Abstract
Pak5 is the most recently identified and least understood member of the p21-activated kinase (Pak) family. This kinase is known to promote neurite outgrowth in vitro, but its localization, substrates, and effects on cell survival have not been reported. We show here that Pak5 has unique properties that distinguish it from all other members of the Pak family. First, Pak5, unlike Pak1, cannot complement an STE20 mutation in Saccharomyces cerevisiae. Second, Pak5 binds to the GTPases Cdc42 and Rac, but these GTPases do not regulate Pak5 kinase activity, which is constitutive and stronger than any other Pak. Third, Pak5 prevents apoptosis induced by camptothecin and C2-ceramide by phosphorylating BAD on Ser-112 in a protein kinase A-independent manner and prevents the localization of BAD to mitochondria, thereby inhibiting the apoptotic cascade that leads to apoptosis. Finally, we show that Pak5 itself is constitutively localized to mitochondria, and that this localization is independent of kinase activity or Cdc42 binding. These features make Pak5 unique among the Pak family and suggest that it plays an important role in apoptosis through BAD phosphorylation.
The p21-activated kinases (Paks) are members of a growing class of Rac/Cdc42-associated Ser/Thr protein kinases, characterized by a highly conserved amino-terminal Cdc42/Rac interactive binding (CRIB) domain and a carboxyl terminal kinase domain (2, 9, 10, 22, 35). Six human Paks, which fall into two subfamilies, have been identified. Group A Paks (Paks 1, 2, and 3) exhibit 80 to 90% sequence identity within their catalytic domains, whereas the recently identified group B Paks (Paks 4, 5, and 6), while highly related to each other, show only about 40 to 50% identity to the kinase domains of the group A Paks (9, 22). The group A Pak subfamily contains additional characteristic motifs, in particular an autoinhibitory domain located downstream of the CRIB domain which inactivates the catalytic function of these enzymes by intramolecular interaction with the kinase domain (6, 28, 30); this motif is absent in group B Paks.
As effectors of Rho family GTPases, Paks play an important role in the regulation of morphology and motility by regulating the actin cytoskeleton (13, 15, 25, 29, 34, 36). While we know in reasonable detail how group A Paks are regulated and what their targets are, we know much less about the group B Paks. Group B Paks are much less closely related to one another than are group A Paks, especially in the regions outside the CRIB and kinase domains. Of the group B Paks, Pak4 is the best characterized. Unlike that of Paks 1, 2, and 3, the interaction of Cdc42 with Pak4 is thought to regulate translocation of this kinase to the Golgi apparatus, rather than its activation (1). Pak4 is involved in reorganization of the actin cytoskeleton, but only one likely substrate (LIM kinase) has been identified thus far (7). Pak6 interacts with the androgen and estrogen receptors and has been reported by one group to translocate into the nucleus with the androgen receptor in response to androgen (27, 41). Pak5 is expressed primarily in neuronal tissues and induces neurite-like projections when overexpressed in NIE-115 cells (8). These data suggest that the group B Paks have disparate cellular functions.
Both group A and B Paks have been implicated in apoptosis, possessing either proapoptotic (Pak2) or antiapoptotic (Pak1 and Pak4) properties (18, 23, 31, 32, 38). Pak2 is cleaved during apoptosis, most likely by caspase 3; this cleavage leads to its activation, and the activated kinases in turn are thought to contribute to morphological and membrane changes that occur during apoptosis (26, 31, 40). In contrast Pak1 and Pak4, which are not cleaved by caspases, have been reported to protect cells from apoptosis (18, 31, 32, 38). The survival signal induced by Paks is thought to be related to phosphorylation of the apoptotic protein BAD on Ser-112 (Pak1 and Pak4) and Ser-136 (Pak1) (18, 31, 32, 38), though it is not known if these phosphorylations are directly catalyzed by Paks in vivo. BAD is a proapoptotic member of the BCL-2 family that has been shown to translocate between cytosol and mitochondrial membrane-based partners such as BCL-2 or BCL-xL. This complex formation inhibits the ability of BCL-2 and BCL-xL to block the release of cytochrome c from mitochondria, a critical step in the activation of the downstream caspase protease cascade (16, 19, 20). BAD is rapidly phosphorylated on two serine residues in response to a survival factor, interleukin-3. Activated Akt has been shown to phosphorylate the proapoptotic BAD protein on Ser-136, while a recent study identified mitochondrion-associated protein kinase A (PKA) as a BAD Ser-112-specific kinase (17). The phosphorylation of BAD results in its dissociation from complexes with BCL-2 and BCL-xL and its subsequent association with the cytosolic adapter protein 14-3-3τ (10, 11). The uncomplexed BCL-xL is then capable of suppressing cell death responses by blocking the release of mitochondrial cytochrome c (18).
Here, we investigate the biochemical and functional properties of Pak5, particularly with regard to its role in apoptosis. We show that Pak5 function differs from that of group A Paks such as Pak1, since it does not complement Ste20 and it is not activated by GTPases. We show that Pak5 induces resistance to apoptosis induced by two distinct stimuli: camptothecin (CA), a topoisomerase I inhibitor, and C2-ceramide (C2C), a well-known analog to a second messenger released from plasma membrane during apoptosis. Expression of Pak5 inhibits PARP and caspase 3 cleavage and promotes BAD phosphorylation on Ser-112, whereas the KD mutant does not protect against apoptosis. Pak5 phosphorylates BAD Ser-112 independently of PKA, and also promotes the phosphorylation of Ser-136, most likely via activation of Akt. Moreover, Pak5 prevents BAD translocation to mitochondria and more importantly, Pak5 localizes to mitochondria, and this localization is independent of kinase activity or the ability to bind Cdc42.
MATERIALS AND METHODS
Reagents and antibodies.
CA and myelin basic protein (MBP) were purchased from Sigma, and C2C, PKA myristoylated inhibitor, and polyclonal anticalnexin were from Calbiochem. Mouse monoclonal anti-c-myc (sc-40, clone 9E10), rabbit polyclonal anti-c-myc (clone A14), and anti-glutathione S-transferase (anti-GST) were from Santa Cruz Biotechnology. Mouse monoclonal antihemagglutinin (anti-HA) (12CA5J) was purchased from Babco. Rabbit polyclonal anti-BAD, anti-phospho-BAD to Ser-112, anti-BCL-xL, and anti-phospho-Akt to serine 473 were from Cell Signaling Inc. Anti-phospho-Ser-136 was from Upstate Biotechnologies. Mouse monoclonal anti-PARP antibody was from Pharmingen, and polyclonal anti-caspase 3 antibody was from Oncogene Research. Monoclonal anti-COX IV was purchased from Molecular Probes, and anti-HSP60 and anti-BCL-2 were from Transduction Laboratories. Antitubulin antibody RB43 specific for Saccharomyces cerevisiae was a gift from V. Guacci (Fox Chase Cancer Center, Philadelphia, Pa.). Polyclonal anti-Pak2 was purchased from Zymed.
Plasmids.
Pak5 cDNA was obtained from the RIKEN cDNA collection and subcloned in pCMV6 containing a Myc epitope tag (36). Kinase-dead (KD) (K478M) mutant of Pak5 (8) was constructed by site-directed mutagenesis (Stratagene QuikChange kit). These Pak5 variants along with the constitutively active allele of Pak5 (PAK5 S573N) were cloned into the S. cerevisiae expression vector pYes2 (Invitrogen). Additionally human Pak2 cloned into pYES2 was also used as a positive control. Plasmids encoding GST-tagged and HA-tagged Cdc42 L61 and N17 and Rac1 L61 and N17 were constructed as described elsewhere (33). The Pak4-encoding vector was obtained from Audrey Minden (Columbia University, New York, N.Y.). pEBG-BAD encoding for GST-BAD was purchased from Cell Signaling. pET vectors encoding His-tagged BAD, BAD S112A, and BAD S136A and pcDNA-HA-BAD were a gift from Michael Greenberg (Harvard Medical School, Boston, Mass.). The BAD S112A/S136A double mutant was obtained by site-directed mutagenesis. HIS-BAD proteins were inducibly expressed in Escherichia coli strain BL21(DE3)pLysS and purified over Ni-nitrilotriacetic acid resin (Qiagen).
Complementation analysis.
