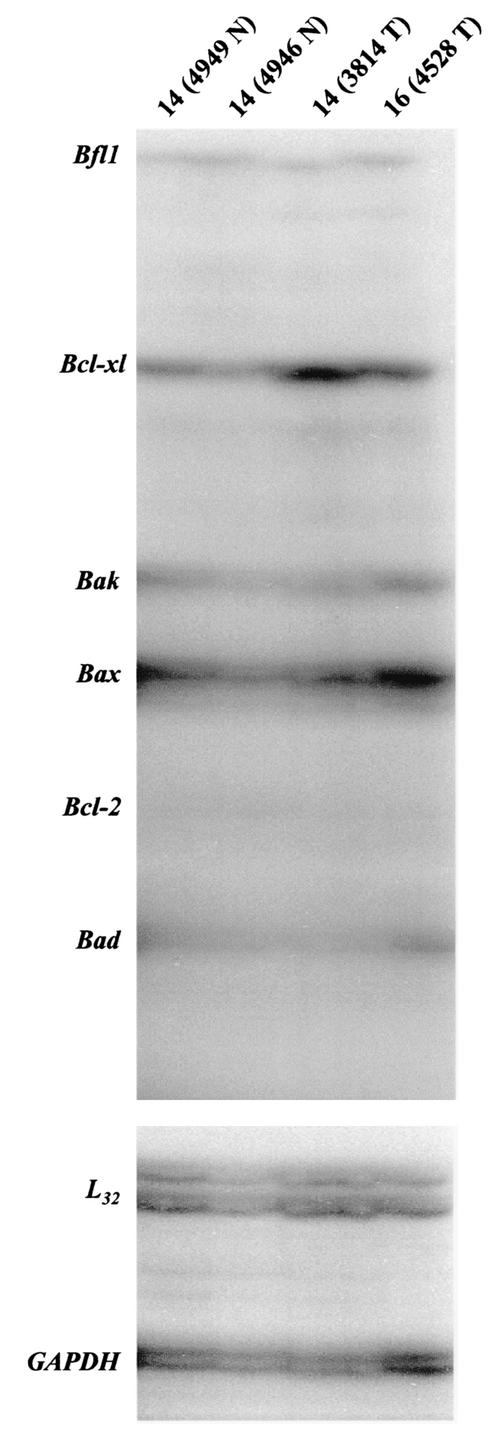
We next evaluated cyclin D1 levels in c-Rel-induced mammary tumors compared to nonmalignant mammary glands in transgenic animals, again using both the semiquantitative RT-PCR assay and Northern blot analyses (Fig. 6B). Total RNA was isolated from mammary carcinomas that had developed in transgenic mice of lines 14 and 16, as well as from grossly normal mammary glands from multiparous age-related transgenic mice of the same lines. As a reference for basal levels of cyclin D1 mRNA in the mammary gland, total RNA was extracted from mammary glands of five virgin adult transgenic mice of lines 16 or 15. In this and a duplicate RT-PCR assay, all of the tumors displayed higher expression levels of cyclin D1 compared to the two grossly normal mammary gland samples tested. Interestingly, the normal sample 14 (5441 N) displayed higher levels of cyclin D1 mRNA than the normal sample 16 (4948 N), which correlated with their respective levels of c-rel transgene expression as seen above (Fig. 3A). In Northern blot analysis, levels of cyclin D1 mRNA were barely detectable in the five virgin mammary gland samples tested. In contrast, one (14 4949 N) of the three grossly normal mammary gland samples and one tumor (14 3814 T) displayed detectable expression levels of cyclin D1 mRNA, while the cyclin D1 mRNA levels were sharply increased in the 3 other tumors (16 4528 T, 14 3996 T, and 14 4936 T). Therefore, MMTV-c-rel mammary glands and carcinomas display a substantial overexpression of cyclin D1 mRNA compared to the WT mammary gland.
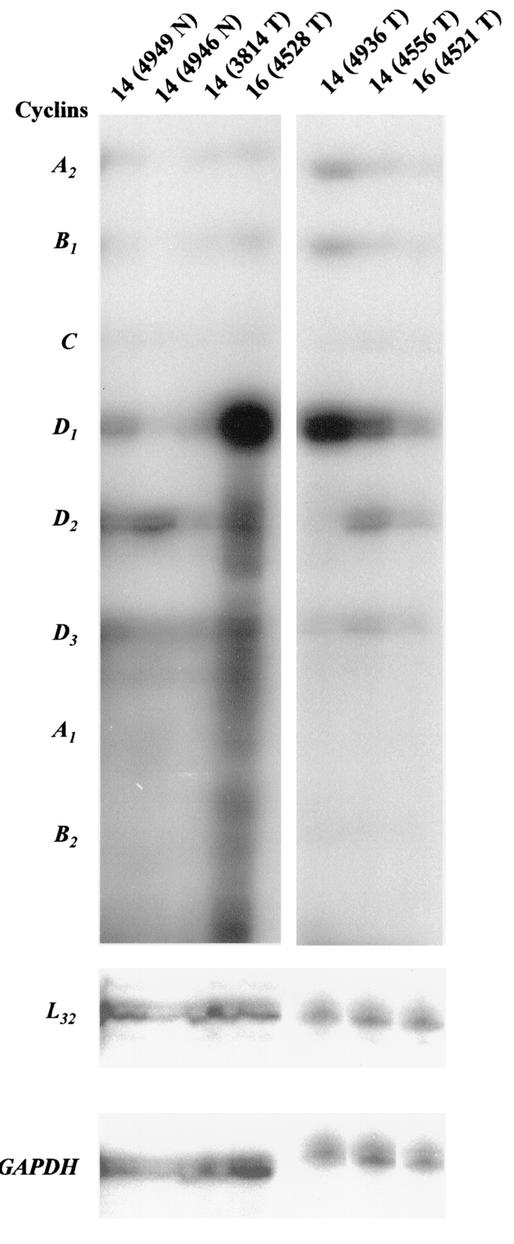
Evidence has also suggested NF-κB mediates regulation of cyclin A (reviewed in reference 32). Thus, we compared the overall cyclin expression profiles in the c-Rel-induced mammary tumors with the grossly normal mammary glands. In the first set, total RNA was prepared from two grossly normal mammary glands from multiparous transgenic mice line 14 (4949 N and 4946 N), and two mammary tumors of line 14 (3814 T) and 16 (4528 T). In a second set, total RNA was prepared from mammary carcinomas of line 14 (4936 T and 4556 T) and line 16 (4521 T). RNA samples were subjected to a multiprobe RPA kit, which assesses mRNA levels for cyclins A1, A2, B1, B2, C, D1, D2, and D3, and L32 and GAPDH housekeeping gene products (Fig. 7). In this and a duplicate experiment, bands were detectable for cyclin A2, B1, D1, D2, and D3 mRNA. The mRNA from the c-Rel-induced tumors displayed increased expression of the cyclin D1 gene compared to the normal mammary glands (Fig. 7, left panel), consistent with the Northern blot analysis above (Fig. 6B). The tumors displayed variable levels of mRNA for cyclins A2, B1, D2, and D3. Analysis of the housekeeping genes L32 and GAPDH confirmed the essentially equal loading of samples within the panels. Thus, RPA showed that MMTV-c-rel tumors display an increase in cyclin D1 gene expression with variable levels of expression of the other cyclins.
