Abstract
Zellweger syndrome is the archetypical peroxisome biogenesis disorder and is characterized by defective import of proteins into the peroxisome, leading to peroxisomal metabolic dysfunction and widespread tissue pathology. In humans, mutations in the PEX13 gene, which encodes a peroxisomal membrane protein necessary for peroxisomal protein import, can lead to a Zellweger phenotype. To develop mouse models for this disorder, we have generated a targeted mouse with a loxP-modified Pex13 gene to enable conditional Cre recombinase-mediated inactivation of Pex13. In the studies reported here, we crossed these mice with transgenic mice that express Cre recombinase in all cells to generate progeny with ubiquitous disruption of Pex13. The mutant pups exhibited many of the clinical features of Zellweger syndrome patients, including intrauterine growth retardation, severe hypotonia, failure to feed, and neonatal death. These animals lacked morphologically intact peroxisomes and showed deficient import of matrix proteins containing either type 1 or type 2 targeting signals. Biochemical analyses of tissue and cultured skin fibroblasts from these animals indicated severe impairment of peroxisomal fatty acid oxidation and plasmalogen synthesis. The brains of these animals showed disordered lamination in the cerebral cortex, consistent with a neuronal migration defect. Thus, Pex13−/− mice reproduce many of the features of Zellweger syndrome and PEX13 deficiency in humans.
The peroxisome biogenesis disorders (PBDs) are neurodegenerative disorders that result from defective biogenesis of the peroxisome, a ubiquitous cellular organelle (17). Peroxisomes are required for a number of essential metabolic functions, including the β-oxidation of very long chain fatty acids (VLCFA), oxidation of phytanic acid, and syntheses of bile acids and ether-phospholipids (28). PBDs are autosomal recessive diseases that arise from mutations in PEX genes that encode proteins, called peroxins, required for the normal biogenesis of the peroxisome (8, 16). Mutations in peroxins either directly disrupt the apparatus required for posttranslational import into the peroxisome of matrix proteins containing either a PTS1 or PTS2 peroxisomal targeting signal or indirectly prevent matrix protein import by disrupting peroxisomal membrane formation (35). The PBDs are thus characterized by the absence, or deficiency, of normal peroxisomes and loss of this organelle's usual complement of proteins and metabolic pathways. Of the PBDs, Zellweger syndrome (ZS), neonatal adrenoleukodystrophy (NALD), and infantile Refsum's disease (IRD) represent a clinical continuum, called the Zellweger spectrum, with ZS the most severe and IRD towards the milder end of the phenotypic spectrum.
Animal models provide a valuable resource for investigating the pathogenesis of human diseases. Three mouse models for ZS have been developed through targeted disruption of the Pex2 (12), Pex5 (1), and Pex11β (22) genes. All three knockout animals exhibit many of the organ abnormalities typical of ZS, including hypotonia and impaired neocortex neuronal migration and maturation. Unfortunately, Pex5−/−, Pex2−/−, and Pex11β−/− pups die at or shortly after birth, limiting their usefulness as models of postnatal disease pathogenesis. In addition, they are not suitable for delineating tissue-specific pathology. To circumvent these obstacles and allow a more widespread and detailed analysis of the cellular and tissue changes that occur in the PBDs, we have initiated the development of a conditional Pex gene mouse model using the Cre/loxP recombination system of gene manipulation in embryonic stem cells (21, 29).
PEX13 is a peroxisomal integral membrane protein with an essential role in both PTS1 and PTS2 protein import. In Saccharomyces cerevisiae, the cytoplasmic SH3 domain of PEX13 binds PEX5, the PTS1 receptor (9, 10, 15). Deletion of PEX13 results in a marked reduction in the level of PEX5 associated with the peroxisomal membrane at steady state and loss of both PTS1 and PTS2 protein import (15). In humans, mutations in PEX13 lead to a Zellweger spectrum disease phenotype and define complementation group 13 of the PBDs. Two mutations in this gene have been reported, a missense mutation in a mildly affected NALD patient (23, 39) and a nonsense mutation in a ZS patient (39).
On the basis of this established function of PEX13 in humans, we propose that targeted disruption of the mouse Pex13 gene would provide a good model of the peroxisome protein import defects and pathogenesis of the PBDs. Here we report on a mouse mutant in which Pex13 has been ubiquitously disrupted using the Cre/loxP system. Pex13-deficient mice reproduce many of the features of ZS patients, including intrauterine growth retardation, hypotonia, abnormal peroxisomal metabolism, and neonatal lethality. This disease phenotype is associated with defective peroxisome biogenesis, confirming the essential role of Pex13 in the molecular mechanisms of peroxisomal matrix protein import in mammals.
MATERIALS AND METHODS
Animal use.
All research described involving animals has been approved by the ethics committees of Griffith University or the Queensland Institute of Medical Research and has complied with the guidelines and policies of the National Health and Medical Council of Australia and the Office of the Gene Technology Regulator.
Generation of Pex13 knockout mice using the Cre-loxP system.
We have previously reported on the genomic organization of the murine Pex13 gene (3) and the generation of mice with a loxP-flanked Pex13 allele (4). The loxP-flanked Pex13 allele incorporates loxP sites (recognized by Cre recombinase) that flank exon 2. The loxP-flanked sequence also incorporates a neomycin resistance gene flanked by FRT sites (4), providing a mechanism for separately removing this cassette by crossing animals with transgenic mice expressing Flp recombinase. Animals heterozygous for a Cre recombinase-mediated disrupted Pex13 allele were obtained by crossing females heterozygous for the loxP-flanked allele with a male Cre-deleter transgenic mouse (X-linked transgene) that expresses Cre recombinase ubiquitously from the two-cell stage onwards under the control of a human cytomegalovirus minimal promoter (37). Female progeny positive for the disrupted Pex13 allele (4) were backcrossed to a male C57BL6/J mouse to generate male and female animals that were heterozygous for the disrupted Pex13 allele and also Cre recombinase negative. Pex13 homozygous knockout animals were generated by crossing progeny heterozygous for the disrupted Pex13 allele. PCR analysis of tail biopsy genomic DNA was undertaken using primers P1 (5′-ATGGCTCCCAAGTTAGTTCTG-3′) and P2 (5′-TCTGTTTCCCTCCCACCTC-3′) to amplify a 492-bp fragment specific to the wild-type allele, primers P3 (5′-TGGCTCCCAAGTTAGTTCTGTC-3′) and P4 (5′-CCTCTCTATTTGTTGCTTACCCC-3′) to amplify a 385-bp fragment specific to the disrupted Pex13 allele (see Fig. 1A), and primers Cre-F (5′-CACCCTGTTACGTATAGC-3′) and Cre-R (5′-CTAATCGCCATCTTCCAG-3′) to detect the Cre recombinase gene.
