Abstract
A nuclear mRNA degradation (DRN) system was identified from analysis of mRNA turnover rates in nup116-Δ strains of Saccharomyces cerevisiae lacking the ability to export all RNAs, including poly(A) mRNAs, at the restrictive temperature. Northern blotting, in situ hybridization, and blocking transcription with thiolutin in nup116-Δ strains revealed a rapid degradation of mRNAs in the nucleus that was suppressed by the rrp6-Δ, rai1-Δ, and cbc1-Δ deletions, but not by the upf1-Δ deletion, suggesting that DRN requires Rrp6p, a 3′-to-5′ nuclear exonuclease, the Rat1p, a 5′-to-3′ nuclear exonuclease, and Cbc1p, a component of CBC, the nuclear cap binding complex, which may direct the mRNAs to the site of degradation. We propose that certain normal mRNAs retained in the nucleus are degraded by the DRN system, similar to degradation of transcripts with 3′ end formation defects in certain mutants.
The rate of synthesis of a protein is determined primarily by the steady-state level of the corresponding mRNA, which, in turn, is determined by the rate of synthesis and degradation of the mRNA. Thus, mRNA stability is an important parameter in the regulation of gene expression, affecting both the steady-state level of the protein and the transient time of translation of the formed transcript.
Normal mature mRNAs of Saccharomyces cerevisiae are degraded through a major 5′-to-3′ (5′→3′) pathway and a minor 3′→5′ pathway, both of which take place in the cytoplasm. Both degradation pathways begin with the shortening of the poly(A) tail to a track of A10 or less, caused by a Pop2p-Ccr4p-Caf1p poly(A) nuclease complex (16, 73). In the major pathway, deadenylation causes disassociation of the Pab1p [poly(A) binding protein] from the cap binding protein eIF4G, followed by the removal of the 5′ cap by the decapping enzyme Dcp1p. Subsequently, the decapped mRNA is rapidly degraded by the 5′→3′ exonuclease Xrn1p and by assistance of Sbp8p and other protein components (4, 5, 17, 32, 47, 57, 58, 71).
In the minor pathway, deadenylated mRNAs are subjected to 3′→5′ degradation by the action of the exosome, a complex of 10 3′→5′ riboexonucleases that also plays a central role in the precise formation of the 3′ ends of several types of RNAs (9), including processing precursors for rRNAs in the nucleus. The exosome may also degrade fragments of mRNA released by endonucleolytic cleavage. Yeast mutants lacking either exonucleolytic pathway degrade their mRNAs more slowly, but the loss of both pathways is lethal (54).
While most mRNAs are slowly deadenylated before rapid degradation, certain normal and mutant mRNAs are rapidly degraded by a third specialized pathway, known as nonsense-mediated mRNA decay (NMD) pathway, or mRNA surveillance, which triggers decapping before deadenylation (26, 29, 55). Substrates of the NMD pathway include not only mRNAs containing nonsense mutations but also wild-type mRNAs that contain the following: inefficiently spliced pre-mRNAs that enter the cytoplasm upstream open reading frames (14, 77) and certain codons subject to leaky scanning (80). In fact, Lelivelt and Culbertson (48) showed that mutation of protein components of NMD could actually lead to the increase in the steady-state levels of a wide spectrum of normal mRNAs. The NMD pathway discriminates between nonsense codons on the basis of downstream sequence elements located 3′ to susceptible nonsense codons (60, 87).
In addition, yeast has the capacity to recognize and degrade mRNAs lacking all termination codons, a process that occurs by a mechanism distinct from NMD and from the major mRNA turnover pathway that requires deadenylation, decapping, and 5′→3′ exonucleolytic decay (20, 76).