FIG. 7.
Profile of cyclin gene expression in MMTV-c-rel mouse mammary glands and tumors. Total RNA was prepared from the indicated mammary tumors (T) or grossly normal mammary glands (N) of age-related multiparous line 14 and 16 MMTV-c-rel mice. RNA samples (5 μg) were subjected to RPA analysis to assess mRNA levels for cyclin A1, A2, B1, B2, C, D1, D2, and D3, and L32 and GAPDH housekeeping genes. Data from two sets of analyses are shown in the left and right panels. The identities of the RNase protected bands were established using the undigested probes as markers and a control RNA for mouse cyclin mRNA expression provided with the kit (data not shown).
MMTV-c-rel mammary tumors overexpress c-myc.
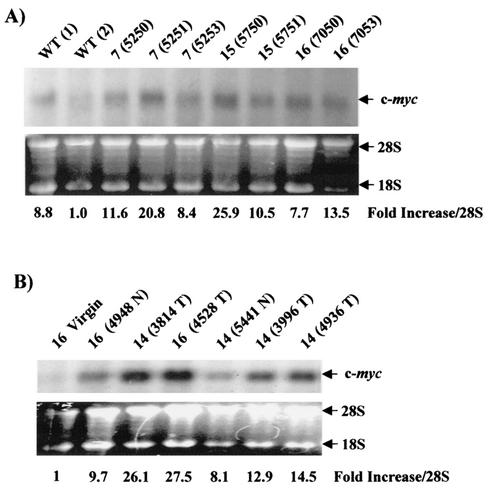
We next tested for changes in c-myc gene expression, another NF-κB target gene (17, 39), which affects cell proliferation and survival (reviewed in reference 5). RNA was isolated at day 18.5 of the first pregnancy from mammary glands of line 7, 15, and 16 MMTV-c-rel transgenic and WT FVB/N mice, all 3 to 6 months old. Samples were subjected to Northern blot analysis for c-myc RNA levels (Fig. 8A). The quality of the RNA was evaluated by ethidium bromide staining of the gel (Fig. 8A) and by Northern blot analysis of GAPDH mRNA expression levels (data not shown). The level of c-myc mRNA appeared higher in most of the mammary glands of the transgenic animals compared to the two WT mice. When results were scanned and normalized to 28S rRNA levels, a (2.4 ± 1.4)-fold increase was observed in c-myc mRNA expression levels in transgenic samples compared to the average of the WT samples. Comparable increase was obtained upon normalization to levels of GAPDH mRNA ([2.8 ± 0.7]-fold, data not shown). These levels of increase did not reach statistical significance.
FIG. 8.
MMTV-c-rel tumors overexpress c-myc mRNA. (A) Mammary glands. Total RNA was prepared from mammary glands and samples (15 μg) subjected to Northern analysis for c-myc mRNA expression. As a control, the gel was stained with ethidium bromide, shown below. The relative values of c-myc signal intensity normalized to levels of 28S rRNA are given relative to the WT (2) virgin sample. (B) Mammary tumors. Total RNA was extracted from the indicated mammary tumors (T) and grossly normal mammary glands (N) of age-related multiparous line 14 and 16 MMTV-c-rel mice. In addition, RNA was isolated from mammary glands of a nulliparous transgenic line 16 mouse (16 Virgin). RNA samples (20 μg) were subjected to Northern analysis for c-myc mRNA expression. As controls for RNA integrity and equal loading, the gel was stained with ethidium bromide, and RNA samples subjected to RT-PCR analysis for β-actin mRNA levels (Fig. 3). The values of c-myc signal intensity normalized to 28S rRNA levels relative to the line 16 virgin sample are given below.
We next compared the levels of c-myc mRNA expression in mammary glands versus carcinomas of multiparous age-related transgenic mice and a virgin transgenic mouse. Quality of the RNA and equal loading was checked by ethidium bromide staining of the gel (Fig. 8B) and by RT-PCR analysis of β-actin mRNA levels, as shown above in Fig. 3B (bottom panel). In Northern blot analysis, all the normal and tumor mammary glands of c-Rel-expressing multiparous transgenic mice demonstrated a substantial increase in c-myc mRNA expression levels compared to the line 16 virgin mouse sample. When the results were scanned and normalized to levels of 28S rRNA, a (16.5 ± 8.3)-fold increase in c-myc mRNA expression levels was observed in mammary glands of multiparous transgenic mice compared to the virgin transgenic mouse sample. In addition, tumors displayed a (2.3 ± 0.8)-fold increase in levels of c-myc RNA expression compared to grossly normal mammary glands of multiparous MMTV-c-rel transgenic mice (P < 0.028) (Fig. 8B). Thus, increased c-rel expression in transgenic mice leads to elevated levels of c-myc mRNA in mammary glands and carcinomas.
MMTV-c-rel mammary glands and tumors overexpress bcl-xl RNA.