FIG. 1.
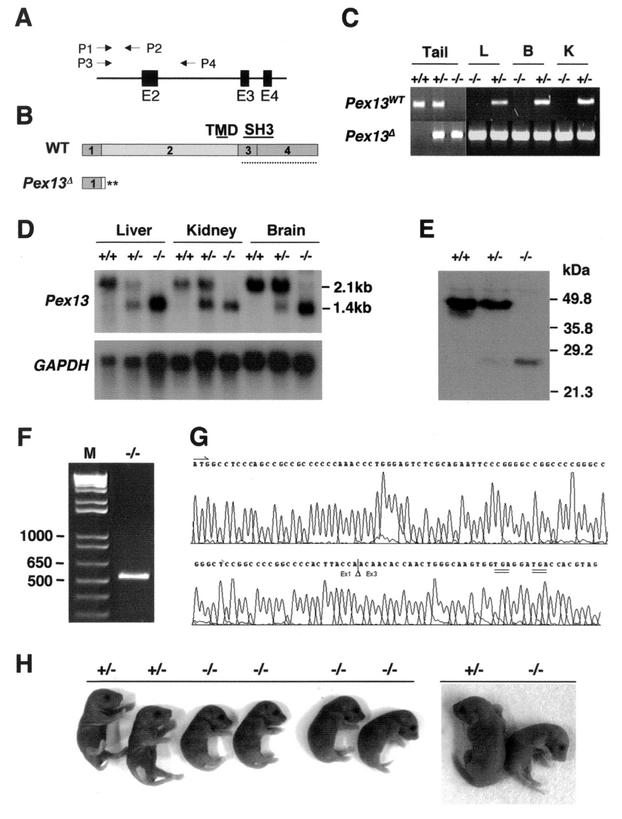
Targeted disruption of Pex13. (A) Schematic representation of the wild-type allele. The positions of exons 2 (E2) to 4 (E4) and the directions and positions of genotyping PCR primers P1 to P4 are indicated. (B) Structures of proteins expressed from the wild-type (WT) Pex13 and disrupted (Pex13Δ) alleles. The numbered boxes depict the sections of the protein encoded by the Pex13 exons and are shown to scale. The positions of the transmembrane domain (TMD) and SH3 domain are indicated by the labeled solid lines. The dotted line represents the portion of the protein used to raise the Pex13 antibody. Two predicted translation termination codons (**) are indicated. (C) Genotyping by PCR screening of tissue genomic DNA. The P1/P2 primer pair generates an amplicon (490 bp) from the wild-type (WT) Pex13 allele, but not from the Pex13Δ allele because of the loss of the P2 primer binding sequence following Cre/loxP excision of exon 2. The P3/P4 primer pair generates an amplicon (385 bp) from the Pex13Δ allele, but not from the wild-type allele because of amplicon size. L, liver; B, brain; K, kidney. (D) Northern blot analysis of 10 μg of total RNA from the liver, kidney, and brain. The blot was probed with a radioactively labeled murine Pex13 cDNA probe, stripped, and reprobed with a labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe. (E) Western blot analysis of total liver homogenate (10 μg of protein) from newborn (0.5 dpp) mice using anti-Pex13 antibody. The mobility of protein mass markers is indicated. (F) RT-PCR of RNA from Pex13−/− mouse liver. Total liver RNA was reverse transcribed and PCR amplified using primers flanking the Pex13 open reading frame. Lane M, DNA ladder with selected markers (in base pairs) indicated. (G) Sequence chromatogram of a portion of the RT-PCR product from Pex13−/− mouse liver RNA. The physiological translation start ATG (arrow) is shown. The position where exon 2 was removed (triangle) and positions of flanking exons 1 and 3 (Ex1 and Ex3, respectively) sequences are indicated. The positions of two in-frame translation termination codons (double underlines) are shown. (H) Appearance of newborn pups. (Left) An entire litter; (Right) one Pex13+/− and one Pex13−/− animal from a second litter. Symbols: +/+, wild-type; +/−, heterozygous for the disrupted Pex13 gene; −/−, homozygous for the disrupted Pex13 gene.
Northern analysis, RT-PCR, and in vitro transcription and translation.
RNA from mouse tissue was prepared using the RNeasy kit (Qiagen, Clifton Hill, Australia) according to the manufacturer's instructions. Northern blot analysis of Pex13 expression using tissue RNA was performed as previously described (3). For reverse transcription (RT), RNA was first treated for 1 h at 37°C with 3 U of RQ1 DNase (Promega, Madison, Wis.), followed by phenol-chloroform extraction and ethanol precipitation. RT reactions were performed using 2 μg of DNase-treated RNA, 0.5 μM T30MN oligonucleotide and 200 U of SuperScript III reverse transcriptase, according to the manufacturer's instructions (Invitrogen, Melbourne, Australia). RT product was PCR amplified using primers that flank the Pex13 open reading frame. The product was gel extracted and sequenced directly using an Applied Biosystems 377 sequencer and BigDye terminator cycle sequencing (Applied Biosystems, Foster City, Calif.). Alternatively, the product was first cloned into pGEM-Teasy vector (Promega) and subclones were sequenced. In vitro transcription and translation of [35S]methionine-labeled protein from the Pex13−/− RT-PCR product, subcloned into pSP64poly(A) (Promega), was performed and analyzed as previously described (3).
Plasmid constructs.
JB221, a pcDNA3-based plasmid encoding the full-length mouse Pex13 protein with a C-terminal myc epitope tag, has been described previously (3). The pPTS2-EGFP plasmid, encoding the minimal rat PTS2 signal fused to the amino-terminal end of enhanced green fluorescent protein (EGFP), was created by blunt cloning of the 0.9-kb BglI-NotI fragment from the PTSwt-GFP plasmid (7) into EcoRV-digested pcDNA3 vector (Invitrogen).
Cell culture and fluorescence microscopy.
Primary mouse skin fibroblast cultures were produced from skin biopsies of newborn mice 0.5 day postpartum (dpp). Cells were cultured in a mixture of Dulbecco's modified Eagle medium and Ham F-12 medium supplemented with 10% fetal bovine serum, 100 μg of penicillin per ml, and 100 U of streptomycin per ml (both antibiotics from Invitrogen, Melbourne, Australia). Cell transfection, using Lipofectamine 2000 (Invitrogen) and 3 μg of relevant plasmid, and indirect immunofluorescence were performed as previously described for human skin fibroblasts (24, 25). The anti-SKL and anti-rat PMP70 antibodies (Zymed Laboratories Inc., San Francisco, Calif.) were used at dilutions of 1/200 and 1/500, respectively.