Previously, Das et al. (15) presented preliminary evidence, based on the analysis of cyc1-512 suppressors, that Cbc1p, the large subunit of nuclear cap binding complex, is involved in a novel mRNA degradation system. The cyc1-512 mutation causes a 90% reduction in the level of iso-1-cytochrome c because of the lack of a proper 3′ end-forming signal, resulting in low levels of eight aberrantly long cyc1-512 mRNAs, which differ in length at their 3′ termini (15, 85). Suppression analysis of cyc1-512 showed that it can be suppressed by deletion of either of the nonessential genes CBC1 or CBC2, which encode, respectively, the CBP80 or CBP20 subunits of the nuclear cap binding complex, or by deletion of the nonessential gene UPF1, which encodes a major component of the mRNA surveillance complex responsible for NMD. Suppression of cyc1-512 by cbc1-Δ occurred by two different mechanisms. The levels of the shorter cyc1-512 transcripts were enhanced in the cbc1-Δ mutants by promoting 3′-end formation at otherwise weak sites; whereas the levels of the longer cyc1-512 transcripts, as well as all mRNAs, were slightly enhanced by diminishing degradation. Furthermore, cbc1-Δ greatly suppressed the degradation of mRNAs and other phenotypes of a rat7-1 strain that is defective in mRNA export. These findings led Das et al. (15) to suggest that Cbc1p possibly defines a novel degradation pathway that acts on mRNAs partially retained in nuclei. However, the interpretation of the results obtained with rat7-1 was complicated by the suppression of the mRNA export defect by cbc1-Δ, allowing growth at the restrictive temperature, and thus preventing meaningful studies with mRNA half-lives and in situ mRNA localization using fluorescence in situ hybridization (FISH). Thus, it remained to be definitively established if the Cbc1p-dependent mRNA decay system was located in the nucleus.
In this study, we definitely established the existence of this novel mRNA degradation pathway which we named the DRN (for decay of RNA in the nucleus) pathway, and we conclusively confirmed the involvement of Cbc1p in this pathway. We have investigated the nature of this degradation pathway, primarily by using a mutation (nup116-Δ) in NUP116 which encodes a nucleoporin that plays a central role in nuclear mRNA export (3, 31). nup116-Δ strains grow slowly at 25°C and are inviable at 37°C (81). The lethal phenotype correlates with defects in mRNA export and perturbations of structures of the nuclear envelope and nuclear pore complexes, resulting in the complete nuclear accumulation of mRNA (82). We show that retention of mRNAs in the nucleus causes accelerated degradation of representative transcripts. Deletions of either CBC1 or RRP6, which encodes a nuclear 3′→5′ exoribonuclease associated with the exosome, suppressed the rapid mRNA degradation phenotype. Deletion of RAI1, which encodes a nuclear protein required for the activity of the nuclear 5′→3′ exoribonuclease Rat1p, also suppressed the rapid degradation, but to a lesser extent. We conclude that DRN involves the Rrp6p and Rat1p nuclear exonucleases, as well as the CBC, the nuclear cap binding complex, which may direct the mRNAs to the site of degradation.
MATERIALS AND METHODS
Strains, media, and yeast genetics.
Standard genetic nomenclature was used to designate wild-type alleles (for example, NUP116, RAT7, CYC1, CYH2, and ACT1), recessive mutant alleles (for example, cyc1-512 and rat7-1, etc.), and disruptants or deletion mutants (for example, cbc1-Δ and cbc1::URA3, etc). The genotypes of S. cerevisiae strains used in this study and the abbreviated genotypes are listed in Table 1. Standard YPD, YPG, SC-Ura (uracil omission), SC-Leu (leucine omission), and other omission media were used for testing and growth of yeast propagation and testing (70). Yeast genetic analysis was carried out by standard procedures described by Sherman (70).
TABLE 1.
List of yeast strains used in this study
| Strain designation | Genotype | Abbreviated genotype | Reference |
|---|---|---|---|
| B-11598 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 | NUP116 or HPR1 | 95 |
| B-11592 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 hpr1::HIS3 | hpr1-Δ | This study |
| B-11599 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 | nup116-Δ | 95 |
| B-13398 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 cbc1::URA3 | nup116-Δ cbc1-Δ | This study |
| B-13755 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 rrp6::URA3 | nup116-Δ rrp6-Δ | This study |
| B-14236 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 upf1::URA3 | nup116-Δ upf1-Δ | This study |
| B-14238 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 rail::LEU2 | nup116-Δ rail-Δ | This study |
| B-14366 | MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100 nup116::HIS3 cbc1::URA3 rrp6::KAN | nup116-Δ cbc1-Δ rrp6-Δ | This study |
| B-10603 | MATahis3-Δ200 ura3-52 leu2-Δ1 (also denoted FY86) | RAT7 | 96 |
| B-10095 | MATahis3-Δ200 ura3-52 leu2-Δ1 rat7-1 | rat7-1 | 25 |
| B-10096 | MATahis3-Δ200 ura3-52 leu2-Δ1 rat7-1 cbc1::URA3 | rat7-1 cbc1-Δ | 18 |
| B-10097 | MATahis3-Δ200 ura3-52 leu2-Δ1 rat7-1 upf1::URA3 | rat7-1 upf1-Δ | 18 |
Transformation, nucleic acid isolation, and manipulation.