The bcl-xl, and bfl-1/a1 genes, which encode two prosurvival members of the Bcl-2 family, have been identified as direct targets of NF-κB (13, 70). Bcl-2 expression has also been found to be upregulated by NF-κB (12, 30, 37), but it is unclear if this is a direct effect of NF-κB. Given the evidence for the role of the Bcl-2 family of cell death regulators in mammary tumorigenesis (29), we next assessed mRNA levels of Bcl-2 family members using a second RPA kit. This kit measures RNA levels of the prosurvival genes bcl-2, bcl-xl, and bfl-1/a1, and of bax, bak, and bad, which promote apoptosis. RNA samples, isolated from two tumors and two grossly normal mammary glands of MMTV-transgenic mice were subjected to RPA (Fig. 9). Results of two independent experiments showed an increase in the expression of bcl-xl RNA levels in the two tumors tested compared to normal mammary glands. When results were scanned and normalized to L32 mRNA levels, an ∼6.6- and 3.2-fold increase in bcl-xl mRNA expression levels were observed in tumor samples 14 (3814 T) and 16 (4528 T), respectively, compared to the two normal samples. By contrast, only low and unchanged expression of bcl-2 and bfl-1/a1, respectively, was observed. The mRNA expression of the pro-apoptotic genes bax, bak, and bad appeared to change only modestly in the tumor samples. Essentially equal loading was confirmed by analysis of the L32 and GAPDH housekeeping genes. These results suggest that c-Rel-induced mammary tumors display increased levels of bcl-xl mRNA expression.
FIG. 9.
Profile of Bcl-2 family member gene expression in MMTV-c-rel mouse mammary glands and tumors. RNA was prepared from the indicated mammary tumors (T) and grossly normal mammary glands (N) of age-related multiparous line 14 and 16 MMTV-c-rel mice. Samples (5 μg) were subjected to RPA analysis to assess mRNA levels of Bcl-2 family member genes, i.e., bfl-1/a1, bcl-xl, bax, bak, bcl-2, and bad, and L32 and GAPDH housekeeping genes. The identity of the RNase protected bands were established using the undigested probes as markers and a control RNA for mouse apoptosis gene expression provided with the kit (data not shown).
To further study the involvement of Bcl-xL in c-Rel-induced mammary tumorigenesis, we measured bcl-xl mRNA expression levels in tumors as well as during pregnancy in the mammary glands of MMTV-c-rel versus WT mice. Samples of total RNA isolated from the mammary glands at day 18.5 of the first pregnancy of lines 7, 14, 15, and 16 MMTV-c-rel mice and age-matched WT FVB/N mice, described above, were subjected to semiquantitative RT-PCR and to Northern blot assays of bcl-xl mRNA levels (Fig. 10A). For the RT-PCR, RNA quality and loading were normalized to β-actin levels, as described above in Fig. 6. The level of bcl-xl mRNA was higher in mammary gland samples from all of the transgenic mice compared with those from the three WT mice. When results of this and a duplicate experiment were scanned a (3.0 ± 0.8)-fold increase (P < 0.02) in normalized bcl-xl mRNA levels was observed in transgenic compared to WT mammary gland samples. In the Northern blotting, RNA quality and loading were normalized to the levels of 28S rRNA (Fig. 10A), as described above. The RNA band detected with the probe comigrated with the transcript seen in NIH 3T3 cells transiently transfected with a human bcl-xl plasmid expression vector. Thus, the transcript is likely bcl-xl rather than the bcl-xs mRNA. In the 5 WT samples tested, bcl-xl mRNA levels were barely detectable, while 4 out of 5 transgenic samples displayed detectable levels of bcl-xl mRNA. Taken together, these findings indicate that expression of bcl-xl mRNA increases by day 18.5 of the first pregnancy in the mammary glands of c-rel transgene mice.
FIG. 10.
MMTV-c-rel mammary glands and carcinomas overexpress bcl-xl mRNA. (A) Mammary glands. Total RNA was prepared from the indicated 3- to 6-month-old transgenic mice and WT FVB/N at day 18.5 of the first pregnancy. RT-PCR: Samples (5 μg) were subjected to RT-PCR analysis of bcl-xl and β-actin mRNA levels, as described above in Fig. 6. The values of bcl-xl signal intensity normalized to β-actin mRNA levels are presented relative to the WT (2) sample. Northern blot: Samples (5 μg) were subjected to Northern blot analysis of bcl-xl mRNA levels, using a mouse bcl-xl cDNA as a probe, as described above in Fig. 6. The values of the bcl-xl signals for the WT samples were below the level of detection. (B) Mammary carcinomas. Total RNA was prepared from the indicated mammary glands (virgin [V] or multiparous grossly normal [N]) or tumors (T), and samples (5 μg) were subjected to either semiquantitative RT-PCR or Northern blot analysis to assess mRNA levels for bcl-xl, as described above. The values of bcl-xl signal intensity normalized to β-actin mRNA levels in the RT-PCR analysis are presented relative to the line 16 (4948) normal sample, while the bcl-xl signals in the Northern blot analysis for the virgin and normal samples were below the level of detection.
To further study the involvement of Bcl-xL in c-Rel-induced mammary tumorigenesis, levels of bcl-x1 mRNA levels were compared in samples of total RNA isolated from mammary carcinomas, grossly normal mammary glands from multiparous age-related transgenic mice, as well as from virgin mice (Fig. 10B). As judged by RT-PCR, all of the tumors displayed higher levels of bcl-xl mRNA expression compared to the normal samples tested, in this and a duplicate experiment. Interestingly, once again the normal line 14 (5441 N) displayed higher levels of bcl-xl mRNA than the normal sample 16 (4948 N), which correlated with their respective levels of c-rel transgene and cyclin D1 mRNA expression seen above (Fig. 3A and 8B). Expression levels of bcl-xl mRNA in all virgin mammary glands as well as all grossly normal mammary glands from multiparous transgenic mice were below levels of detection by Northern blot. In contrast, expression of bcl-xl mRNA was readily detected in the four tumors tested. Altogether, these results indicate that enforced expression of c-Rel in mammary epithelial cells leads to increased mRNA levels of cyclin D1, c-myc, as well as bcl-xl in the mammary glands and derived tumors.
c-Rel-p52 and p50 heterodimer complexes induce cyclin D1 promoter activity in mammary epithelial cells.