Liver fractionation and Western blot analysis.
Livers were removed from newborn mouse pups (0.5 dpp), weighed, frozen in liquid nitrogen, and stored at −80°C. Liver portions (approximately 20 mg) were subsequently thawed on ice and homogenized in 200 μl of cold 0.25 M sucrose-0.1% ethanol containing protease inhibitors (50 μg of leupeptin per ml, 10 μg of pepstatin per ml, 10 μg of chymostatin per ml, and 200 μM phenylmethylsulfonyl fluoride) (SEPI buffer) using one stroke of a Potter-Elvehjem homogenizer rotating at 1,000 rpm. A sample of homogenate was added to 0.5 volume of 3× sodium dodecyl sulfate (SDS) sample buffer (3× SDS sample buffer is 30% glycerol, 6% SDS, 15% β-mercaptoethanol, 0.2% bromophenol blue, and 187.5 mM Tris-HCl buffer [pH 6.8]) and immediately heated for 5 min in a boiling water bath prior to SDS-polyacrylamide gel electrophoresis. Western blotting was performed as previously described, using antibodies to mouse liver catalase, peroxisomal bifunctional protein, and PMP70 (5), or mouse Pex13 (3). The antibody to mouse Pex13 was raised against recombinant maltose-binding protein fused to the region of Pex13 encompassing the SH3 domain to the C terminus (see Fig. 1B). The remaining liver homogenate was centrifuged using a Sorvall RC-5B centrifuge for 10 min at 1,000 × g to sediment nuclei and unbroken cells. The supernatant from this step was centrifuged for 10 min at 16,000 × g to produce an organelle pellet and postorganellar supernatant. These fractions were suspended to equivalent volumes in SEPI buffer and assayed for catalase activity or mixed with SDS sample buffer as described above for SDS-polyacrylamide gel electrophoresis.
Biochemical analyses.
Catalase activity in mouse liver fractions was assayed by a kinetic spectrophotometric procedure (6). The identities and levels of VLCFA in a total lipid extract from tissue samples were determined as previously described (33). Plasmalogens were assayed essentially as described previously (34) but using cis-3-O-nonadec-1′-enylglycerol 1,2-carbonate as the internal standard. Measurements of the rate of phytanic acid and pristanic acid oxidation and of the activities of alkyl-dihydroxyacetone phosphate (DHAP) synthase and DHAP acyltransferase (DHAP-AT) in cultured skin fibroblasts were performed as previously described (32).
Histology and electron microscopy.
For routine necroscopy and tissue histology, newborn mice (0.5 dpp) were killed by carbon dioxide asphyxiation, perfused with Bouin's fixative by injection into the peritoneal and thoracic cavities, and immersed in Bouin's fixative for at least 7 days. Coronal brain sections, cut at 1-mm intervals rostral to caudal, were embedded in paraffin wax, and 6-μm-thick sections were cut and stained with hematoxylin and eosin. The body was then cut longitudinally into two equal halves and embedded in paraffin in order to examine visceral organs. Preliminary neuropathological examination of these mice was confined to a small number of coronal sections taken from directly comparable levels of the brain in Pex13−/− and wild-type mice. For routine electron microscopy, tissue was removed from animals, thinly sliced, and fixed for 2 h at 4°C in 3% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.3. Specimens were postfixed in 1% osmium tetroxide and 1% aqueous uranyl acetate, dehydrated by immersion in a series of graded ethanol solutions, and embedded in Spurr epoxy. Ultrathin sections (approximately 50 to 100 nm thick) were treated with 1% aqueous uranyl acetate and Reynold's lead citrate for contrast and viewed using a JEOL 1200EX electron microscope operating at 80 kV. For catalase cytochemistry, liver slices were fixed for 2 h at 4°C in 0.1 M sodium cacodylate buffer (pH 7.3) (19, 20) containing 4% formaldehyde and 1% glutaraldehyde, incubated at 25°C for 1 h in Teorell & Stenhagen buffer (pH 10.5) (11) containing 3,3′-diaminobenzidine (DAB) tetrahydrochloride (2 mg/ml) (Sigma Chemicals, St. Louis, Mo.) and 0.05% H2O2, and washed in 0.1 M sodium cacodylate buffer (pH 7.3). Subsequent processing was performed as described above except that ultrathin sections were examined without further treatment for contrast.
RESULTS
Targeted disruption of Pex13.
The Cre/loxP site-specific recombination approach was adopted to develop mouse lines in which the Pex13 gene could be disrupted ubiquitously or in a cell- or developmental stage-specific manner by using appropriate Cre recombinase-expressing transgenic mice. Construction of the Pex13 targeting vector, generation of chimeras, and production of animals carrying a Pex13 allele in which exon 2 is flanked by loxP sites have been described previously (4). For generation of mice with ubiquitous disruption of Pex13, animals heterozygous for the exon 2-deleted Pex13 allele were produced by crossing animals carrying the loxP-modified Pex13 allele with transgenic mice that express Cre recombinase ubiquitously from the two-cell stage on (37). Pex13−/− pups were then generated by crossing animals heterozygous for the disrupted Pex13 allele. Genotyping of pups was performed by Southern analysis as previously described (4) (data not shown) and then routinely by PCR analysis of genomic DNA using primers to amplify sequence-specific fragments of the wild-type or disrupted Pex13 alleles (Fig. 1A). Exon 2 removal predicts a protein that lacks the essential transmembrane and SH3 domains; the effect on Pex13 protein structure is represented diagrammatically in Fig. 1B.
PCR analyses of genomic DNA confirmed homozygous disruption of the Pex13 allele in the tail and in the other tissues analyzed (Fig. 1C), viz., liver, brain, and kidney, consistent with the reported ubiquitous tissue expression of the Cre recombinase in the strain used. Disruption of Pex13 was demonstrated by Northern blot analysis, which showed that the normal mRNA transcript (approximately 2.1 kb) was not expressed in tissues of Pex13−/− animals (Fig. 1D). Western blot analysis confirmed that the Pex13 protein (apparent molecular mass of approximately 47 kDa) was not detectable in liver homogenates of these animals (Fig. 1E). Unexpectedly, these analyses also indicated that the disrupted allele produced a shorter, stable mRNA transcript, whose size (approximately 1.4 kb) is compatible with exon 2 (694-bp) deletion (Fig. 1D). The nature of this transcript was investigated by RT-PCR of mRNA from the livers of Pex13−/− mice. A single product was obtained (Fig. 1F). The size (approximately 520 bp) and sequence (Fig. 1G) of the cloned cDNA product were identical to those of the wild-type transcript, except for the absence of exon 2. The sequence also confirmed that exon 2 removal resulted in the introduction of immediate downstream translation termination codons, consistent with the structure and sequence of the targeting construct (Fig. 1B).