An S. cerevisiae strain bearing a deletion in STE20 (MATa ade2 his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ste20::KanMX) or the parental isogenic strain BY4741 (Research Genetics) was transformed by a lithium acetate procedure (17) with a galactose-inducible expression vector, pYES2 bearing either no insert or a cDNA encoding Pak-2, Pak5, Pak5 K478M (KD), or S573N (hyperactivated) mutants. Transformants were selected for growth on minimal medium lacking uracil. To perform qualitative mating assays, the transformants were serially diluted onto minimal raffinose plates lacking uracil (URA− plates). After the colonies had grown sufficiently the plates were replica plated onto either yeast nutrient broth (YNB) or Gal URA− plates and allowed to express the transgene for 5 h at 30°C. These plates were then replica plated onto YEPD plates that contained a lawn of the mating tester strain RSY16 (MATα ade2-1 leu1-2 lys2-1 trp5-20 ura1) (a gift from R. Strich, Fox Chase Cancer Center), and the cells were allowed to mate for 6 h before being replicated plated onto GN plates (0.17% YNB, 2% glucose, 0.5% ammonium sulfate). Mating efficiency was scored by the formation of prototrophic strains on the GN plates.
Protein isolation from S. cerevisiae and immunoprecipitation kinase assays.
Fifty-milliliter cultures of the transformants were grown in Raff URA− medium. Galactose was added to 2% and the cells were allowed to express the transgene for 5 h. At time zero and 5 h, 25-ml fractions were collected by centrifugation and frozen on dry ice. The pellets were resuspended in 1.0 ml of Tris-EDTA and divided as follows: 100 μl was transferred to an Eppendorf tube, collected by centrifugation, and lysed by boiling in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 10 min. The remaining 900 μl was collected by centrifugation and lysed with YPER buffer (Pierce) (5 ml/g of cell pellet) for 25 min at room temperature. The lysates were centrifuged, and the supernatant was transferred to an Eppendorf tube. The remaining pellet was resuspended in 500 μl of radioimmunoprecipitation (RIPA) buffer and subjected to four rounds of freeze-thaw in liquid nitrogen. The lysates were centrifuged for 10 min at 14,000 rpm, and the supernatant was transferred to the tube containing the YPER lysate supernatant. The combined lysates were used for immunoprecipitation using the anti-Myc polyclonal antibody A14 or the anti-Pak2 specific antibody overnight at 4°C. Protein A/G was added for 2 h and the immunoprecipitates were washed five times with lysis buffer (66% YPER buffer-33% RIPA buffer), then three times with kinase assay buffer. Although the YPER-RIPA lysis was effective, we could not immunologically detect expression of the transgenes in the lysates or in the immunoprecipitations suggesting a problem of limited solubility of the mammalian Paks in S. cerevisiae. Multiple extraction procedures failed to resolve this limitation. Kinase assays were performed as described below.
Cell culture, transfection and treatment.
All cell lines were grown at 37°C in 5% CO2 and cultured in Dulbecco's modified Eagle's medium for COS cells and Dulbecco's modified Eagle's medium-F-12 for CHO cells containing 10% fetal bovine serum, and puromycin (2 μg/ml; Sigma) for CHO stable cell lines transfected with either empty vector (pCMV6, control cells), wild-type (WT) Pak5, or KD Pak5 K478M. HMN1 cells were kindly provided by R. Pittman (University of Pennsylvania, Philadelphia). Cells were transfected using Lipofectamine (Gibco), and lysates were collected 48 h after transfection. CHO stable cell lines transfected with pcDNA-HA-BAD were treated 24 h after transfection with CA (10 μM) or C2C (50 μM).
GST pull down assay.
To perform Rac and Cdc42 binding assays, 50 μg of purified GST fusion proteins coupled to glutathione-Sepharose beads (Amersham Pharmacia Biotech) were incubated with buffer A (40 mM HEPES [pH 7.4], NP-40 1%, 1 mM EDTA, 150 mM NaCl) supplemented with 10 mM EDTA for 15 min at room temperature to release any nucleotide, washed with buffer A, incubated in buffer B (buffer A supplemented with 1 mM GTP-γS and 10 mM MgCl2) for 30 min at room temperature, and then washed with buffer B. Next, the proteins were incubated with 250 μl of Cos lysates for 3 h at 4°C. Precipitates were washed three times with lysis buffer (described below). Bound proteins were eluted in SDS sample buffer and subjected to immunoblotting with anti-Myc monoclonal antibody 9E10.
Immunoblot and immunoprecipitation.
Cos cells were transfected with appropriate vectors. Cell extracts were obtained in lysis buffer (50 mM Tris [pH 8], 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride, aprotinin [1 μg/ml]), protein concentration was assessed using bicinchoninic acid (Pierce) and equal amounts of proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore Corp.). Blots were blocked 1 h in Tris-buffered saline-0.1% Tween 20 supplemented with 5% nonfat milk, and incubated with primary antibodies for an hour or overnight at 4°C. After washing, blots were incubated with alkaline phosphatase-conjugated secondary antibodies (Jackson Laboratories), which were detected using the CPD Star reagent (New England Nuclear). The immunoprecipitation assay was carried out as follows: whole-cell lysates were incubated with polyclonal anti-Myc A14 overnight at 4°C and then 1 h at 4°C with protein A agarose beads (Pierce). Beads were collected by centrifugation and washed with the lysis buffer described above. Proteins were eluted by boiling in SDS sample buffer and subjected to immunoblotting, and probed with anti-HA (12CA5J).
Protein kinase assay.
Cos cells were transfected with appropriate expression vectors. Transfected cells were collected in lysis buffer 48 h after transfection and equal amounts of the Myc-tagged proteins, as assayed by Western blotting, were then immunoprecipitated with anti-Myc antibody A14 and protein A-agarose. After incubation, the immunoprecipitates were washed twice in lysis buffer and twice in a kinase buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 1 mM MnCl2, 10 mM MgCl2, 1 mM dithiothreitol) and then were incubated with 1.5 μg of MBP or His-tagged BAD recombinant proteins in the kinase buffer supplemented with 20 μM ATP and 5 μCi of [γ-32P]ATP for 30 min at 30°C. The reaction was terminated with SDS-PAGE sample buffer, followed by SDS-PAGE and autoradiography.
Apoptosis assays.
To estimate apoptosis of stably transfected cell lines, equal numbers of cells were seeded in six-well plates. Cells were treated 16 h with CA (10 μM) or C2C (50 μM), and collected for flow cytometry or used to prepare lysates. Cells were collected by trypsinization and, after combination with the floating cells, were labeled with annexin V and propidium iodide according to the manufacturer's recommendations (Clontech) for fluorescence-activated cell sorting analysis (Becton Dickinson). When the PKA inhibitor (PKI) was used, cells were pretreated 30 min with 1 μM before inducers were added for 16 h.
Cellular fractionation.
CHO cell lines were transfected with pcDNA-HA-BAD and after 24 h were treated for 16 h with CA (10 μM) or C2C (50 μM). Cellular fractionation was then performed as previously described (14). Briefly, cells were trypsinized and resuspended in isotonic mitochondrial buffer (MB) (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 10 mM HEPES [pH 7.5] supplemented with proteases inhibitors). Cells were broken by passages through a 25-gauge needle fitted on a syringe, and the suspension was centrifuged at 2,000 × g at 4°C. Supernatants containing heavy membranes were subjected to a 13,000 × g centrifugation at 4°C, the supernatant corresponding to the cytosolic fraction was centrifuged at 100,000 × g for 30 min, and the pellet containing mitochondria was resuspended in MB and submitted to a 500 × g centrifugation. The resultant supernatant was centrifuged 10 min at 10,000 × g to collect the mitochondrial fraction. A further purification step was performed for Pak's subcellular localization (24). The latter mitochondrial fraction was resuspended in MB and applied on a 1-to-2 M linear sucrose gradient and centrifuged 30 min at 60,000 × g using an SW28 Beckman rotor. The mitochondria, which banded at a region corresponding to 1.5 M were collected and diluted in MB to 0.25 M and pelleted at 15,000 × g for 15 min. The mitochondrial pellets were resuspended in MB. The S100 cytosolic fraction also generated the light microsomal pellet after centrifugation, which was resuspended in buffer H (20 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 6 mM β-mercaptoethanol [pH 7.5], 1% SDS).