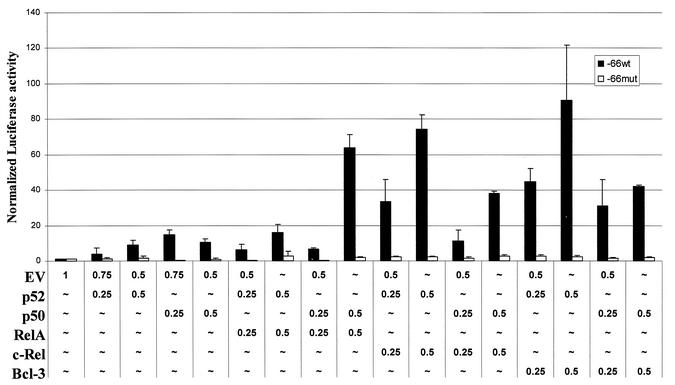
While the c-myc and bcl-xl genes have been shown to be direct targets of c-Rel (13, 24, 39, 70), the cyclin D1 promoter has been shown to be activated by classical p50/RelA complexes (25, 26), and even more potently by Bcl-3/p52 (65). Since no studies have fully investigated the role of c-Rel in the regulation of the cyclin D1 promoter, we compared the activation of the cyclin D1 promoter by different NF-κB complexes in mammary epithelial cells. NMuMG mouse untransformed mammary epithelial cells were transfected with vectors expressing cyclin D1 promoter luciferase reporter constructs containing either a wt (−66 wt) or mutated (−66 mut) proximal NF-κB element, plus pSV40-β-gal, for normalization, in the absence or presence of various NF-κB expression vectors (Fig. 11). Expression of p50 or p52 alone results in only a modest increase in cyclin D1 promoter activity. Consistent with previous reports (65), coexpression of p52 and Bcl-3 resulted in a sharp increase in cyclin D1 promoter activity, while p50 and Bcl-3 complexes were less potent. At the higher dose tested, RelA plus p50 caused an increase in cyclin D1 promoter activity. Interestingly, coexpression of c-Rel and p52 resulted in an activation of the cyclin D1 promoter activity comparable to that seen with p52 plus Bcl-3, whereas c-Rel/p50 complexes were again less potent. As controls for protein expression levels, NIH 3T3 cells were similarly transfected, and similar 5 to 10-fold increases in levels of c-Rel, RelA, p50, p52, and Bcl-3 were observed compared to cells transfected with the empty vector DNA (data not shown). These findings indicate that c-Rel can potently induce the cyclin D1 promoter, in particular when present with the p52 subunit.
FIG. 11.
c-Rel heterodimer complexes induce the cyclin D1 promoter. NMuMG cells were transfected, in duplicate, with −66 WT-cyclin D1 or −66 Mut-cyclin D1 luciferase gene reporter constructs and 0.5 μg of pSV40-β-gal in the presence of the indicated amounts of NF-κB or Bcl-3 plasmid expression vectors. After 48 h, cultures were harvested, normalized for β-Gal activity, and assayed for luciferase activity. Normalized values of luciferase activity are presented (error bars, standard deviations).
Ectopic c-Rel expression increases the levels of cyclin D1, p52, and p50 in mammary epithelial cells.
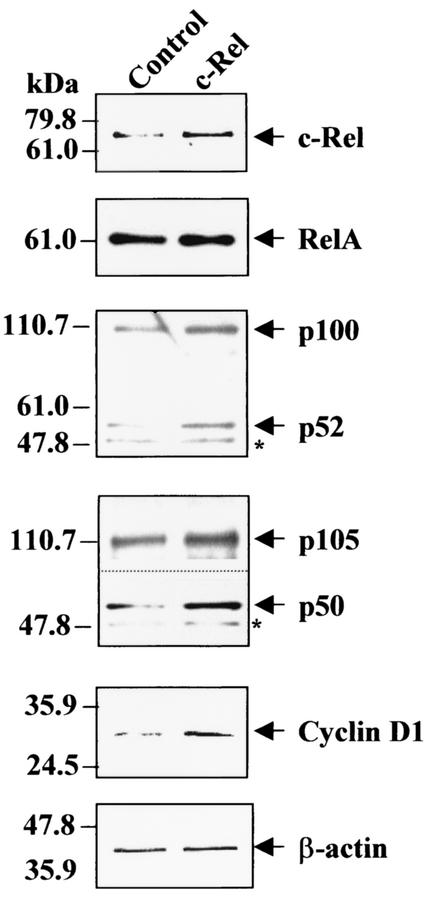
Next, we investigated the effects of ectopic stable c-Rel overexpression on cyclin D1 and NF-κB subunit levels using MCF-10F human untransformed breast epithelial cells. Cells were transfected with a mouse c-Rel expression vector along with a puromycin selection plasmid, and grown in the presence of puromycin. Mixed populations of selected cells were used for analysis. As controls, we used either MCF-10F cells transfected with the pBluescript and puromycin selection plasmids and selected with puromycin or MCF-10F untransfected parental cells grown under the same conditions. (All populations of cells grew at the same rate, data not shown). We first confirmed overexpression of c-Rel in WCEs of c-Rel-transfected cells compared to parental MCF-10F cells, using β-actin as control for equal loading (Fig. 12). Scanning of this and a duplicate experiment indicated that c-Rel-transfected MCF-10F cells displayed a (2.5 ± 1.2)-fold increase in total level of c-Rel compared to control cells. Importantly, we observed that c-Rel-transfected MCF-10F cells displayed significant increases in the levels of cyclin D1, as well as in the NF-κB subunits p52 and p50 (and their precursors p100 and p105) compared to control cells (Fig. 12). Densitometry analysis indicated that c-Rel-transfected MCF-10F cells displayed a 3.1 and 2.7-fold increase in levels of p52 and p50, respectively. Densitometry of this and two other immunoblots (data not shown) indicated that c-Rel-transfected MCF-10F cells displayed a (1.8 ± 0.1)-fold increase in levels of cyclin D1. Essentially no change was observed in levels of RelA in c-Rel transfected cells. Control puromycin-selected cells displayed levels of c-Rel and cyclin D1 comparable to the parental untransfected cells MCF-10F cells, indicating that the selection was not responsible for this observed increase (data not shown). Thus, sustained overexpression of c-Rel in untransformed mammary epithelial cells leads to increased levels of p52, p50, and cyclin D1 expression. Overall, these findings indicate sustained c-Rel expression in mammary epithelial cells is sufficient to lead to the induction in NF-κB subunits and target genes, responsible for mammary tumorigenesis.
FIG. 12.