Western blotting of liver homogenate proteins also indicated that a low-abundance protein of approximately 26 kDa appeared to segregate with the disrupted Pex13 allele (Fig. 1E). The identity of this immunoreactive protein is not clear; the disrupted Pex13 mRNA predicts a protein lacking the C-terminal third of Pex13, the segment of the protein recognized by the Pex13 antibody (Fig. 1B and G), and does not contain an alternative, downstream translation start codon that could generate a protein of this molecular mass. Also, in vitro transcription and translation from Pex13−/− cDNA template produced a single radioactive product of the predicted mass (approximately 5 kDa) (cf. Fig. 1B and G) (data not shown) and no immunoreactive bands, as expected. We have also shown that the 26-kDa protein is found in the supernatant (after centrifugation at 100,000 × g for 60 min) when the livers from Pex13−/− animals are subfractionated by differential centrifugation (data not shown). These findings, together with the fact that the Pex13+/− heterozygous mice did not display a disease phenotype (see below), suggest that this 26-kDa protein is not acting in a dominant-negative manner and has no Pex13-related function.
Pex13-deficient mice display growth and developmental abnormalities.
Pex13−/− pups died soon after birth (6 to 12 h) and displayed severe hypotonia and hypoactivity. The pups showed no obvious dysmorphia but maintained a contracted “C” posture that appeared to particularly affect the hind limbs (Fig. 1H). The body weight of these animals was 73% ± 9% (mean ± standard deviation [SD]; n = 14) of that of littermate wild-type and heterozygote pups at the same stage (0.5 dpp), suggesting severe intrauterine growth retardation. The pups did not feed, as ascertained by the absence of milk in their stomachs, and in rare cases displayed respiratory distress. In contrast, none of the Pex13+/− pups displayed a disease phenotype. Inheritance of the disrupted Pex13 allele was compatible with normal Mendelian genetics, indicating that there was no intrauterine loss of homozygotes.
Defective peroxisome biogenesis in Pex13-deficient mice.
The essential role of PEX13 in peroxisomal matrix protein import in yeast (9, 10, 15), CHO cells (41), and humans (23, 39) predicts that Pex13 deficiency in mice should lead to impaired peroxisomal matrix protein import. To test this, skin fibroblast cells from newborn Pex13−/− pups were cultured and processed for indirect immunofluorescence microscopy. Using an anti-SKL antibody, which recognizes the prototypic signal required for peroxisomal import of PTS1-containing matrix proteins, peroxisomes were visualized as punctate cytoplasmic fluorescence in cells from Pex13+/+ mice but were completely absent from Pex13−/− cells, where there was diffuse fluorescence in the cytoplasm instead (Fig. 2A). PTS2 protein import was assessed by transfecting cells with a plasmid that encodes GFP fused to the rat PTS2 signal. In Pex13+/+ cells, GFP was detected as punctate cytoplasmic fluorescence. Double indirect fluorescence of this same cell using an antibody to the peroxisomal integral membrane protein PMP70 indicated peroxisomal colocalization of GFP and PMP70. In Pex13−/− cells, in contrast, peroxisomal import of PTS2-EGFP was deficient, as demonstrated by the lack of punctate GFP fluorescence, but some peroxisomal membranes appeared to be formed normally, as indicated by PMP70-positive vesicles in the same cell. PTS1 and PTS2 protein import defects in Pex13−/− cells could be rescued by transfection with the plasmid JB221, which expresses wild-type Pex13 protein containing a C-terminal myc tag (Fig. 2B). The results demonstrate colocalization of all fluorescence signals in the rescued Pex13−/− cells, consistent with the detected cytoplasmic vesicles representing peroxisomes. These results also confirm that the molecular basis of the biogenesis defects in these animals is Pex13 deficiency.
FIG. 2.
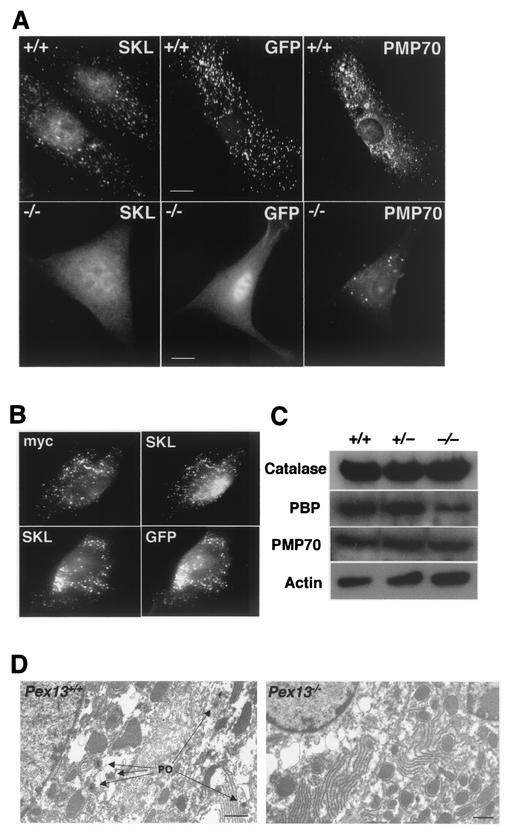
Pex13 deficiency disrupts protein import into peroxisomes. (A) Peroxisomal protein import in cultured skin fibroblasts from wild-type (+/+) and Pex13-deficient (−/−) mice. PTS1 protein import assessed by indirect immunofluorescence using an anti-SKL antibody (SKL panels), PTS2 protein import assessed by GFP autofluorescence in cells transfected with a plasmid that expresses a PTS2-EGFP fusion protein (GFP panels), and peroxisomal membranes detected by indirect immunofluorescence using an antibody to the peroxisomal integral membrane protein PMP70 (PMP panels) are shown. Note that GFP fluorescence and PMP70 immunofluorescence are from the same cell in each case. Bars, 10 μm. (B) Transfection of Pex13−/− cells with a plasmid that expresses wild-type, myc-tagged Pex13 rescues import of PTS1 and PTS2-EGFP protein import. myc and SKL immunofluorescence in the same cell (top) and GFP fluorescence and SKL immunofluorescence in the same cell (bottom) are shown. (C) Western blot analysis of liver homogenates to determine the levels of the peroxisomal proteins catalase, peroxisomal bifunctional protein (PBP), and PMP70. Actin levels indicate protein loading. Liver homogenates from wild-type (+/+) and heterozygous (+/−) and homozygous (−/−) knockout animals were used. (D) Electron microscopic analysis of DAB-stained liver for detection of peroxisomes (PO) in wild-type and Pex13−/− mice. Bars, 1 μm.