Immunofluorescence.
HMN1 cells seeded on coverslips were transiently transfected with empty vector or vectors encoding Myc-tagged WT Pak5 or KD Pak5 (K478M). HMN1 and CHO cells were stained 15 min at 37°C with a 5-ng/ml concentration of the mitochondrion-specific dye MitoTracker Red CMXRos (Molecular Probes Inc.) 24 h after transfection and then fixed in 4% paraformaldehyde. Cells were fixed, permeabilized in PBS-0.1% NP-40, and blocked 30 min with 10% bovine serum albumin. Cells were stained for the presence of Pak5 with rabbit polyclonal anti-Myc antibody (A14) and incubated with anti-rabbit immunoglobulin G conjugated with fluorescein isothiocyanate (FITC) (Jackson Laboratories). Cells were then washed, mounted and photomicrographs were obtained using a Bio-Rad radiant 2000 laser scanning confocal microscope with a Plan APO 60× water immersion objective (1.20 numerical aperture; Nikon E800 Eclipse).
RESULTS
Complementation analysis of Pak5.
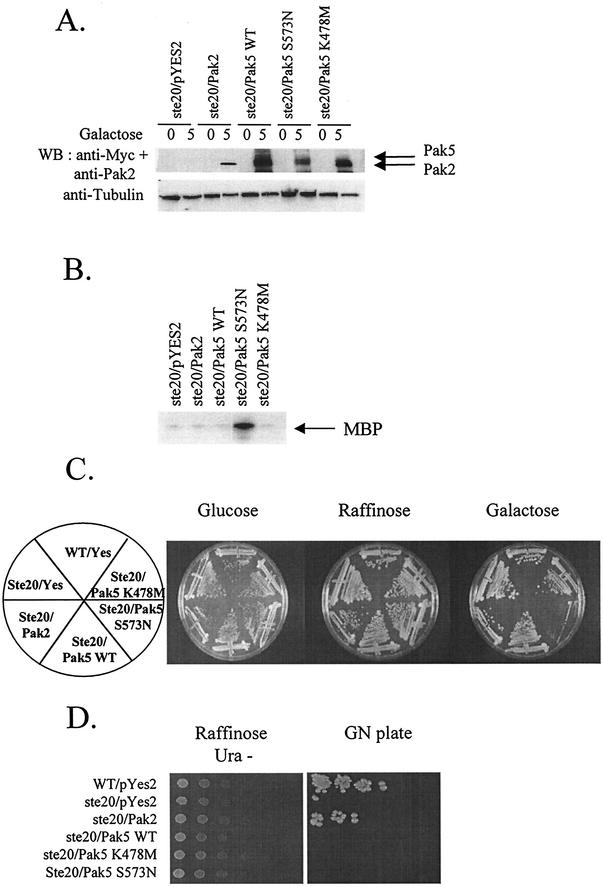
Group A mammalian Paks are able to replace the function of Ste20p in S. cerevisiae (3, 4). The mammalian group B Paks, though clearly distinct in structure and biochemical properties from group A Paks (see below), are nevertheless as closely related as group A Paks to Ste20p. It was therefore of interest to determine if group B Paks can functionally substitute for Ste20p. We transformed a haploid STE20-null strain of S. cerevisiae with empty vector or vectors designed for inducible expression of human Pak2 or Pak5 alleles. Immunoblot analysis revealed that the transgenes are expressed when cells are grown in galactose medium (Fig. 1A). We then carried out immunocomplex-kinase assays to assess the activity of the Pak transgenes. As expected, Pak5 S573N was far more active than either WT Pak5 or Pak2 (Fig. 1B), and expression of this hyperactivated allele was cytostatic (Fig. 1C). This effect is similar to the effect of constitutively active alleles of S. pombe Pak1 when expressed in S. cerevisiae (33, 39). A mating assay was then carried out to determine complementation (Fig. 1D). WT and STE20-null cells expressing Pak2 are competent for mating, whereas cells expressing any of the Pak5 alleles are not. These results indicate that the functions of human Pak2 and Pak5 are distinct.
FIG. 1.
Complementation analysis of Pak5. ste20-null or a control strain of S. cerevisiae was transformed with a galactose-inducible expression vector bearing either no insert or a cDNA encoding Pak2, Myc-tagged WT Pak5, hyperactivated mutant (S573N), or K478 M (KD) mutant. Transformants were selected for growth in YNB URA− raffinose medium, galactose was added to 2% at time zero, and the cultures were induced for 5 h before harvesting. (A) Expression of the transgenes. Cells were lysed before and 5 h after induction, then assayed for protein expression by immunoblotting with monoclonal anti-Myc 9E10 or anti-Pak2. Equal protein levels was assessed using an antitubulin antibody. (B) Kinase assay. Cultures tested in panel A and obtained after 5 h of induction were lysed and immunoprecipitated with a polyclonal anti-Myc antibody or polyclonal anti-Pak2 and assayed for kinase activity as described in Materials and Methods. (C) Transformants were streaked onto YNB glucose, YNB raffinose, and YNB galactose URA− plates and incubated at 30°C before being photographed. (D) To perform semiquantitative mating assays, the transformants were serially diluted onto YNB raffinose URA− plates. After the colonies had grown sufficiently, the plates were replicated onto YNB Gal URA− for 5 h to allow expression of the transgenes. The cells were then replica-plated onto YEPD plates containing a lawn of the mating tester strain, and allowed to mate for six hours before being replicated onto GN plates. Mating efficiency was scored by the formation of prototrophic strains on the GN plates.
Biochemical properties of Pak5.
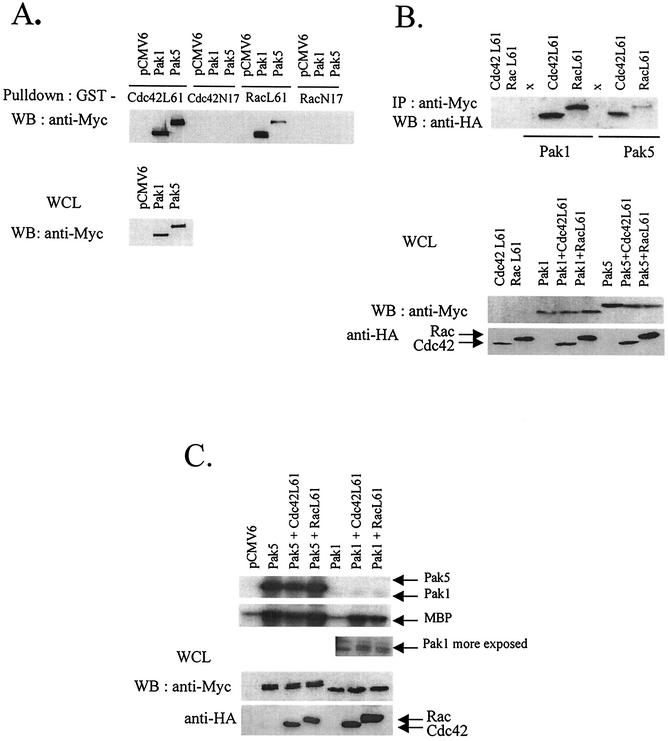
Sequence analysis indicates that Pak5 has a putative GBD/CRIB motif similar to that of Pak4. To determine whether Pak5 interacts with the GTPases, a pull-down assay was performed in which lysates from Cos cells transfected with empty vector or a Myc-Pak5 or a Myc-Pak1 expression vectors were incubated with GST-Cdc42 L61 (constitutively active) or -N17 (dominant-negative) or GST-Rac1 L61 or -N17. As shown in Fig. 2A, Pak5 interacts with both Cdc42 and Rac1, suggesting that Pak5 is a target for these GTPases. However, contrary to Pak1, but similar to Pak4 (1), Pak5 binds more strongly to Cdc42 than to Rac1.
FIG. 2.