Ectopic c-Rel expression in human MCF-10F untransformed mammary epithelial cells induces levels of cyclin D1, and p52 and p50 expression. Cells were cotransfected with pSVSport-c-Rel (c-Rel) and pGKpuro plasmid expression vectors, and a mixed population of cells was selected with puromycin. WCEs were prepared from transfected cells and parental untransfected cells (Control), and samples (10 μg) were subjected to immunoblot analysis for c-Rel, RelA, p52/p100, p50/p105, cyclin D1, and β-actin levels using two identical blots that were successively reprobed with the different antibodies. To better visualize the p105 versus p50 band, a longer exposure was used.
DISCUSSION
Here we demonstrate for the first time that c-Rel plays a causal role in tumorigenesis of the mammary gland in an MMTV-LTR-driven mouse model. Overall, one or more mammary tumors were detected in 31.6% of MMTV-c-rel transgenic mice at an average age of 19.9 months. Histological analysis of the mammary tumors in four independent lines provided evidence for a wide spectrum of tumor subtypes, including adenocarcinomas, adenosquamous carcinomas, squamous carcinomas, a spindle cell carcinoma and a papillary carcinoma. One mouse developed pulmo-nary metastasis in addition to multiple mammary adenocarcinomas. In addition to mammary carcinomas, several mice had enlarged spleen or other abnormalities including lymphoid or myeloid hyperplasia or centrocytic lymphomas in the spleen, which can be correlated to the expression of the transgene in splenocytes (Fig. 1B), as also shown in previous studies of mammary tumors in transgenic mice using the same promoter (55, 67). By contrast, only rare (<1%) spontaneous cases of mammary tumors were reported in the FVB/N strain, consisting of squamous carcinomas or keratoacanthomas (42). Although the types of some tumors seen here are similar to those found in elderly FVB/N mice, the range and types of mammary tumors seen in the MMTV-c-rel mice are entirely similar to those induced by the wnt pathway (51). The MMTV-c-rel mammary tumors displayed sustained expression of the c-rel transgene mRNA. Tumors were also typified by overexpression of c-Rel protein, and displayed elevated mRNA levels of cyclin D1, c-myc, and bcl-xl, three NF-κB target genes implicated in control of growth and cell survival. These increases were detected in normal mammary glands during and after the first cycle of pregnancy. Stable ectopic c-Rel expression in untransformed mammary epithelial cells led to elevated levels of p50/p105, p52/p100, and cyclin D1. Furthermore, c-Rel complexes with p50 or p52 activated the cyclin D1 promoter. While the v-rel oncogene has been shown to be highly tumorigenic, our findings represent the first in vivo demonstration of the transforming ability of the c-Rel NF-κB subunit.
As expected, expression of c-rel transgene mRNA was detected in all the c-Rel-induced tumors tested and also in some grossly normal transgenic mammary glands from multiparous mice. Total c-Rel protein and c-rel transgene mRNA levels did not always correlate (Fig. 3 and 5), suggesting that endogenous c-Rel levels increased in some MMTV-c-rel mammary tumors, as observed previously in patient primary tumor samples (15, 59). Importantly, supershift analysis confirmed the presence of c-Rel in the NF-κB DNA binding complexes (Fig. 4 and data not shown). EMSA and immunoblot analysis demonstrated the induction of other NF-κB subunits in the MMTV-c-rel tumor mammary gland samples, including p50, p52, RelB, and RelA and the Bcl-3 protein in addition to the expected c-Rel. The vast majority of primary breast cancer specimens from patients display activation of multiple NF-κB proteins, including c-Rel, p50, p52, RelA, and RelB subunits and Bcl-3 protein (15, 59). The genes encoding the p105/p50, p100/p52 and RelB proteins are known NF-κB targets (reviewed in reference 44), and ectopic c-Rel expression in MCF-10F cells induced p50 and p52 expression. These findings suggest that direct activation by c-Rel may be responsible for the induction of these NF-κB subunits. In the case of Bcl-3, we have found that it is normally expressed in the mouse mammary epithelial cell during development (data not shown). A modest increase in the RelA subunit level was observed in MMTV-c-rel tumors by immunoblot, but not in the c-Rel transfected mammary epithelial cells. It is likely that the elevated level of this subunit observed in the tumors results indirectly from overexpression of the c-Rel transgene, e.g., activation of a cytokine such as tumor necrosis factor. If true, posttranslational modifications might explain the relatively high levels of RelA-containing complexes observed in EMSA in many of the mammary tumor samples. Lastly, it appeared that the level of c-Rel-containing complexes was relatively low, compared to the RelA subunit, suggesting that c-Rel may need to be modified or to interact with other proteins to efficiently bind the DNA. We are currently investigating factors that regulate c-Rel binding in mammary epithelial cells.
NF-κB appears to promote cell proliferation and survival of mammary epithelial cells in culture (34, 58, 59) and development of the normal mammary gland (8, 10, 14). Thus, we hypothesized that c-Rel activation could contribute to tumor cell growth and survival through the induction of a number of NF-κB target genes, including regulators of cell cycle, proliferation, and survival (reviewed in references 33 and 47). The cyclin D1 promoter, which contains several κB elements, can be activated by classical NF-κB (25, 26), although a more recent study has found Bcl-3/p52-mediated activation of the cyclin D1 promoter more potent (65). Here, we demonstrate the ability of c-Rel/p52 and c-Rel/p50 complexes to induce the cyclin D1 promoter. Female mice null for the cyclin D1 gene fail to develop normal mammary glands (18, 54). Overexpression of cyclin D1 has been implicated in breast cancer in humans and in rodent models (reviewed in reference 32). Importantly, MMTV-cyclin D1 mice develop mammary cancers (64), and an increase in levels of cyclin D1 was seen in mammary tumors that develop in mice with enforced expression of Neu or Ras (68). Female mice deficient for cyclin D1 are totally resistant to breast cancers induced by Neu and Ras, but not to those induced by Myc and Wnt-1 (68). Interestingly, several groups, including our own, have shown that activated Neu and Ras induce functional NF-κB in mammary and liver epithelial and fibroblast cells (4, 19, 45, 69). The human and mouse c-myc promoters have been shown to be targets of NF-κB complexes, including those containing c-Rel, in breast epithelial and other cell types (17, 31, 34, 39, 40). The c-myc gene is overexpressed in breast neoplasia samples from patients (reviewed in reference 41). Targeted c-myc expression in the mammary gland of mice using either the MMTV-LTR or the whey acidic protein promoter leads to mammary tumor development (53, 61). The promoter of the bcl-xl cell death antagonist gene is regulated by p50-p65- or p50-c-Rel-containing complexes (13, 70). In the present study, we observed that many of the MMTV-c-rel tumors display elevated levels of expression of cyclin D1, bcl-xl, and c-myc RNA. No correlation was noted between tumor latency and cyclin D1 levels; while, some correlation was seen with c-myc mRNA levels, although, tumors numbers were too low to draw any firm conclusions. Change in expression profiles of these transcripts were also observed in normal mammary glands of transgenic mice, starting the first cycle of gestation. Transfection analysis suggested that the genes encoding cyclin D1, p52 and p50 are c-Rel targets. These findings are consistent with a direct role of c-Rel in the events leading to mammary gland tumorigenesis in the MMTV-c-rel mice.