Peroxisome biogenesis defects in Pex13−/− mice were also assessed by determining the levels of specific peroxisomal proteins in liver homogenates. Catalase is the most abundant peroxisomal matrix protein and is imported relatively inefficiently via a nonconsensus PTS1 signal (36, 38). Total catalase levels in the liver were relatively unchanged in Pex13-deficient animals compared to wild-type controls (Fig. 2C). To determine the subcellular distribution of catalase in these animals, liver homogenates were subfractionated by differential centrifugation to produce an organellar pellet and a postorganellar supernatant. The proportion of catalase in the postorganellar supernatant, expressed as a percentage (mean ± SD; n = 3) of the total catalase activity (organellar pellet plus postorganellar supernatant), was 18% ± 1% for Pex13+/+ animals and 81% ± 6% for Pex13−/− animals. These results indicate that catalase is predominantly organelle associated in Pex13+/+ mice but cytoplasmic in Pex13−/− mice; the latter distribution is consistent with a defect in the peroxisomal import of this protein and the known stability of this protein in the cytoplasm. Western blot analysis was also used to test for abundance of another peroxisomal matrix protein, the peroxisomal bifunctional protein of the β-oxidation pathway, which is also imported by the PTS1 pathway. The amount of this protein in the liver was reduced in Pex13−/− mice, consistent with defective peroxisomal import and instability or degradation of the missorted protein. In contrast, levels of the peroxisomal membrane protein PMP70, whose peroxisomal targeting does not involve either the PTS1 or PTS2 pathway, were not significantly affected in Pex13−/− mice.
To confirm that peroxisomes were depleted in the Pex13−/− animals, livers were processed for DAB histochemistry and electron microscopy to detect peroxisomal catalase. In contrast to livers from Pex13+/+ mice which contained numerous DAB-positive peroxisomes with crystalline cores, no DAB-positive peroxisome-like structures were observed in Pex13−/− animals (Fig. 2D). When considered together, these results indicate normal peroxisomal membrane synthesis but defective matrix protein import in the Pex13-deficient animals.
Biochemical abnormalities of Pex13-deficient mice.
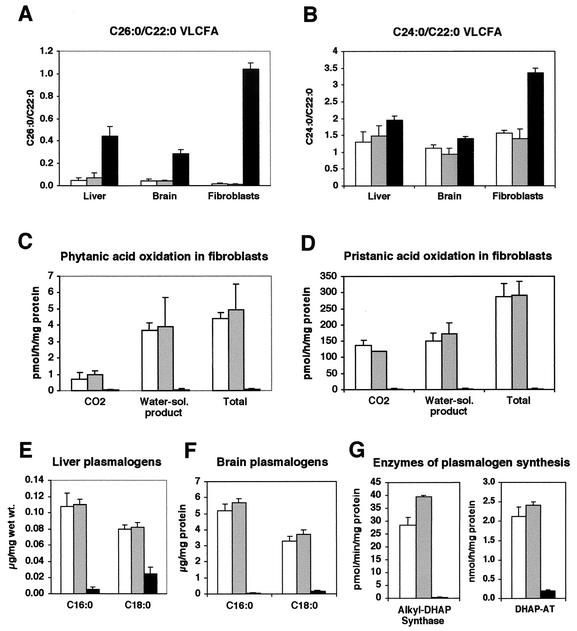
ZS patients display widespread metabolic abnormalities due to the absence of functional peroxisomes (17). To assess whether peroxisomal metabolic function was similarly compromised in Pex13-deficient mice, the levels of VLCFA, rates of oxidation of branched-chain fatty acids (i.e., phytanic acid and pristanic acid), and levels of plasmalogens and peroxisomal enzymes required for plasmalogen synthesis were measured. VLCFA levels, expressed as the C26:0/C22:0 ratio, were significantly elevated in the livers (9-fold), brains (6.5-fold), and cultured skin fibroblasts (50-fold) of Pex13−/− pups compared to wild-type and Pex13+/− animals (Fig. 3A). VLCFA levels in these tissues, when expressed as the C24:0/C22:0 ratio, were up to twofold elevated (Fig. 3B). The rates of oxidation of the branched-chain fatty acid phytanic acid (Fig. 3C) and its α-oxidation product, pristanic acid (Fig. 3D), were measured in cultured skin fibroblasts as the production of radiolabeled CO2 and water-soluble product from 1-14C-labeled substrate. Oxidation rates of both these branched-chain fatty acids were decreased by between 50- and 100-fold in Pex13−/− pups compared with wild-type and Pex13+/− animals. Plasmalogen metabolism was assessed by measuring liver and brain plasmalogen levels and the activity in cultured skin fibroblasts of peroxisomal enzymes required for plasmalogen synthesis. The levels of C16:0 and C18:0 plasmalogen in the livers of Pex13−/− pups were 20-fold and 3-fold lower, respectively, than in wild-type and Pex13+/− animals (Fig. 3E). The relative levels of these plasmalogen molecular species were even lower in the brains of Pex13−/− pups (Fig. 3F). The tissue plasmalogen levels were paralleled by marked reductions in the activities of alkyl-DHAP synthase and DHAP-AT in fibroblasts (Fig. 3G). These results therefore indicate severe impairment of a number of peroxisomal metabolic pathways in Pex13-deficient animals, consistent with a generalized metabolic deficiency that would result from defective peroxisome biogenesis.
FIG. 3.
Peroxisomal biochemical abnormalities in Pex13-deficient mice. VLCFA measurements in the liver, brain, and cultured skin fibroblasts expressed as a ratio of C26:0/C22:0 (A) or C24:0/C22:0 (B) fatty acids. Oxidation of [1-14C]phytanic acid (C) and [1-14C]pristanic acid (D) by cultured skin fibroblasts, expressed as the production of [1-14C]CO2 and 1-14C-water-soluble (Water-sol.) products. Levels of C16:0 and C18:0 plasmalogen in liver (E) and brain (F). (G) Activities of peroxisomal enzymes required for plasmalogen synthesis in cultured fibroblasts. Values are the means of three or four measurements on Pex13+/+ (white bars), Pex13+/− (grey bars), and Pex13−/− (black bars) animals. Each error bar represents one SD.