S. cerevisiae cultures were transformed with linear DNA for gene disruption (66), using the lithium acetate method (33), followed by selection on SC-Ura (uracil omission) or SC-Leu (leucine omission) media. The yeast chromosomal gene CBC1 was disrupted by transforming the appropriate yeast strains with DNA fragments that were prepared by digesting the plasmid pAB1100 with SalI and BamHI. Similarly, the UPF1 gene on the chromosome was disrupted by transforming the suitable yeast strains with DNA fragments that was prepared by digesting the plasmid YCpPL51 (47) with BamHI and EcoRI. The RRP6, RAI1, and XRN1 chromosomal genes were similarly disrupted by digesting plasmids pAB2755 with BamHI and PvuII (7), pAB2806 with SstI and HindIII (84), and pAB2809 with XhoI and SalI (46), respectively, and transforming yeast strains with the appropriate DNA fragments. Escherichia coli strains DH5α and XL1-Blue were transformed by the protocol of Hanahan (22). Standard techniques of DNA manipulation such as cloning, subcloning, and sequencing, etc., used in this study are described by Sambrook et al. (68).
Analysis of mRNA steady-state levels and stability.
The stability of the various mRNAs and pre-mRNAs were determined by the inhibition of transcription with thiolutin (4 μg/ml) at 37°C unless mentioned otherwise, as described previously (15). Total RNA was isolated as described by Russo et al. (67) from approximately 108 cells. Northern blot analysis of different mRNAs was conducted as outlined by Russo et al. (67). mRNA levels were quantified by storage phosphorimager analysis (model 425E; Molecular Dynamics) and normalized against the 18S rRNA signals.
The decay rates and half-lives were estimated with the SigmaPlot (version 4.0) regression analysis program, using either a single exponential decay formula, y = 100 e−bx, or a four-parameter double-exponential decay formula, y = ae−bx + ce−dx (where a + c = 100).
FISH analysis.
Cells for FISH analysis were grown in YPD medium to early log phase at 23°C. Half of the culture was then mixed with an equal volume of prewarmed medium and shifted to 37°C. Aliquots of 108 cells were removed both from the mock-shifted as well as from shifted culture at different time intervals after temperature shift and mixed with fresh 4% formaldehyde. The cultures were immediately centrifuged at 3,500 × g for 5 min and fixed in 1/10 volume of freshly prepared solution of 0.1 M potassium phosphate buffer (pH 6.5), 3.7% formaldehyde, and 10% methanol for 1 h at room temperature. The cells were centrifuged, and the cell pellets were washed three times with 0.1 M potassium phosphate, pH 6.5, and once with SCP buffer (which contains 0.1 M dipotassium hydrogen phosphate, 0.033 M citric acid, and 1.2 M sorbitol) and were subsequently resuspended in 100 μl of SCP. Spheroplasts were generated by incubating 108 cells in 100 μl SCP containing 1/40 volume of glusulase (NEN) and 100 μg of zymolase T-20 (U.S. Biologicals) for 1 h at 30°C. Spheroplasts were washed three time with SCP and adhered to coverslips precoated with 0.01% poly-lysine and plunged into ice-cold methanol for 5 min followed by rinsing in acetone for 30 s at room temperature and dry at same temperature for 20 s.
Each coverslip for in situ hybridization was rehydrated in 5 ml of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min and prehybridized in a solution containing 2× SSC, 50% formamide, 1% bovine serum albumin, 10 mM VRC (Gibco-BRL), 10% dextran sulfate, salmon sperm DNA (500 μg/ml), and E. coli tRNA (125 μg/ml) for 1 h at 37°C. Coverslips were inverted on 24 μl of this solution containing10 ng of a Cy3-labeled 43-mer oligo(dT), and hybridizations were performed overnight at 37°C. Following hybridization, each coverslip was washed twice at 37°C for 15 min in a solution of 10% formamide and 2× SSC, once in a solution of 2× SSC and 0.1% Triton X-100 for 15 min, twice in 1× SSC for 15 min, and once in 1× phosphate-buffered saline (1 mM KH2PO4, 10 mM Na2HPO4, 140 mM NaCl, 3 mM KCl [pH 7.4]) for 15 min. Coverslips were mounted in phenylenediamine containing glycerol and DAPI (4′,6-diamino-2-phenylindole).