Aberrant activation of nuclear NF-κB/Rel has been found to correlate with oncogenesis in several other systems, including thyroid carcinoma, non-small cell lung carcinoma, colon carcinoma, ovarian carcinoma, prostate cancer (62), Hodgkin's disease (6), and various types of lymphomas (reviewed in reference 47). In hematopoietic tumors, amplification, overexpression, or rearrangement of the c-rel, nf-kb1, nf-kb2, or relA genes or the bcl-3 gene; and mutations inactivating the IκB-α protein have been noted (reviewed in reference 47). Therefore, increased activation of NF-κB can occur by multiple mechanisms in tumor cells. Similarly, in breast cancer cells, multiple mechanisms that lead to aberrant activation of NF-κB have also been described. We observed that many human breast tumor specimens and cell lines in culture display constitutive IKK and protein kinase CK2 activity, which correlated with elevated levels of NF-κB binding activity (50). Products of several oncogenes induce NF-κB activity in mammary epithelial cells, such as Her-2/neu (45, 69). Presumably these oncogenes require downstream effectors that activate either an IκB kinase or CK2, thereby increasing the rate of IκB turnover, and basal NF-κB nuclear translocation and binding to the DNA. Recently, we have demonstrated that activation of NF-κB by Her-2 can be reduced upon inhibition of CK2 (49). Interestingly, in vitro studies have suggested that Her-2/neu, Ras, and Raf preferentially induce classical p50/RelA complexes (4, 19, 45, 69). The late-onset of tumor development and the variety in the tumor histological patterns in MMTV-c-rel mice implies the requirement of additional pathways in the mammary gland tumorigenesis, in addition to overexpression of c-Rel. This phenotype is very reminiscent of the MMTV-CK2α subunit transgenic mice, which we have described recently (38). These mice developed late-onset mammary carcinomas of heterogeneous histological profiles. Consistent with the biological properties of CK2, the MMTV-CK2 tumors expressed high levels of NF-κB activity, c-Myc, and activated β-catenin (38). Altogether, our findings indicate that cellular c-Rel is able to transform mammal epithelial cells in vivo and thus represents a potential therapeutic target.
Acknowledgments
R.R.-M. and D.W.K. contributed equally to this work.
We thank T. Gilmore, S. Hann, N. Rice, and M. Ernst for generously providing clones and antibodies. The excellent technical assistance of Judy E. Walls, Lee Gazourian, Alan Lau, and Aundrea Oliver is acknowledged. The excellent assistance of Keisha Bratton in preparation of the manuscript is acknowledged.
This work was supported by grants from the Massachusetts Department of Public Health Breast Cancer Program (to E.L.-B. and R.R.-M.), the Department of Army (DAMD 17-01 10158 to R.R.-M.), and the UCD Mutant Mouse Regional Resource Center (NIH/NCRR U42-RR14905 to R.D.C) and by grants NIH RO1 CA82742 and PO5 ES11624 (G.E.S. and D.C.S.).
REFERENCES
- 1.Abbadie, C., N. Kabrun, F. Bouali, J. Smardova, D. Stehelin, B. Vandenbunder, and P. J. Enrietto. 1993. High levels of c-rel expression are associated with programmed cell death in the developing avian embryo and in bone marrow cells in vitro. Cell 75:899-912. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, L. M. Staudt, and et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503-511. [DOI] [PubMed] [Google Scholar]
- 3.Arsura, M., M. J. FitzGerald, N. Fausto, and G. E. Sonenshein. 1997. Nuclear factor-κB/Rel blocks transforming growth factor beta1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 8:1049-1059. [PubMed] [Google Scholar]
- 4.Arsura, M., F. Mercurio, A. L. Oliver, S. S. Thorgeirsson, and G. E. Sonenshein. 2000. Role of the IκB kinase complex in oncogenic Ras- and Raf-mediated transformation of rat liver epithelial cells. Mol. Cell. Biol. 20:5381-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arsura, M., and G. E. Sonenshein. 2001. The roles of the c-myc and c-myb oncogenes in hematopoiesis and leukemogenesis, p. 521-549. In K. Ravid and J. Licht (ed.), Transcription factors: normal and malignant development of blood cells. Wiley-Liss Inc., New York, N.Y.