Tissue ultrastructural changes in Pex13-deficient mice.
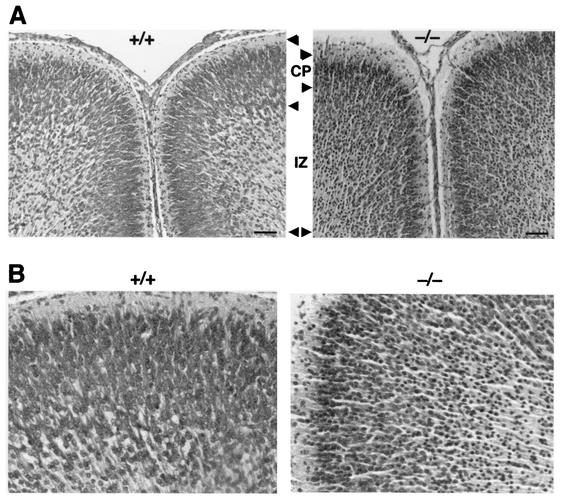
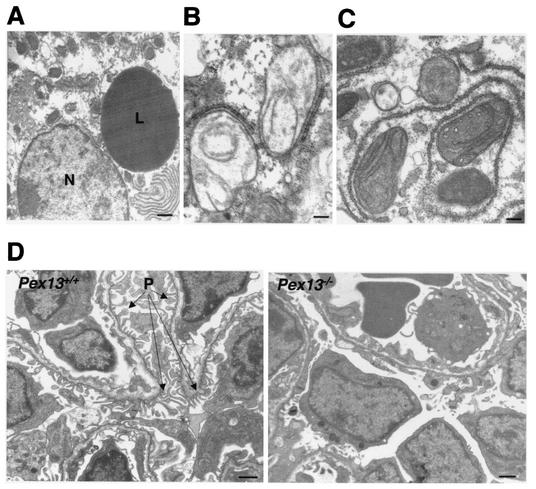
One of the hallmarks of ZS is defective neuronal migration in brain. In comparison with wild-type mice, there was reduced thickness and disorganization of the neocortex in Pex13−/− animals, and much of the cortical mantle was densely populated by neurons with small, round, hyperchromatic (pyknotic) nuclei, often with a thin rim of amphophilic cytoplasm (Fig. 4). These degenerate neurons also imparted increased cellularity to the subcortical zone of radial glia, the prospective white matter, and were present in smaller numbers in the periventricular germinative region. These results are consistent with abnormal neuronal migration and neuronal degeneration in Pex13-deficient mice. Visceral organs were unremarkable by light microscopy. However, analysis of the ultrastructure of hepatocytes of Pex13−/− mice by electron microscopy revealed an abundance of large lipid droplets (Fig. 5A), which is also a feature of ZS patients and Pex5−/− mice (1). The presence of mitochondria with abnormal structure, in particular abnormal crista morphology, was another distinctive feature. Typically, the appearance of cristae was curvilinear and looped (Fig. 5B) or elongated and parallel (Fig. 5C). Similar mitochondrial ultrastructural changes have been reported for Pex5−/− mice (1, 2), and mitochondrial abnormalities are a reported feature of ZS patients (14). Ultrastructural analysis of kidneys revealed delayed differentiation of glomeruli in Pex13−/− mice, evident as reduced size and capillary structure and paucity of podocyte foot processes (Fig. 5D). In addition, myelin-like lipid structures were identified in some adrenocortical cells of Pex13−/− mice (data not shown). Thus, Pex13 deficiency leads to a number of tissue ultrastructural changes in the mouse that model many of those observed for ZS patients.
FIG. 4.
Pex13 deficiency leads to abnormal lamination of the cerebral cortex. (A) Matched coronal sections of wild-type (+/+) and Pex13-deficient (−/−) mice stained with hematoxylin and eosin. Corresponding cortical layers are delimited by arrowheads. CP, cortical plate; IZ, intermediate zone. Bars, 200 μm. (B) Higher-magnification views of the cortical plate zone from the sections shown in panels A.
FIG. 5.
Tissue ultrastructural changes in Pex13-deficient mice. (A) Large lipid droplet (L) in a hepatocyte. N, nucleus. Bar, 1 μm. (B and C) Hepatic mitochondria with abnormal cristae. Bars, 200 nm. (D) Section through kidney glomeruli of Pex13+/+ and Pex13−/− mice. P, podocyte foot processes. Bars, 1 μm.
DISCUSSION
Ubiquitous inactivation of Pex13 in mice represents the first stage in the development of a group of PBD animal models based on conditional inactivation of this gene using the Cre-loxP recombination system. The Pex13−/− mouse exhibits the expected peroxisome biogenesis deficiencies, given the known function of the PEX13 protein in peroxisomal matrix protein import through the PTS1 and PTS2 pathways in eukaryotes. In yeast, PEX13 is required for both PTS1 and PTS2 protein import into the peroxisome. The role of PEX13 in PTS1 protein import involves interaction with PEX5, the PTS1 receptor. This interaction is thought to involve a step subsequent to binding of PTS1 cargo-laden receptor with another peroxisomal membrane protein, PEX14, the primary docking protein, and may require displacement of this cargo (42). The involvement of PEX13 in PTS2 protein import reflects a primary docking event with the PTS2 receptor, PEX7 (40). A similar role for PEX13 in protein import in mammalian cells is indicated by the disruption of both PTS1 and PTS2 protein import in patients with PEX13 mutations (23, 39), although the mechanisms involved may differ slightly (31) and include a direct interaction between cargo-laden PEX7 and PEX5 prior to docking at the peroxisomal membrane (30).
The organelle biogenesis defects in the Pex13−/− mouse, coupled with the absence of morphologically distinguishable peroxisomes in the liver and the presence of peroxisome ghosts in cultured fibroblasts, are paralleled by peroxisomal metabolic deficiency. The VLCFA levels in livers, brains, and skin fibroblasts are elevated and branched-chain fatty acid oxidation in fibroblasts is severely reduced in Pex13−/− mice compared to wild-type mice. In addition, the markedly reduced levels of plasmalogens in the brain and liver and of alkyl-DHAP synthase and DHAP-AT activities in fibroblasts are indicative of a severe impairment in plasmalogen synthesis. Defects in these unrelated peroxisomal metabolic pathways indicate generalized impairment of peroxisomal metabolism in these animals, consistent with a primary deficiency of the peroxisomal enzymes required for these metabolic pathways.