Biochemical properties of Pak5. (A) COS cells were transfected with equal amounts of expression vectors encoding WT Myc-tagged Pak1 and Pak5 or empty vector. After transient expression, Myc-Pak1 and Myc-Pak5 were pulled down from whole-cell lysate (WCL) with GST-Cdc42 and GST-Rac loaded with GTP, Western blotting was then performed using an anti-Myc antibody. The expression level of Pak1 and Pak5 in the lysates used for the pull-down is shown in the bottom panel, expression was detected by Western blotting using an anti-Myc antibody. (B) COS cells were cotransfected with equal amounts of expression vectors encoding HA-tagged Cdc42 L61 or HA-Rac L61 or Myc-tagged Pak1 or -Pak5 alone or plus HA-tagged Cdc42 L61 or HA-Rac L61. After transient expression, Myc-Pak1 and Myc-Pak5 were immunoprecipitated from total lysates with an anti-Myc antibody and protein A-agarose. Western blotting was then performed and the complexes were revealed with an anti-HA antibody. The bottom panel shows the expression of Pak1 and Pak5, revealed with an anti-Myc and Cdc42 and Rac, revealed with an anti-HA in the WCL. (C) COS cells were transfected with expression vectors for Myc-tagged Pak1 or -Pak5 or cotransfected with Pak1 or Pak5 cDNAs and the HA-tagged GTPases Cdc42 and Rac. Myc-tagged Pak1 and Pak5 were immunoprecipitated with an anti-Myc antibody and the kinase assay was performed as described in Materials and Methods using [γ-32P]ATP and MBP as a substrate. Reaction products were separated with SDS-PAGE and subjected to autoradiography. The bottom panel shows the expression levels of proteins in the WCL prior to immunoprecipitation.
To confirm these results in cells, COS cells were transfected with Pak5 or Pak1 alone or in combination with Cdc42 L61 or Rac L61 and immunoprecipitations were performed on the lysates (Fig. 2B). As in the pull-down assay, the coprecipitation experiments show that Pak5 binds to both GTPases, but with a marked preference for Cdc42. These results are consistent with those reported by Dan et al. (7).
The group A Paks requires binding of GTPases to be activated whereas Pak4 has been shown in vitro to autophosphorylate and to phosphorylate Histone H4 (5). We assessed Pak5 kinase activity using MBP as a substrate (Fig. 2C). COS cells were transfected with Myc-tagged Pak5 expression vector or Myc-tagged Pak1, used as a positive control. GTPases expression vectors were also cotransfected. Equal amounts of Pak5 and Pak1 were immunopurified from cell lysates and incubated with MBP and [γ-32P]ATP in kinase buffer. Autophosphorylation of Pak5 and transphosphorylation of MBP in Fig. 2C shows that Pak5 has a strong autophosphorylation activity compared to Pak1 and phosphorylates MBP in the absence of GTPases. On the contrary, Pak1 requires activation by GTPases in order to phosphorylate MBP, and does so at orders of magnitude less than Pak5 (Fig. 2C, inset).
Pak5 induces resistance to apoptosis induced by ceramide or CA.
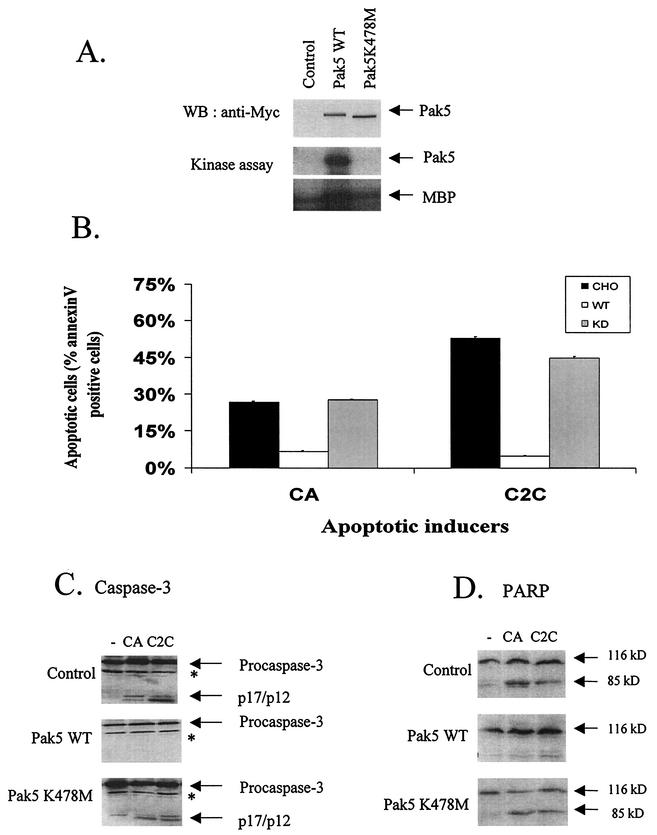
As many of the Paks have previously been shown to either induce (Pak2, once cleaved by caspase 3) (26, 31, 40) or inhibit (Pak1 and Pak4) (18, 32, 38) apoptosis, we sought to determine if Pak5 also plays a role in this process. In order to examine the role of Pak5 in cell survival, CHO stable cell lines were generated expressing either WT Pak5 or KD Pak5 (K478M) (8) or empty vector (control). At least three independent stable clones were analyzed for each construct. Representative data for protein expression and kinase activity data are shown (Fig. 3A). Consistent with the data from transiently transfected COS cells, WT Pak5 in CHO cells exhibits considerable basal activity, whereas the K478 M mutant is inactive.
FIG. 3.
Pak5 induces resistance to apoptosis. (A) Expression and activity of Pak5 in CHO stable cell lines. Stable CHO cell lines were established to study apoptosis. Equal amounts of lysates from stable CHO cells containing empty vector, Myc-WT Pak5, or Myc-KD Pak5 were analyzed for the expression of Pak5 proteins by Western blotting using anti-Myc antibody. Kinase activity was assessed by performing a kinase assay. Myc-tagged proteins were immunoprecipitated with anti-Myc and using MBP as a substrate. (B) Stable cell lines expressing Pak5 are resistant to apoptosis. Cells were treated with C2C (50 μM) and CA (10 μM) for 16 h and were collected and stained with annexin V and propidium iodide. Apoptosis was assessed by flow cytometry. The percentages of total cells positive for annexin V are shown on the graph, and represent the average of three experiments. (C and D) Caspase 3 and PARP cleavage. Cells were treated as indicated above with C2C and CA. Cells were lysed and Western blotting was performed using anti-caspase 3 and anti-PARP antibodies. *, nonspecific band.
To induce apoptosis, CHO cells were treated with two inducers: CA, a topoisomerase I inhibitor, and C2C, a well-known analog to a second messenger released from plasma membrane during apoptosis. Cell survival was assessed with annexin V staining after a 16-h treatment with CA or C2C. As shown in Fig. 3B, CA and C2C induce 35 and 50% apoptosis, respectively, in control cells, whereas no apoptosis is detectable in WT Pak5 CHO cells. In contrast, expression of KD Pak5 does not protect against apoptosis, and we observe a percentage of apoptotic cells that is similar to the CHO cells when cells are treated with CA, and to a lesser extent with C2C. These data show that Pak5 kinase activity is responsible for the resistance to apoptosis.
We examined PARP and caspase 3 cleavage, two biochemical markers of apoptosis, in the stable cell lines. After a 16-h treatment with C2C and CA, PARP and caspase 3 were cleaved in control and KD Pak5 cell lines (Fig. 3C and D), whereas WT Pak5-expressing cell lines, which are resistant to apoptosis, retain full-length PARP and caspase 3 in both untreated and induced cells. This result indicates that the apoptotic signal leading to the release of cytochrome c from the mitochondrion, subsequently activating caspase 9 and caspase 3, is blocked downstream of the mitochondrion when Pak5 is expressed.
Pak5 phosphorylates BAD.