Cells were examined with a Nikon Diaphot inverted epifluorescence microscope, using a 100× objective. Digital images were captured using a Princeton Instruments (Princeton, N.J.) Micromax camera and analyzed with MetaFluor software from Universal Imaging (Downingtown, Pa.). The images were processed with Adobe Photoshop 5.5 software.
Poly(A) tail lengths.
Poly(A) tail lengths were analyzed as described by Butler et al. (10).
RESULTS
Experimental approach.
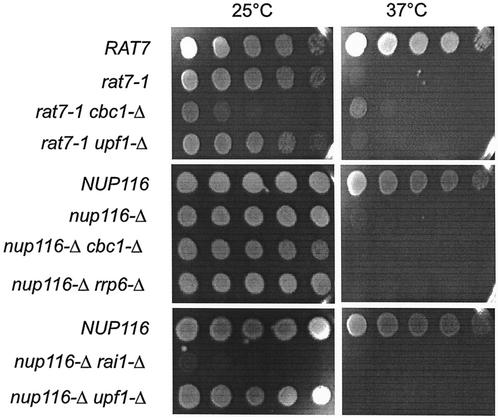
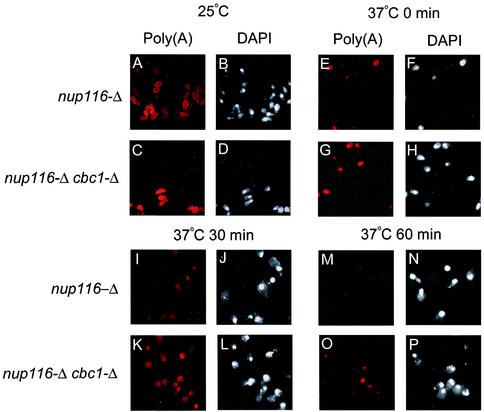
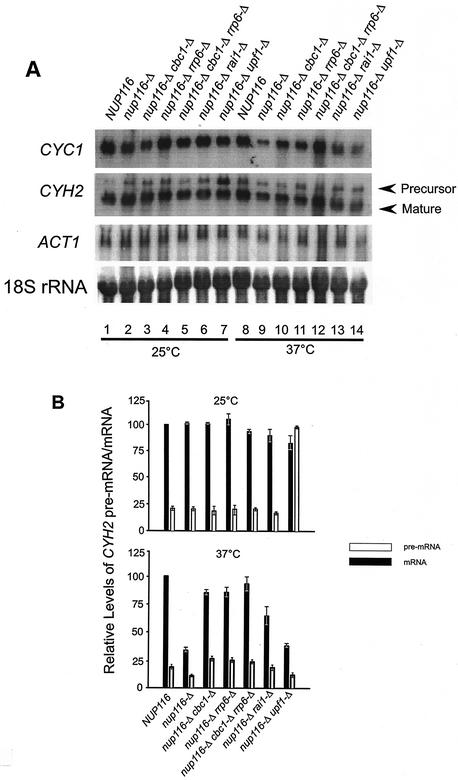
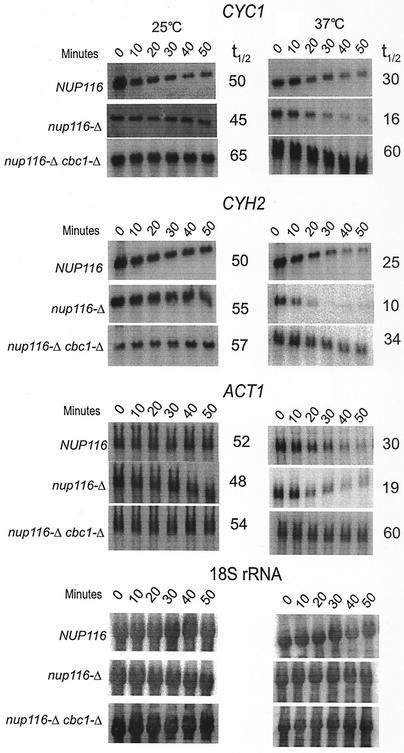
In this investigation, we have demonstrated the existence of DRN, a novel nuclear mRNA decay pathway, primarily by using strains containing the nup116-Δ mutation that was previously reported to prevent the export of RNA at the restrictive condition of 37°C (3, 31, 81). We first demonstrated that lethal effect of nup116-Δ was not suppressed by cbc1-Δ or rrp6-Δ, etc., which are mutations in putative components of DRN (Fig. 1), thus suggesting that mRNA is retained in the nucleus in these mutant strains. Subsequently, we used the FISH procedure to directly verify that total poly(A) RNA is retained in the nucleus at the restrictive condition of 37°C in the nup116-Δ, as well as in the cbc1-Δ nup116-Δ strain (Fig. 2). Furthermore, Northern blot analysis was used to verify the cytological results by determining the steady-state levels (Fig. 3) and half-lives (Fig. 4) of the representative CYC1, CYH2, and ACT1 mRNAs in nup116-Δ strains under the restrictive condition. Finally the effect of the cbc1-Δ, rrp6-Δ, and other mutations on the degradation of the representative mRNAs in a set of isogenic nup116-Δ strains were tested by examining steady-state levels (Fig. 3) and half-lives (Fig. 4 and 5). Graphical representation of some of the half-lives are presented in Fig. 6.
FIG. 1.
The growth of 1/10 serial dilutions of suspensions of various strains (Table 1) at 25°C for 4 days or 37°C for 2 days on YPD medium, demonstrating that the growth defect at 37°C of nup116-Δ strains is not suppressed by cbc1-Δ, rrp6-Δ, or other mutations, whereas the growth defect at 37°C of rat7-1 strains are partially suppressed by cbc1-Δ.
FIG. 2.
FISH analyses revealing that total poly(A)+ RNA in nup116-Δ and nup116-Δ cbc1-Δ strains are retained in the nucleus and that there is less degradation in the nup116-Δ cbc1-Δ strain. The isogenic pairs of strains were grown at 25°C to the mid-logarithmic phase of growth. Subsequently, one-half of each culture was transferred to the restrictive temperature of 37°C; the cultures were further incubated