The clinical phenotype of Pex13−/− mice indicates that these animals are a good model of the PBDs and more specifically for the severe ZS phenotype, as seen in the human patient homozygous for a PEX13 nonsense mutation at W234 (39). This finding is compatible with loss of function of a protein that is highly conserved, in both structure and function, between these species. In addition to the peroxisome protein import defects and metabolic abnormalities seen in ZS patients (17, 28), the mice reproduce a subset of the pathological features of ZS patients, including neonatal lethality, intrauterine growth retardation, hypotonia, tissue pathology, and impaired neuronal migration in the cerebral cortex. However, some ZS pathology was not reproduced; these features included facial dysmorphia, renal cysts, and specific changes to the cerebellum, liver, and retina, key targets of PBD pathogenesis (27). In this respect, the Pex13−/− animals are similar to the existing Pex2, Pex5, and Pex11β knockout animals. Many of the features of ZS patients not modeled by the Pex13−/−animals may involve postnatal developmental changes that are precluded by the neonatal lethality of these animals. This limitation is also true for the Pex2−/−, Pex5−/−, and Pex11β−/− pups, although placing the Pex2 null mutation on a different genetic background has been shown to extend postnatal survival up to 18 days and allowed investigation of cerebellar changes over this period (13). However, none of the existing knockout Pex gene mutants is suitable for delineating the tissue-specific pathology seen in PBD patients. In this context, the development of the loxP-modified Pex13 mouse provides a means of circumventing these limitations by allowing for conditional inactivation of Pex13 using transgenic animals that express Cre recombinase in a tissue-specific or developmental stage-specific manner (29). The presumptive central roles of the liver and brain in PBD pathogenesis provide two immediate target tissues for conditional inactivation of Pex13.
The molecular basis of the tissue pathogenesis of ZS and of its milder variants, NALD and IRD, is not known. A number of causative agents have been proposed, including elevated VLCFA, elevated bile acid precursors, decreased plasmalogen, and decreased docosahexaenoic acid, all of which can be related back to abnormal peroxisomal metabolism (26). Recent investigations of the neuronal migration defects in Pex5−/− animals have implicated abnormal glutamate receptor-mediated calcium signaling, resulting from decreased levels of platelet-activating factor, a plasmalogen (18). However, the phenotype of Pex11β−/− animals, which includes neonatal lethality, hypotonia, developmental delay, abnormal neuronal migration, and increased neuronal apoptosis, but not abnormal peroxisomal matrix protein import or significant peroxisomal metabolic dysfunction, appears to be incompatible with the tenet that disease pathogenesis correlates to impaired metabolic pathways involving peroxisomal matrix enzymes (22). The explanations for these seemingly disparate findings will require a molecular dissection of the pathogenic processes involved. In this context, Pex13 conditional mutants represent a potentially valuable tool and their proposed use for modeling the human PBDs is validated by the pathogenic findings observed for the Pex13−/− mouse.
Acknowledgments
This research was supported in part by grant 123107 from the National Health and Medical Research Council of Australia.
We thank Stephen Gould for continued interest in this project, Ann Trezise for early advice on the Cre-loxP system, and Suresh Subramani for the kind gift of the PTSwt-GFP plasmid. We also thank Deb Stenzel, Judith Newman, and Rob Parton for assistance with electron microscopic analyses.
REFERENCES
- 1.Baes, M., P. Gressens, E. Baumgart, P. Carmeliet, M. Casteels, M. Fransen, P. Evrard, D. Fahimi, P. E. Declercq, D. Collen, P. P. van Veldhoven, and G. P. Mannaerts. 1997. A mouse model for Zellweger syndrome. Nat. Genet. 17:49-57. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart, E., I. Vanhorebeek, M. Grabenbauer, M. Borgers, P. E. Declercq, H. D. Fahimi, and M. Baes. 2001. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am. J. Pathol. 159:1477-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkman, J., S. J. Gould, and D. I. Crane. 2002. Pex13, the mouse ortholog of the human peroxisome biogenesis disorder PEX13 gene: gene structure, tissue expression, and localization of the protein to peroxisomes. Genomics 79:162-168. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkman, J., I. Tonks, M. A. Maxwell, C. Paterson, G. F. Kay, and D. I. Crane. 2002. Conditional inactivation of the peroxisome biogenesis Pex13 gene by Cre-loxP excision. Genesis 32:179-180. [DOI] [PubMed] [Google Scholar]
- 5.Chen, N., and D. I. Crane. 1992. Induction of the major integral membrane protein of mouse liver peroxisomes by peroxisome proliferators. Biochem. J. 283:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crane, D. I., A. C. Hemsley, and C. J. Masters. 1985. Purification of peroxisomes from livers of normal and clofibrate-treated mice. Anal. Biochem. 148:436-445. [DOI] [PubMed] [Google Scholar]
- 7.Dammai, V., and S. Subramani. 2001. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105:187-196. [DOI] [PubMed] [Google Scholar]
- 8.Distel, B., R. Erdmann, S. J. Gould, G. Blobel, D. I. Crane, J. M. Cregg, G. Dodt, Y. Fujiki, J. M. Goodman, W. W. Just, J. A. Kiel, W. H. Kunau, P. B. Lazarow, G. P. Mannaerts, H. W. Moser, T. Osumi, R. A. Rachubinski, A. Roscher, S. Subramani, H. F. Tabak, T. Tsukamoto, D. Valle, I. van der Klei, P. P. van Veldhoven, and M. Veenhuis. 1996. A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elgersma, Y., L. Kwast, A. Klein, T. Voorn-Brouwer, M. van den Berg, B. Metzig, T. America, H. F. Tabak, and B. Distel. 1996. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J. Cell Biol. 135:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann, R., and G. Blobel. 1996. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135:111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahimi, H. D. 1979. An assessment of the DAB methods for cytochemical detection of catalase and peroxidase. J. Histochem. Cytochem. 27:1365-1366. [DOI] [PubMed] [Google Scholar]
- 12.Faust, P. L., and M. E. Hatten. 1997. Targeted deletion of the PEX2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J. Cell Biol. 139:1293-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faust, P. L., H. M. Su, A. Moser, and H. W. Moser. 2001. The peroxisome deficient PEX2 Zellweger mouse: pathologic and biochemical correlates of lipid dysfunction. J. Mol. Neurosci. 16:289-297. [DOI] [PubMed]
- 14.Goldfischer, S., C. L. Moore, A. B. Johnson, A. J. Spiro, M. P. Valsamis, H. K. Wisniewski, R. H. Ritch, W. T. Norton, I. Rapin, and L. M. Gartner. 1973. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science 182:62-64. [DOI] [PubMed] [Google Scholar]
- 15.Gould, S. J., J. E. Kalish, J. C. Morrell, J. Bjorkman, A. J. Urquhart, and D. I. Crane. 1996. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J. Cell Biol. 135:85-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould, S. J., and D. Valle. 2000. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 16:340-345. [DOI] [PubMed] [Google Scholar]
- 17.Gould, S. J., D. Valle, and G. V. Raymond. 2001. The peroxisome biogenesis disorders, p. 3181-3217. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 8th ed., vol. 2. McGraw-Hill, New York, N.Y.