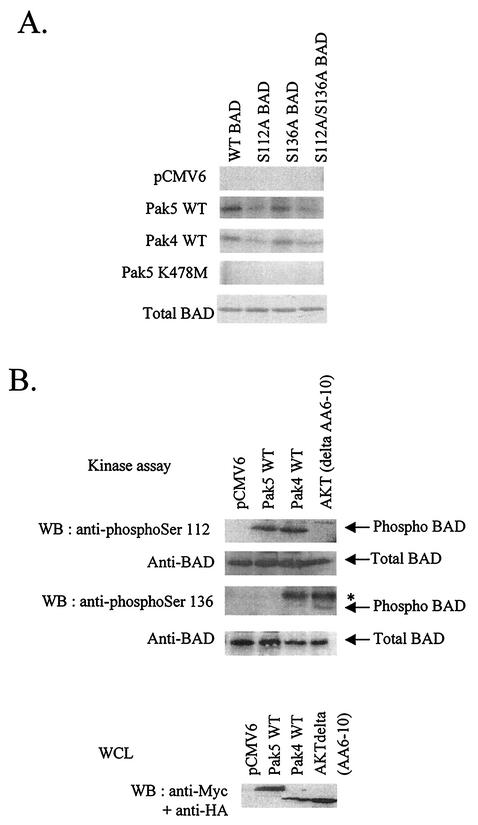
Activated Pak1 and Pak4 have been shown to phosphorylate BAD on Ser-112 and Ser-136 and Ser-112, respectively (18, 32). We addressed whether BAD is a target for Pak5, BAD being a key proapoptotic molecule. To analyze BAD phosphorylation by Pak5, COS cells were transfected with empty vector, WT Pak5, KD Pak5 or WT Pak4 as a positive control. In vitro kinase assay was carried out in which His-tagged recombinant WT BAD, S112A or S136A mutants or double mutant S112A/S136A proteins were used as substrates for immunopurified Pak proteins (Fig. 4A). These results indicate that WT Pak5 and Pak4 cannot phosphorylate BAD S112A mutant or the double mutant S112A/S136A, whereas they are able to phosphorylate WT BAD or the S136A mutant. Therefore, Pak5 phosphorylates BAD mainly on Ser-112, although BAD mutation on Ser-112 does not completely abolish its phosphorylation, indicating that Pak5 could target to a lesser extent additional residues. In order to confirm this result, we used phosphospecific antibodies directed against Ser-112 or Ser-136 to determine the status of the 112 and 136 sites in an in vitro non radioactive kinase assay (Fig. 4B). These data show that Pak5 phosphorylates BAD on Ser-112, in agreement with the radioactive kinase activity assay.
FIG. 4.
Pak5 phosphorylates BAD on serine 112 in vitro. (A) COS cells were transfected with empty vector or Myc-Pak5 or HA-Pak4. An equal amount of cell extracts were used for in vitro kinase assay. Myc-tagged proteins were immunoprecipitated with anti-Myc, the immunocomplexes were incubated in the presence of [γ-32P]ATP and equal amounts of recombinant His-tagged BAD (WT, S112A, S136A, and S112A/S136A) as substrates as shown on the bottom panel. After SDS-PAGE, phosphorylated BAD was detected by autoradiography. (B) The kinase assay was performed using His-tagged WT BAD, and immunocomplexes from lysates of COS cells transfected with Myc-Pak5, HA-Pak4 and HA-Akt (activated mutant, del AA6-10). After Western blotting, phosphorylated BAD was detected with anti-phospho-Ser 112 and anti-phospho-Ser 136, while total BAD was detected with an anti-BAD antibody. Bottom panel shows the expression of the different proteins in the whole-cell lysate (WCL). * shows cross-reaction of anti-phospho-Ser 136 BAD with anti-HA immunoglobulin G light chain.
Pak5 prevents BAD dephosphorylation during apoptosis.
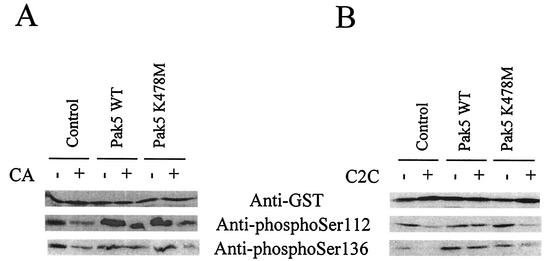
As BAD is phosphorylated by Pak5 in vitro, we determined whether Pak5 expression causes sustained BAD phosphorylation in cells treated with apoptotic agents. CHO cell lines were transfected with a vector encoding a GST-BAD protein. After a 16-h treatment with CA (A) or C2C (B), a GST pull down was performed on lysates to retrieve GST-BAD. The expression level of BAD was assessed by Western blotting with an anti-GST antibody and BAD phosphorylation status was assessed by Western blotting using phosphospecific antibodies. As shown in Fig. 5A and B, in treated cells, BAD is dephosphorylated on Ser-112 in control cells as well as in cells expressing KD Pak5, whereas no dephosphorylation on Ser-112 is observed in cells expressing WT Pak5. Notably, BAD Ser-136 is also dephosphorylated in control and KD Pak5 treated cells, but not in WT Pak5 cells. Because Pak5 does not phosphorylate BAD at Ser-136 in vitro (Fig. 4), these results suggest that Pak5 influences BAD phosphorylation by both direct and indirect mechanisms.
FIG. 5.
Pak5 prevents BAD dephosphorylation in vivo. CHO control cells and stable cell lines expressing WT Pak5 or KD Pak5 were transfected with pEBG-BAD and were treated with CA (10 μM) or C2C (50 μM) 24 h after transfection. Equal amounts of cell extracts were analyzed by Western blotting after pull-down assay of GST-BAD, using an anti-GST antibody to assess BAD expression level, and antibodies directed against phospho-Ser 112 and phospho-Ser 136 to assess BAD phosphorylation status.
Analysis of Pak5-mediated BAD phosphorylation in cells.
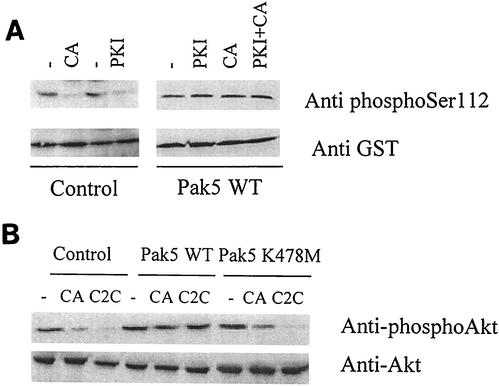
Although Pak5 phosphorylates BAD at Ser-112 in vitro, and maintains BAD phosphorylation in cells treated with apoptotic agents, it is possible that Pak5 acts indirectly by activating other kinases that phosphorylate BAD. As PKA has been reported to catalyze BAD phosphorylation at Ser-112 (21), we sought to determine if this kinase is required for Pak5-mediated BAD phosphorylation in cells. Control and Pak5-expressing CHO cells were pretreated with a PKI and then treated with CA, and BAD phosphorylation on Ser-112 was assessed with a phospho-specific antibody. As shown in Fig. 6A, in control cells, CA and PKI both induce BAD dephosphorylation at Ser-112. In contrast, basal levels of BAD Ser-112 phosphorylation are elevated in cells expressing Pak5, and neither CA nor PKI reduces Ser-112 phosphorylation. Identical results were obtained using ceramide as the apoptotic stimulus (data not shown). These results show that Pak5 does not act through PKA to phosphorylate BAD at Ser-112.
FIG. 6.
Pak5 phosphorylates BAD independently of PKA and prevents Akt dephosphorylation on serine 473. (A) Stable cell lines were transfected with pEBG-BAD and after 24 h were pretreated 30 min with a specific myristoylated protein kinase A inhibitor (1 μM) before CA (10 μM) was added for 16 h. GST-BAD was then retrieved from the lysates and an anti-phospho-Ser-112 BAD antibody was used for the Western blot to assess BAD phosphorylation status. (B) Stable cell lines were treated for 16 h with CA (10 μM) or C2C (50 μM). After lysis, Western blotting was performed using an anti-phospho-Ser 473 specific antibody.
We also assessed the role of Pak5 on Akt phosphorylation, since Akt has been reported to phosphorylate BAD at Ser-136 (11, 12). Control cells, or cells expressing WT or KD Pak5 were treated with vehicle, CA or C2C, and Akt activity was measured by assessing phosphorylation on Akt Ser-473. Figure 6B shows that both apoptotic inducers reduce Akt activity in CHO and KD Pak5-expressing cells. However, expression of WT Pak5 prevents Akt dephosphorylation. These results suggest that the Pak5-mediated phosphorylation of BAD at Ser-136 may be mediated through Akt.
Pak5 prevents BAD translocation to mitochondria.