for one additional hour at both temperatures. Transcription of the cells in the culture shifted to 37°C was inhibited by the addition of thiolutin (4 μg/ml); the cells were harvested at various times after transcription block as indicated on top of each panel in the figure and subsequently fixed and processed for FISH and DAPI analysis as described in Materials and Methods. Left panels of mock shifted as well as each time point after transcription block show the localization and decay of the total poly(A)+ RNA as visualized using Cy3-labeled oligo(dT) are denoted as Poly(A), whereas right panels of respective time points show the nuclear DNA as visualized using DAPI staining denoted as DAPI. The time indicated at the top of each panel represents the time after transcription block to after shift to 37°C. See the Results (“Existence of DRN, a Cbc1p-dependent nuclear mRNA degradation pathway: cytological evidence”) for details of each panel.
FIG. 3.
Comparison of the steady-state levels of CYC1, ACT1, and CYH2 precursor and matured mRNAs at 25°C (mock-shifted) and 37°C (shifted) in NUP116 (normal), nup116-Δ, nup116-Δ cbc1-Δ, nup116-Δ rrp6-Δ, nup116-Δ cbc1-Δ rrp6-Δ, nup116-Δ rai1-Δ, and nup116-Δ upf1-Δ deletion strains. (A) Northern blots of steady-state levels of CYC1, ACT1, and CYH2 mRNAs and pre-mRNA in different strains are indicated at the top of each lane. All the strains were grown at 25°C until mid-log phase, half of the culture of each strain was then shifted to 37°C. Both cultures of each strain at 25 and 37°C were incubated for 1 h at the respective temperature and harvested. Subsequently, the steady-state levels of the each mRNA and pre-mRNA were determined by Northern blot analysis with the total RNA isolated from each of these strains and probing for respective pre-mRNA and mRNAs isolated from strains grown at 25°C (lanes 1 to 7) and shifted to 37°C (lanes 8 to 14). The signal for each mRNA in each lane was quantified as described in Materials and Methods and normalized against the 18S rRNA signals (shown at the lowest panel of A) for loading errors and the relative steady-state levels of each mRNA are presented in Table 2. (B) Quantification of the CYH2 pre-mRNA and mRNA signals. The intensity of each band of mature and precursor mRNA were determined by scanning the blots with a PhosphorImager and by normalizing for loading differences with respect to 18S rRNA signals. The relative levels of pre-mRNA and mRNA in each strain were expressed with respect to that in the NUP116 (normal) strain at each temperature, which was considered to be 100%. The error bar represents the range of three independent experiments.