- 6.Bargou, R. C., C. Leng, D. Krappmann, F. Emmerich, M. Y. Mapara, K. Bommert, H. D. Royer, C. Scheidereit, and B. Dorken. 1996. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood 87:4340-4347. [PubMed] [Google Scholar]
- 7.Bours, V., G. Franzoso, V. Azarenko, S. Park, T. Kanno, K. Brown, and U. Siebenlist. 1993. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell 72:729-739. [DOI] [PubMed] [Google Scholar]
- 8.Brantley, D. M., C. L. Chen, R. S. Muraoka, P. B. Bushdid, J. L. Bradberry, F. Kittrell, D. Medina, L. M. Matrisian, L. D. Kerr, and F. E. Yull. 2001. Nuclear factor-κB (NF-κB) regulates proliferation and branching in mouse mammary epithelium. Mol. Biol. Cell 12:1445-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calaf, G., and J. Russo. 1996. Transformation of human breast epithelial cells by chemical carcinogens. Carcinogenesis 14:483-492. [DOI] [PubMed] [Google Scholar]
- 10.Cao, Y., G. Bonizzi, T. N. Seagroves, F. R. Greten, R. Johnson, E. V. Schmidt, and M. Karin. 2001. IKKα provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell 107:763-775. [DOI] [PubMed] [Google Scholar]
- 11.Cardiff, R. D., and R. J. Munn. 1995. Comparative pathology of mammary tumorigenesis in transgenic mice. Cancer Lett. 90:13-19. [DOI] [PubMed] [Google Scholar]
- 12.Catz, S. D., and J. L. Johnson. 2001. Transcriptional regulation of bcl-2 by nuclear factor kappa B and its significance in prostate cancer. Oncogene 20:7342-7351. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C., L. C. Edelstein, and C. Gelinas. 2000. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol. Cell. Biol. 20:2687-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarkson, R. W., J. L. Heeley, R. Chapman, F. Aillet, R. T. Hay, A. Wyllie, and C. J. Watson. 2000. NF-κB inhibits apoptosis in murine mammary epithelia. J. Biol. Chem. 275:12737-12742. [DOI] [PubMed] [Google Scholar]
- 15.Cogswell, P. C., D. C. Guttridge, W. K. Funkhouser, and A. S. Baldwin, Jr. 2000. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 19:1123-1131. [DOI] [PubMed] [Google Scholar]
- 16.Dean, M., R. B. Kent, and G. E. Sonenshein. 1983. Transcriptional activation of immunoglobulin alpha heavy-chain genes by translocation of the c-myc oncogene. Nature 305:443-446. [DOI] [PubMed] [Google Scholar]
- 17.Duyao, M. P., A. J. Buckler, and G. E. Sonenshein. 1990. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc. Natl. Acad. Sci. USA 87:4727-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fantl, V., G. Stamp, A. Andrews, I. Rosewell, and C. Dickson. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 9:2364-2372. [DOI] [PubMed] [Google Scholar]
- 19.Finco, T. S., J. K. Westwick, J. L. Norris, A. A. Beg, C. J. Der, and A. S. Baldwin, Jr. 1997. Oncogenic Ha-Ras-induced signaling activates NF-κB transcriptional activity, which is required for cellular transformation. J. Biol. Chem. 272:24113-24116. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, T., G. P. Nolan, H. C. Liou, M. L. Scott, and D. Baltimore. 1993. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 7:1354-1363. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109:S81-96. [DOI] [PubMed] [Google Scholar]
- 22.Gilmore, T. D. 1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene 18:6925-6937. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore, T. D., C. Cormier, J. Jean-Jacques, and M. E. Gapuzan. 2001. Malignant transformation of primary chicken spleen cells by human transcription factor c-Rel. Oncogene 20:7098-7103. [DOI] [PubMed] [Google Scholar]
- 24.Grumont, R. J., A. Strasser, and S. Gerondakis. 2002. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-κB regulated c-myc transcription. Mol. Cell 10:1283-1294. [DOI] [PubMed] [Google Scholar]
- 25.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin, Jr. 1999. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinz, M., D. Krappmann, A. Eichten, A. Heder, C. Scheidereit, and M. Strauss. 1999. NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol. Cell. Biol. 19:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hrdlickova, R., J. Nehyba, and E. H. Humphries. 1994. In vivo evolution of c-rel oncogenic potential. J. Virol. 68:2371-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, D. B., Y. Q. Chen, M. Ruetsche, C. B. Phelps, and G. Ghosh. 2001. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure (Cambridge) 9:669-678. [DOI] [PubMed] [Google Scholar]
- 29.Jager, R., U. Herzer, J. Schenkel, and H. Weiher. 1997. Overexpression of Bcl-2 inhibits alveolar cell apoptosis during involution and accelerates c-myc-induced tumorigenesis of the mammary gland in transgenic mice. Oncogene 15:1787-1795. [DOI] [PubMed] [Google Scholar]
- 30.Jeay, S., G. E. Sonenshein, M. C. Postel-Vinay, and E. Baixeras. 2000. Growth hormone prevents apoptosis through activation of nuclear factor-κB in interleukin-3-dependent Ba/F3 cell line. Mol. Endocrinol. 14:650-661. [DOI] [PubMed] [Google Scholar]
- 31.Ji, L., M. Arcinas, and L. M. Boxer. 1994. NF-κ B sites function as positive regulators of expression of the translocated c-myc allele in Burkitt's lymphoma. Mol. Cell. Biol. 14:7967-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joyce, D., C. Albanese, J. Steer, M. Fu, B. Bouzahzah, and R. G. Pestell. 2001. NF-κB and cell-cycle regulation: the cyclin connection. Cytokine Growth Factor Rev. 12:73-90. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 34.Kim, D. W., M. A. Sovak, G. Zanieski, G. Nonet, R. Romieu-Mourez, A. W. Lau, L. J. Hafer, P. Yaswen, M. Stampfer, A. E. Rogers, J. Russo, and G. E. Sonenshein. 2000. Activation of NF-κB/Rel occurs early during neoplastic transformation of mammary cells. Carcinogenesis 21:871-879. [DOI] [PubMed] [Google Scholar]
- 35.Kindy, M. S., and G. E. Sonenshein. 1986. Regulation of oncogene expression in cultured aortic smooth muscle cells. Post-transcriptional control of c-myc mRNA. J. Biol. Chem. 261:12865-12868. [PubMed] [Google Scholar]
- 36.Kontgen, F., R. J. Grumont, A. Strasser, D. Metcalf, R. Li, D. Tarlinton, and S. Gerondakis. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965-1977. [DOI] [PubMed] [Google Scholar]
- 37.Kurland, J. F., R. Kodym, M. D. Story, K. B. Spurgers, T. J. McDonnell, and R. E. Meyn. 2001. NF-κB1 (p50) homodimers contribute to transcription of the bcl-2 oncogene. J. Biol. Chem. 276:45380-45386. [DOI] [PubMed] [Google Scholar]