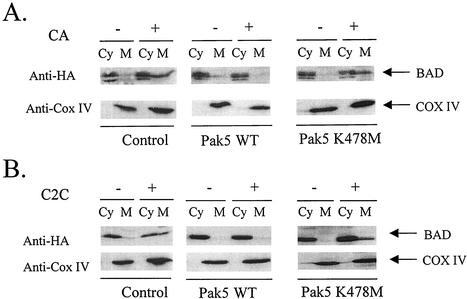
Under normal condition, phosphorylated BAD is sequestered by 14-3-3τ in the cytosol (42). In response to apoptotic stimuli, BAD is dephosphorylated and translocates to the mitochondrial membrane where it binds to the antiapoptotic proteins BCL-2 and BCL-xL (18). Having demonstrated that Pak5 phosphorylates BAD on serine 112 and indirectly on serine 136, we determined if this activity of Pak5 was sufficient to maintain BAD in the cytosol and particularly if it prevents BAD translocation to mitochondria, and therefore the apoptotic cascade. We transfected control, WT Pak5, and KD Pak5 stable cell lines with a pcDNA-BAD plasmid, and treated cells with inducers, CA or C2C, in order to induce apoptosis, and subsequent BAD translocation to mitochondria. We found that BAD translocates to mitochondria in both control and KD Pak5 CHO cells, as shown by the presence of BAD in the mitochondrial fraction, whether apoptosis is induced by CA (Fig. 7A) or C2C (Fig. 7B). In contrast, no translocation occurs in the WT Pak5 cell lines, as suggested by the absence of BAD in the mitochondrial fraction of these cells. This is consistent with the phosphorylation of serine 112 by Pak5 and shows that Pak5, by its phosphorylation activity, can prevent BAD translocation to mitochondria.
FIG. 7.
Pak5 prevents BAD translocation to mitochondria. CHO control cells and stable cell lines expressing WT Pak5 and KD Pak5 were transfected with HA-BAD and 24 h later treated with CA or C2C for 16 h. Cells were collected by trypsinization and a cellular fractionation was performed to isolate mitochondria as described in Materials and Methods. Equal amount of proteins were separated by SDS-PAGE. BAD was detected by Western blotting using an anti-HA antibody. Equal loading of the mitochondrial pellet was assessed with an anti-COX IV antibody.
Pak5 localizes to mitochondria.
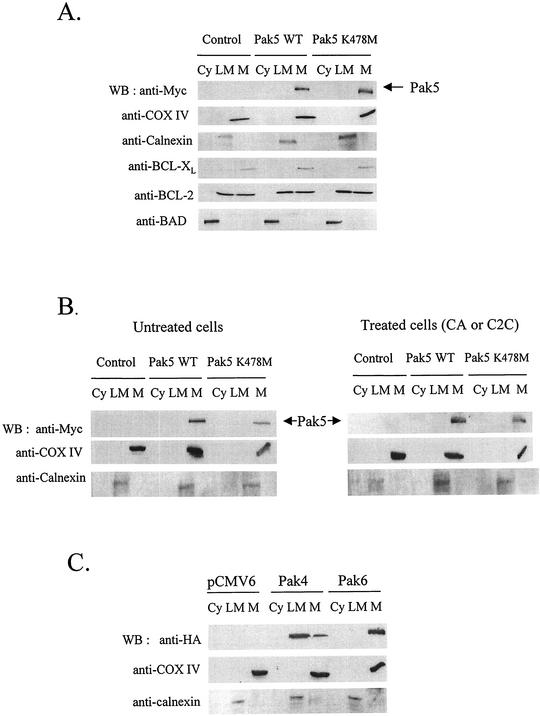
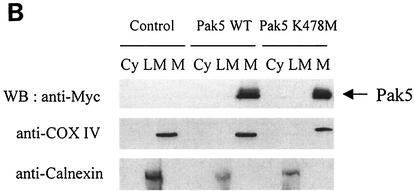
As Pak5 stimulates BAD phosphorylation and prevents its translocation to mitochondria, we investigated whether Pak5 acts in the cytosol or on mitochondria. We assessed Pak5 localization by performing cellular fractionation. As shown in Fig. 8A, we found that both WT and KD Pak5 are found exclusively in the mitochondrial fraction. Immunoblots with specific marker antibodies (COX IV for mitochondria, calnexin for endoplasmic reticulum/light microsomes) show that the subcellular fractions are reasonably pure. Nuclear fractions are not shown because they are contaminated with unbroken cells and aggregated mitochondria, and therefore are not representative of a pure nuclear fraction. The purity of the fractions was confirmed by reprobing the blots with antibodies against proteins known to be cytosolic (BAD [42]), mitochondrial and endoplasmic reticulum (BCL-2 [24]) and mitochondrial (BCL-xL [24]).
FIG. 8.
Pak5 is present in the mitochondrial fraction. A cellular fractionation was performed on CHO stable cell lines as described in Materials and Methods to assess Pak5 presence in the cellular compartments. (A) Equal amount of proteins from cytosolic (Cy) S100, light microsomes (LM) and mitochondrial (M) fractions were loaded on a gel, and Western blotting was performed using anti-Myc antibody. The mitochondrial fraction purity was assessed using an anti-COX IV and anticalnexin antibodies. The fractions were also subjected to immunoblotting with anti-BAD, anti-BCL-2 and anti-BCL-xL antibodies. (B) CHO cells expressing vector alone, WT Pak5, or KD Pak5 were treated with CA (10 μM) or C2C (50 μM) for 16 h. A cellular fractionation was then performed. Equal amount of proteins from cytosolic (Cy) S100, light microsomes (LM) and mitochondrial (M) fractions were loaded on a gel, and Western blotting was performed using anti-Myc antibody. The mitochondrial fraction purity was assessed using an anti-COX IV and anticalnexin antibodies. (C) CHO cells were transfected with pCMV6, HA-Pak4 or HA-Pak6, and a subcellular fractionation was performed 48 h after transfection as described above.
We next treated the CHO stable cell lines with CA or C2C to see if Pak5 relocalizes during the apoptotic process. As shown in Fig. 8B, apoptosis does not modify WT Pak5 and KD Pak5 localization. Thus, Pak5 is a constitutive resident of the mitochondria.
We next studied the localization of other group B Paks in order to determine if either of these could also localize to mitochondria. We transfected CHO cell lines with vectors encoding HA-Pak4 and HA-Pak6. The results show that Pak4 is predominantly localized in the light microsome fraction, and a smaller fraction is also found in the mitochondrial fraction (Fig. 8C). Surprisingly, Pak6, like Pak5, was found exclusively in the mitochondrial fraction.
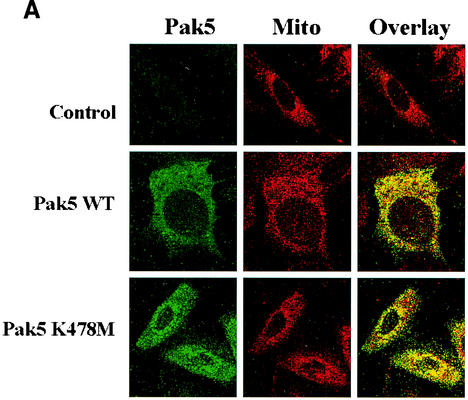
To confirm the mitochondrial localization of Pak5, immunofluorescent staining was performed on the CHO stable cell lines. As shown in Fig. 9A, there is substantial colocalization of Myc-WT Pak5 with mitochondria, as assessed using MitoTracker. Similar results were obtained when other mitochondrial markers, such as HSP60 and COX-IV, were examined in Pak5-transfected cells (data not shown). As in the cell fractionation studies, kinase activity is not required for this colocalization, as a substantial proportion of the KD mutant also localized to mitochondria. We also tested in CHO cells the localization of a mutant of Pak5 that bears two point mutations (K19L and K23L) in the CRIB domain where Cdc42 binds (1, 31). These mutations do not affect Pak5 mitochondrial localization (data not shown).
FIG. 9.