FIG. 4.
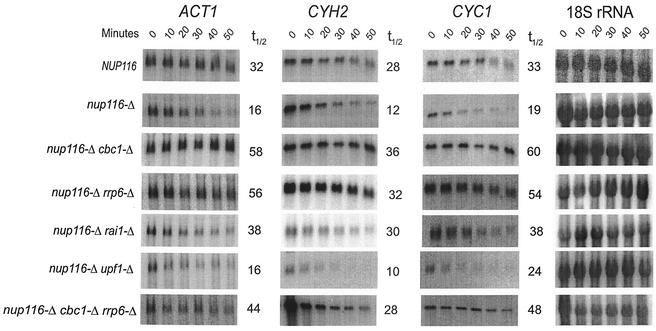
Northern blot analysis revealing an increased degradation of CYC1, CYH2, and ACT1 mRNAs that are retained in the nucleus because of the export deficiency caused by nup116-Δ mutation. Furthermore, the Northern blot analysis also revealed that the degradation is suppressed by cbc1-Δ at 37°C. The NUP116 (normal), nup116-Δ, and nup116-Δ cbc1-Δ strains were grown at 25°C to the mid-logarithmic phase of growth. Subsequently, one-half of each culture was transferred to the restrictive temperature of 37°C. Both the cultures of each strain at 25 and 37°C were further incubated for one additional hour at both temperatures, and transcription was inhibited by the addition of thiolutin (4 μg/ml), as described in Materials and Methods. Cells of each strain from both the temperatures, mock shifted and shifted, were harvested after various times of thiolutin addition; Northern blots were prepared with total RNA; the half-lives of CYC1, CYH2, and ACT1 mRNA were determined as described in Materials and Methods and normalized against 18S rRNA shown at the bottom of the figure. The half-lives are presented beside each panel as well as in Table 3.
FIG. 5.
A comparison of the decay of ACT1, CYH2, and CYC1 mRNAs in NUP116 and various nup116-Δ strains by Northern blot analysis. The analysis was performed as described in the legend of Fig. 4 after normalizing each signal against that from 18S rRNA internal control (as shown on the rightmost panels) for each strain and the half-lives are presented beside each panel as well as in Table 4.
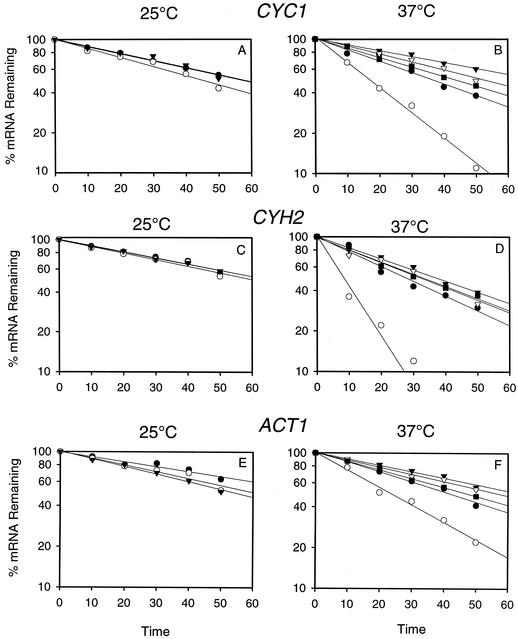
FIG. 6.
Graphical representation of decay of CYC1, CYH2 and ACT1 mRNAs at 25°C (A, C, and E) and at 37°C (B, D, and F) from thiolutin treated cells of NUP116 (•), nup116-Δ (○), nup116-Δ cbc1-Δ (▾), nup116-Δ rrp6-Δ (▿), and nup116-Δ cbc1-Δ rrp6-Δ (▪). The decay was determined by Northern blot analysis of the RNA extracted from the strains mentioned above treated with thiolutin from 0 to 50 min. The result from one typical experiment from each strain at different temperatures are presented as the percentage of mRNA remaining versus time of incubation of thiolutin.
cbc1-Δ and other mutations do not suppress the growth defect of nup116-Δ.