- 38.Landesman-Bollag, E., R. Romieu-Mourez, D. H. Song, G. E. Sonenshein, R. D. Cardiff, and D. C. Seldin. 2001. Protein kinase CK2 in mammary gland tumorigenesis. Oncogene 20:3247-3257. [DOI] [PubMed] [Google Scholar]
- 39.La Rosa, F. A., J. W. Pierce, and G. E. Sonenshein. 1994. Differential regulation of the c-myc oncogene promoter by the NF-κB rel family of transcription factors. Mol. Cell. Biol. 14:1039-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, H., M. Arsura, M. Wu, M. Duyao, A. J. Buckler, and G. E. Sonenshein. 1995. Role of Rel-related factors in control of c-myc gene transcription in receptor-mediated apoptosis of the murine B cell WEHI 231 line. J. Exp. Med. 181:1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao, D. J., and R. B. Dickson. 2000. c-Myc in breast cancer. Endocr. Relat. Cancer 7:143-164. [DOI] [PubMed] [Google Scholar]
- 42.Mahler, J. F., W. Stokes, P. C. Mann, M. Takaoka, and R. R. Maronpot. 1996. Spontaneous lesions in aging FVB/N mice. Toxicol. Pathol. 24:710-716. [DOI] [PubMed] [Google Scholar]
- 43.Nakshatri, H., P. Bhat-Nakshatri, D. A. Martin, R. J. Goulet, Jr., and G. W. Sledge, Jr. 1997. Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 17:3629-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pahl, H. L. 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 45.Pianetti, S., M. Arsura, R. Romieu-Mourez, R. J. Coffey, and G. E. Sonenshein. 2001. Her-2/neu overexpression induces NF-κB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IκB-alpha that can be inhibited by the tumor suppressor PTEN. Oncogene 20:1287-1299. [DOI] [PubMed] [Google Scholar]
- 46.Ravid, K., D. L. Beeler, M. S. Rabin, H. E. Ruley, and R. D. Rosenberg. 1991. Selective targeting of gene products with the megakaryocyte platelet factor 4 promoter. Proc. Natl. Acad. Sci. USA 88:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 48.Rice, N. R., and M. K. Ernst. 1993. In vivo control of NF-kappa B activation by I kappa B alpha. EMBO J. 12:4685-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romieu-Mourez, R., E. Landesman-Bollag, D. C. Seldin, and G. E. Sonenshein. 2002. Protein kinase CK2 promotes aberrant activation of NF-κB, transformed phenotype and survival of breast cancer cells. Cancer Res. 62:6770-6778. [PubMed] [Google Scholar]
- 50.Romieu-Mourez, R., E. Landesman-Bollag, D. C. Seldin, A. M. Traish, F. Mercurio, and G. E. Sonenshein. 2001. Roles of IKK kinases and protein kinase CK2 in activation of nuclear factor-κB in breast cancer. Cancer Res. 61:3810-3818. [PubMed] [Google Scholar]
- 51.Rosner, A., K. Miyoshi, E. Landesman-Bollag, X. Xu, D. C. Seldin, A. R. Moser, C. L. MacLeod, G. Shyamala, A. E. Gillgrass, and R. D. Cardiff. 2002. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am. J. Pathol. 161:1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider, T. J., D. Grillot, L. C. Foote, G. E. Nunez, and T. L. Rothstein. 1997. Bcl-x protects primary B cells against Fas-mediated apoptosis. J. Immunol. 159:4834-4839. [PubMed] [Google Scholar]
- 53.Schoenenberger, C. A., A. C. Andres, B. Groner, M. van der Valk, M. LeMeur, and P. Gerlinger. 1988. Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumours with constitutive milk protein gene transcription. EMBO J. 7:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 55.Sinn, E., W. Muller, P. Pattengale, I. Tepler, R. Wallace, and P. Leder. 1987. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell 49:465-475. [DOI] [PubMed] [Google Scholar]
- 56.Sonenshein, G. E. 1997. Rel/NF-kappa B transcription factors and the control of apoptosis. Semin. Cancer Biol. 8:113-119. [DOI] [PubMed] [Google Scholar]
- 57.Song, D. H., D. J. Sussman, and D. C. Seldin. 2000. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J. Biol. Chem. 275:23790-23797. [DOI] [PubMed] [Google Scholar]
- 58.Sovak, M. A., M. Arsura, G. Zanieski, K. T. Kavanagh, and G. E. Sonenshein. 1999. The inhibitory effects of transforming growth factor beta1 on breast cancer cell proliferation are mediated through regulation of aberrant nuclear factor-κB/Rel expression. Cell Growth Differ. 10:537-544. [PubMed] [Google Scholar]
- 59.Sovak, M. A., R. E. Bellas, D. W. Kim, G. J. Zanieski, A. E. Rogers, A. M. Traish, and G. E. Sonenshein. 1997. Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Investig. 100:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanton, L. W., R. Watt, and K. B. Marcu. 1983. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature 303:401-406. [DOI] [PubMed] [Google Scholar]
- 61.Stewart, T. A., P. K. Pattengale, and P. Leder. 1984. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38:627-637. [DOI] [PubMed] [Google Scholar]
- 62.Sumitomo, M., M. Tachibana, J. Nakashima, M. Murai, A. Miyajima, F. Kimura, M. Hayakawa, and H. Nakamura. 1999. An essential role for nuclear factor kappa B in preventing TNF-alpha-induced cell death in prostate cancer cells. J. Urol. 161:674-679. [PubMed] [Google Scholar]
- 63.Thiery, J. P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2:442-454. [DOI] [PubMed] [Google Scholar]
- 64.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 65.Westerheide, S. D., M. W. Mayo, V. Anest, J. L. Hanson, and A. S. Baldwin, Jr. 2001. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G1 transition. Mol. Cell. Biol. 21:8428-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, M., H. Lee, R. E. Bellas, S. L. Schauer, M. Arsura, D. Katz, M. J. FitzGerald, T. L. Rothstein, D. H. Sherr, and G. E. Sonenshein. 1996. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. EMBO J. 15:4682-4690. [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, Y. A., O. Dukhanina, B. Tang, M. Mamura, J. J. Letterio, J. MacGregor, S. C. Patel, S. Khozin, Z. Y. Liu, J. Green, M. R. Anver, G. Merlino, and L. M. Wakefield. 2002. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. J. Clin. Investig. 109:1607-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, B. P., M. C. Hu, S. A. Miller, Z. Yu, W. Xia, S. Y. Lin, and M. C. Hung. 2000. HER-2/neu blocks tumor necrosis factor-induced apoptosis via the Akt/NF-κB pathway. J. Biol. Chem. 275:8027-8031. [DOI] [PubMed] [Google Scholar]
- 70.Zong, W. X., L. C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 13:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]