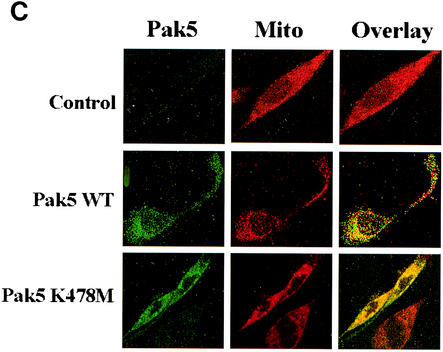
Pak5 colocalizes with mitochondria. CHO stable cell lines (A) expressing WT Pak5 or KD Pak5 were labeled as described in Materials and Methods. The fields shown were analyzed independently by digital confocal fluorescence microscopy at the appropriate wavelength for FITC (Pak5) and MitoTracker Red CMXRos (Mito), and the two images were overlaid (Overlay). (B) HMN1 cells were transfected with the empty vector (control) or with a vector carrying WT Pak5 or Pak5 K478M cDNA. A cellular fractionation was then performed 48 h after transfections as described in Materials and Methods to assess Pak5 presence in the cellular compartments. Equal amount of proteins from cytosolic (Cy) S100, light microsomes (LM) and mitochondrial (M) fractions were loaded on a gel, and Western blotting was performed using anti-Myc antibody. The mitochondrial fraction was assessed using anti-COX IV and anticalnexin antibodies. (C) HMN1 cells were transfected with empty vector or Myc-tagged WT Pak5 or KD Pak5 were stained as described in Materials and Methods. The fields shown were analyzed independently by fluorescence microscopy at the appropriate wavelength for FITC (Pak5) and MitoTracker Red CMXRos (Mito), and the two images were overlaid (Overlay).
As Pak5 is mainly expressed in the brain, we asked whether the localization to mitochondria is also present in neuronal cells. We transiently transfected HMN1 cells with empty vector or Pak5 WT- or K478M mutant-encoding vectors and a mitochondrial isolation was then performed. As shown in Fig. 9B, Pak5 is exclusively localized in the mitochondrial fraction. Immunofluorescent staining was performed on the transfected cells using MitoTracker and shows a colocalization of Pak5, both WT and KD, with mitochondria (Fig. 9C). Same results were obtained with COX IV and HSP60 (data not shown). Therefore, we conclude that the mitochondrial localization is not dependent on the cell type, and occurs in cell types that are expected to express high levels of endogenous Pak5.
DISCUSSION
In this work, we show that a group B Pak, Pak5, has properties that are distinct from all other Paks described to date. Pak5 is distinguished from group A Paks by its inability to complement STE20 function in budding yeast, by its high basal activity that is not regulated by GTPases, and by its marked preference for binding Cdc42 over Rac. While Pak5 shares some common properties with the group B enzyme Pak4, such as conferring protection from apoptotic stimuli and the ability to phosphorylate BAD on Ser-112, Pak5 is localized to mitochondria. These features suggest that Pak5 plays a different role than Pak4 in regulating apoptosis.
Consistent with the absence of apoptosis in cells expressing Pak5, we observed an absence of caspase 3 cleavage and the subsequent PARP cleavage, two key features of programmed cell death. In contrast, expression of a KD mutant of Pak5 does not protect against apoptosis, and does not prevent caspase 3 and PARP cleavage. Therefore, we conclude that Pak5 kinase activity is responsible for resistance to apoptosis. Interestingly, activation of the group A enzyme, Pak2, is associated with proapoptotic rather than antiapoptotic effects. One key difference between proapoptotic kinases such as Pak2 and antiapoptotic kinases such as Pak1, Pak4, and Pak5 is that the former contains a caspase site and is sensitive to cleavage by caspase 3 (26, 31), whereas the latter lack such sites and are not known to be cleaved. Possibly, the generation of a separated kinase domain, such as occurs following Pak2 cleavage, results in relocalization and acquisition of new substrates that contribute to cell death. Therefore, we suspect that the N terminal portion of Pak5 regulates the localization and substrate specificity of this kinase, and is required for its pro-survival effects.
One of the mechanisms by which Pak5 induces resistance to apoptosis is the phosphorylation of BAD. We show that Pak5 phosphorylates BAD in vitro and in vivo, as it has also been observed for Pak1 and Pak4 (18, 31, 32, 38). These phosphorylations occur at Ser-112, and perhaps, to a lesser extent, on additional Ser residues. While Pak5 may have additional targets that are relevant to its antiapoptotic effects, the phosphorylation of BAD is likely to be germane in this setting. Although Pak5 can phosphorylate BAD in vitro and induce BAD phosphorylation in cells, there are several other kinases that also exhibit these properties. To date, PKA has been identified as a major BAD Ser-112 kinase, while Akt has been reported to be a major Ser-136 kinase. Our data suggest that the Pak5-stimulated phosphorylation of BAD at Ser-112 is direct, as Pak5 acts on this site in vitro and inhibitors of PKA do not affect the ability of Pak5 to maintain BAD Ser-112 phosphorylation in cells treated with inducers of apoptosis. In contrast, the Pak5-stimulated phosphorylation of BAD at Ser-136 may be mediated through Akt, as Pak5 expression is associated with elevated activity of Akt. These results suggest that Pak5 and PKA are independent mediators of BAD phosphorylation (Fig. 10). They also suggest that Pak5 signaling differs from Pak1 which appears to act downstream of Akt (38).
FIG. 10.
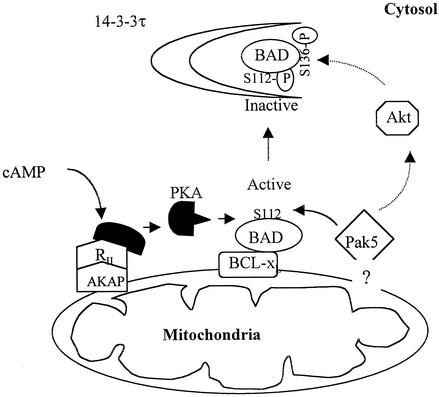
Model of Pak5 in cell survival signaling.
Pak5 is localized to mitochondria, either directly or via interactions with a tethering protein. There, it encounters BAD, which it phosphorylates at Ser-112. The PKA complex (comprising a catalytic subunit, a regulatory subunit [RII], and a mitochondrial-tethering subunit [AKAP]) independently targets the Ser-112 site. Pak5 also induces, through an unknown mechanism, the activation of Akt, which phosphorylates BAD at Ser-136. Phosphorylated BAD complexes with 14-3-3τ in the cytosol.
The phosphorylation of BAD by Pak5 is likely to occur at the mitochondrion, as Pak5 appears to be a constitutive resident of this organelle, and BAD translocates to mitochondria following apoptotic stimuli. The predominantly mitochondrial localization of Pak5 is independent of cell type, as we were able to see that same colocalization of Pak5 in CHO, HMN1 and NIH-3T3 cells (data not shown). Pak5 is not recruited to mitochondria specifically during apoptosis, and its residence there is independent of kinase activity or its ability to bind Cdc42. In this respect, Pak5 resembles the mitochondrion-anchored PKA, although we do not know if Pak5 is anchored to the mitochondrion directly or bound to a mitochondrion-anchored protein.
To date, localization of other group B Pak isoforms has not been studied in detail; therefore, we also assessed Pak4 and Pak6 localization. Surprisingly, we found that Pak6, and, to a lesser extent, Pak4, also localized to mitochondria. The bulk of Pak4, however, is detected in the light microsome fraction (Fig. 8C), in agreement with the findings of Abo et al. (1), who showed that Pak4 translocates to Golgi apparatus when coexpressed with Cdc42. Pak6 was previously shown to be cytosolic and to shuttle to the nucleus when coexpressed with androgen and estrogen receptors (27), but has not been previously reported to localize to mitochondria. Our data suggest that mitochondrial localization is a common feature of group B Paks.
Mammalian group B Paks appear to localize to many distinct cellular compartments, unlike group A Paks such as Pak1, which are found mostly in areas of active actin dynamics, such as leading edge lamellipodia (37) or the group B X-Pak5 from Xenopus, which has been found in association with microtubules (5). The mammalian group B Paks are in general less similar to one another than are the group A Paks, and it is likely that this diverged sequence reflects diverged cellular function. While we do not yet know what domain on Pak5 targets the mitochondrion, the N terminus is probably involved. Paks 4, 5, and 6 differ substantially in this noncatalytic N-terminal domain, whereas the C-terminal kinase domains are very similar. Moreover preliminary data from our lab shows that the isolated N terminus of Pak5 localizes to mitochondria. Whether the Cdc42 binding domain, which is located at the extreme N terminus of the protein, is involved is currently unknown but appears unlikely as Cdc42 is not known to localize to this organelle.