- 18.Gressens, P., M. Baes, P. Leroux, A. Lombet, P. Van Veldhoven, A. Janssen, J. Vamecq, S. Marret, and P. Evrard. 2000. Neuronal migration disorder in Zellweger mice is secondary to glutamate receptor dysfunction. Ann. Neurol. 48:336-343. [PubMed] [Google Scholar]
- 19.Hughes, J. L., D. I. Crane, E. Robertson, and A. Poulos. 1993. Morphometry of peroxisomes and immunolocalization of peroxisomal proteins in the liver of patients with generalised peroxisomal disorders. Virchows Arch. A Pathol. Anat. 423:459-468. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, J. L., A. Poulos, D. I. Crane, C. W. Chow, L. J. Sheffield, and D. Sillence. 1992. Ultrastructure and immunocytochemistry of hepatic peroxisomes in rhizomelic chondrodysplasia punctata. Eur. J. Pediatr. 151:829-836. [DOI] [PubMed] [Google Scholar]
- 21.Kwan, K.-M. 2002. Conditional alleles in mice: practical considerations for tissue specific knockouts. Genesis 32:49-62. [DOI] [PubMed] [Google Scholar]
- 22.Li, X., E. Baumgart, J. C. Morrell, G. Jimenez-Sanchez, D. Valle, and S. J. Gould. 2002. PEX11β deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 22:4358-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., J. Bjorkman, A. Urquhart, R. J. Wanders, D. I. Crane, and S. J. Gould. 1999. PEX13 is mutated in complementation group 13 of the peroxisome-biogenesis disorders. Am. J. Hum. Genet. 65:621-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell, M. A., T. Allen, P. Solley, T. Svingen, B. C. Paton, and D. I. Crane. 2002. Novel PEX1 mutations and genotype-phenotype correlations in Australasian peroxisome biogenesis disorder patients. Hum. Mut. 20:342-351. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell, M. A., P. V. Nelson, S. J. Chin, B. C. Paton, W. F. Carey, and D. I. Crane. 1999. A common PEX1 frameshift mutation in patients with disorders of peroxisome biogenesis correlates with the severe Zellweger syndrome phenotype. Hum. Genet. 105:38-44. [DOI] [PubMed] [Google Scholar]
- 26.Moser, H. W. 1996. Pathogenetic mechanisms in peroxisomal disorders. Curr. Opin. Neurol. 9:473-476. [DOI] [PubMed] [Google Scholar]
- 27.Moser, H. W. 1993. Peroxisomal diseases. Adv. Hum. Genet. 21:1-106. [PubMed] [Google Scholar]
- 28.Moser, H. W. 1999. Peroxisomal disorders: classification and overview of biochemical abnormalities. Rev. Neurol. 28(Suppl. 1):S45-S48. [PubMed]
- 29.Nagy, A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99-109. [PubMed] [Google Scholar]
- 30.Otera, H., T. Harano, M. Honsho, K. Ghaedi, S. Mukai, A. Tanaka, A. Kawai, N. Shimizu, and Y. Fujiki. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275:21703-21714. [DOI] [PubMed] [Google Scholar]
- 31.Otera, H., K. Setoguchi, M. Hamasaki, T. Kumashiro, N. Shimizu, and Y. Fujiki. 2002. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22:1639-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paton, B. C., P. C. Sharp, D. I. Crane, and A. Poulos. 1996. Oxidation of pristanic acid in fibroblasts and its application to the diagnosis of peroxisomal beta-oxidation defects. J. Clin. Investig. 97:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulos, A., R. Gibson, P. Sharp, K. Beckman, and P. Grattan-Smith. 1994. Very long chain fatty acids in X-linked adrenoleukodystrophy brain after treatment with Lorenzo's oil. Ann. Neurol. 36:741-746. [DOI] [PubMed] [Google Scholar]
- 34.Poulos, A., L. Sheffield, P. Sharp, G. Sherwood, D. Johnson, K. Beckman, A. J. Fellenberg, J. E. Wraith, C. W. Chow, S. Usher, and H. Singh. 1988. Rhizomelic chondrodysplasia punctata: clinical, pathologic and biochemical findings in two patients. J. Pediatr. 113:685-690. [DOI] [PubMed] [Google Scholar]
- 35.Purdue, P. E., and P. B. Lazarow. 2001. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17:701-752. [DOI] [PubMed] [Google Scholar]
- 36.Purdue, P. E., and P. B. Lazarow. 1996. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J. Cell Biol. 134:849-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheikh, F. G., K. Pahan, M. Khan, E. Barbosa, and I. Singh. 1998. Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc. Natl. Acad. Sci. USA 95:2961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimozawa, N., Y. Suzuki, Z. Zhang, A. Imamura, R. Toyama, S. Mukai, Y. Fujiki, T. Tsukamoto, T. Osumi, T. Orii, R. J. Wanders, and N. Kondo. 1999. Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum. Mol. Genet. 8:1077-1083. [DOI] [PubMed] [Google Scholar]
- 40.Stein, K., A. Schell-Steven, R. Erdmann, and H. Rottensteiner. 2002. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 22:6056-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyama, R., S. Mukai, A. Itagaki, S. Tamura, N. Shimozawa, Y. Suzuki, N. Kondo, R. J. Wanders, and Y. Fujiki. 1999. Isolation, characterization and mutation analysis of PEX13-defective Chinese hamster ovary cell mutants. Hum. Mol. Genet. 8:1673-1681. [DOI] [PubMed] [Google Scholar]
- 42.Urquhart, A. J., D. Kennedy, S. J. Gould, and D. I. Crane. 2000. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 275:4127-4136. [DOI] [PubMed] [Google Scholar]