Pak5 expression is restricted to the brain and overexpression of this kinase has been shown to induce neurite outgrowth in N1E-115 cells (8). It is likely that such kinase activity is tightly controlled in central nervous system, as 95% of neurons undergo apoptosis during embryogenesis. How is this kinase regulated? Pak5 has unusually high basal activity compared to other group A or group B Paks. While Pak5 binds Cdc42, this interaction has no effect on kinase activity in vitro. This finding suggests that the binding to Cdc42 could have a role other than direct activation. However, it is also possible that there are endogenous inhibitors of Pak5 that regulate its activity under physiological conditions. Provided that adequate sera can be raised to the endogenous protein, we plan to examine these and related issues in the near future.
Acknowledgments
We thank Audrey Minden, Michael Greenberg, Randall Pittman, and Randy Strich for their generous donation of reagents and Edna Cukierman and Jonathan Boyd for assistance in imaging.
This work was supported by grants from the National Institutes of Health (RO1 GM-54168 and CA-09035-27) and the American Cancer Society (CB-189), as well as CORE Grant CA-06927 and an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Abo, A., J. Qu, M. S. Cammarano, C. Dan, A. Fritsch, V. Baud, B. Belisle, and A. Minden. 1998. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17:6527-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagrodia, S., and R. A. Cerione. 1999. PAK to the future. Trends Cell Biol. 9:350-355. [DOI] [PubMed] [Google Scholar]
- 3.Bagrodia, S., S. Taylor, C. L. Creasy, J. Chernoff, and R. A. Cerione. 1995. Identification of a murine p21Cdc42/Rac activated protein kinase (PAK). J. Biol. Chem. 270:22731-22738. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. L., L. Stowers, M. Maer, J. Trejo, S. Coughlin, and J. Chant. 1996. Human Ste20 homologue hPak1 links GTPases to the JNK MAP kinase pathway. Curr. Biol. 6:598-605. [DOI] [PubMed] [Google Scholar]
- 5.Cau, J., S. Faure, M. Comps, C. Delsert, and N. Morin. 2001. A novel p21-activated kinase binds the actin and microtubule networks and induces microtubule stabilization. J. Cell Biol. 155:1029-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong, C., L. Tan, L. Lim, and E. Manser. 2001. The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J. Biol. Chem. 276:17347-17353. [DOI] [PubMed] [Google Scholar]
- 7.Dan, C., A. Kelly, O. Bernard, and A. Minden. 2001. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276:32115-32121. [DOI] [PubMed] [Google Scholar]
- 8.Dan, C., N. Nath, M. Liberto, and A. Minden. 2002. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol. Cell. Biol. 22:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dan, I., N. M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220-230. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, R. H., and G. M. Bokoch. 1999. p21-activated protein kinase: a crucial component of morphological signaling? Trends Biochem. 24:350-355. [DOI] [PubMed] [Google Scholar]
- 11.Datta, S. R., H. Dudek, X. Tao, S. Masters, H. Fu, Y. Gotoh, and M. E. Greenberg. 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91:231-241. [DOI] [PubMed] [Google Scholar]
- 12.del Peso, L., M. Gonzalez-Garcia, C. Page, R. Herrera, and G. Nunez. 1997. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278:687-689. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, D. C., L. C. Sanders, G. M. Bokoch, and G. N. Gill. 1999. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1:253-259. [DOI] [PubMed] [Google Scholar]
- 14.Eskes, R., S. Desagher, B. Antonsson, and J. C. Martinou. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20:929-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost, J. A., A. Khokhlatchev, S. Stippec, M. A. White, and M. H. Cobb. 1998. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem. 273:28191-28198. [DOI] [PubMed] [Google Scholar]
- 16.Gajewski, T. F., and C. B. Thompson. 1996. On a BAD influence. Cell 87:589-592. [DOI] [PubMed] [Google Scholar]
- 17.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnesutta, N., J. Qu, and A. Minden. 2001. The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J. Biol. Chem. 276:14414-14419. [DOI] [PubMed] [Google Scholar]
- 19.Golstein, P. 1997. Controlling cell death. Science 275:1081-1082. [DOI] [PubMed] [Google Scholar]
- 20.Green, D., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 21.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 22.Jaffer, Z. M., and J. Chernoff. 2002. p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell Biol. 34:713-717. [DOI] [PubMed] [Google Scholar]
- 23.Jakobi, R., E. Moertl, and M. A. Koeppel. 2001. p21-activated protein kinase gamma-PAK suppresses programmed cell death of BALB3T3 fibroblasts. J. Biol. Chem. 276:16624-16634. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann, T., S. Schlipf, J. Sanz, K. Neubert, R. Stein, and C. Borner. 2003. Characterization of the signal that directs Bcl-XL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiosses, W. B., R. H. Daniels, C. Otey, G. M. Bokoch, and M. A. Schwartz. 1999. A role for p21-activated kinase in endothelial cell migration. J. Cell Biol. 147:831-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, N., H. MacDonald, C. Reinhard, R. Halenbeck, A. Roulston, T. Shi, and L. T. Williams. 1997. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc. Natl. Acad. Sci. USA 94:13642-13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. R., S. M. Ramos, A. Ko, D. Masiello, K. D. Swanson, M. L. Lu, and S. P. Balk. 2002. AR and ER interaction with a p21-activated kinase (PAK6). Mol. Endocrinol. 16:85-99. [DOI] [PubMed] [Google Scholar]
- 28.Lei, M., W. Lu, W. Meng, M.-C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 29.Manser, E., H.-Y. Huang, T.-H. Loo, X. Q. Chen, T. Leung, and L. Lim. 1997. Expression of constitutively active a-Pak reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17:1129-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morreale, A., M. Venkatesan, H. R. Mott, D. Owen, D. Nietlispach, P. N. Lowe, and E. D. Laue. 2000. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 7:384-388. [DOI] [PubMed] [Google Scholar]
- 31.Rudel, T., and G. M. Bokoch. 1997. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276:1571-1574. [DOI] [PubMed] [Google Scholar]
- 32.Schurmann, A., A. F. Mooney, L. C. Sanders, M. A. Sells, H.-G. Wang, J. C. Reed, and G. M. Bokoch. 2000. p21-activated kinase 1 (PAK1) phosphorylates the death agonist BAD and protects cells from apoptosis. Mol. Cell. Biol. 20:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sells, M. A., J. T. Barratt, J. Caviston, S. Ottilie, E. Leberer, and J. Chernoff. 1998. Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J. Biol. Chem. 273:18490-18498. [DOI] [PubMed] [Google Scholar]
- 34.Sells, M. A., J. T. Boyd, and J. Chernoff. 1999. p21-activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J. Cell Biol. 145:837-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sells, M. A., and J. Chernoff. 1997. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell. Biol. 7:162-167. [DOI] [PubMed] [Google Scholar]
- 36.Sells, M. A., U. G. Knaus, S. Bagrodia, D. M. Ambrose, G. M. Bokoch, and J. Chernoff. 1997. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7:202-210. [DOI] [PubMed] [Google Scholar]
- 37.Sells, M. A., A. Pfaff, and J. Chernoff. 2000. Temporal and spatial distribution of activated Pak1 in fibroblasts. J. Cell Biol. 151:1449-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, Y., H. Zhou, A. Chen, R. N. Pittman, and J. Field. 2000. The Akt proto-oncogene links Ras to Pak and cell survival signals. J. Biol. Chem. 275:9106-9109. [DOI] [PubMed] [Google Scholar]
- 39.Tu, H., and M. Wigler. 1999. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter, B. N., Z. Huang, R. Jakobi, P. T. Tuazon, E. S. Alnemri, G. Litwack, and J. A. Traugh. 1998. Cleavage and activation of p21-activated protein kinase gamma-PAK by CPP32 (caspase 3). Effects of autophosphorylation on activity. J. Biol. Chem. 273:28733-28739. [DOI] [PubMed] [Google Scholar]
- 41.Yang, F., X. Li, M. Sharma, M. Zarnegar, and Z. Lim. 2001. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 276:15345-15353. [DOI] [PubMed] [Google Scholar]
- 42.Zha, J., H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87:619-628. [DOI] [PubMed] [Google Scholar]