We previously demonstrated that cbc1-Δ suppressed both the growth defect as well as rapid decay of specific mRNAs in the rat7-1 strain at 37°C (15). The suppression of rat7-1 by cbc1-Δ complicated our efforts to study the fate of mRNAs retained in the nucleus, so we have extended our studies of the Cbc1p-dependent decay of mRNAs by investigating a number of mutants defective in mRNA export, including rat7-1, nup116-Δ, and hpr1-Δ (Table 1). These mRNA export defective mutants were tested for suppression of their growth defect by cbc1-Δ, rrp6-Δ, and several other mutants. As shown in Fig. 1, cbc1-Δ and all other tested mutations did not suppress the growth defects of nup116-Δ at 37°C, a finding that is critical for the studies described below. The lack of growth of nup116-Δ cbc1-Δ and other double mutant strains (Table 1) at the restrictive temperature implies that total poly(A) RNA in these strains is not exported to the cytoplasm and still remains in the nucleus. This allowed us to directly investigate the effect of cbc1-Δ and other mutations on mRNAs retained in the nucleus.
Existence of DRN, a Cbc1p-dependent nuclear mRNA degradation pathway: cytological evidence.
The inability of the cbc1-Δ deletion mutant and the several other mutants to suppress the temperature sensitive mRNA export defective nup116-Δ deletion mutant (82) prompted us to test the nuclear retention of total poly(A) RNA in a nup116-Δ cbc1-Δ strain. FISH analysis was used to verify if total poly(A) RNA accumulates in the nucleus under our experimental conditions and to demonstrate the existence of the DRN system. This technique distinguishes between the nuclear and cytoplasmic distribution of poly(A) RNA. A steady-state FISH analysis of poly(A) RNA, using Cy3-labeled 43-mer oligo(dT) probe, revealed that after 1 h of a shift from the permissive condition of 25°C to restrictive condition of 37°C, the fluorescent signal is predominantly nuclear in both the nup116-Δ and nup116-Δ cbc1-Δ strains when transcription is blocked, thus confirming that under this condition the vast majority of the cellular total poly(A) RNA is nuclear (data not shown; compare Fig. 2E and G and with Fig. 2F and H, which represents the position of the nucleus counterstained with DAPI). However, we sometimes observed a weak cytoplasmic background signal in a fraction of cells of both nup116-Δ and nup116-Δ cbc1-Δ strains. These observations justify the method employed and the condition used to investigate the degradation of total poly(A) RNA, as well as specific mRNAs by determining their half-lives, as described below.
The rates of degradation in situ of poly(A) RNA in the nucleus of both nup116-Δ and nup116-Δ cbc1-Δ strains were similarly investigated by using the Cy3-labeled 43-mer oligo(dT) probe under the following conditions: (i) the control culture at 25°C (Fig. 2A to D; designated 25°C) before the temperature shift, when both the export and transcription are still progressing; (ii) 1 h after the shift to 37°C and just before transcription is blocked (Fig. 2E to H; designated 37°C, 0 min); and (iii) 30 min (Fig. 2I to L; designated 37°C, 30 min) and (iv) 60 min (Fig. 2M to P; designated 37°C, 60 min) after the transcription is blocked. The left half of each pair of panels for each time point shows the poly(A) RNA localization and the right half shows the counter staining of the nucleus by DAPI. As shown in Fig. 2, the poly(A) RNA signal was diffuse over both the nucleus and cytoplasm of both nup116-Δ and nup116-Δ cbc1-Δ strains at the permissive condition of 25°C (Fig. 2A to D). After 1 h at 37°C and before blocking transcription (37°C, 0 min), the signal was predominantly nuclear in both strains, as revealed by the colocalization of the DAPI and poly(A) signals (Fig. 2E to H). On continued incubation at 37°C after blocking transcription, the nuclear fluorescence of the poly(A) RNA decreases more rapidly in the nup116-Δ strain compared to the nup116-Δ cbc1-Δ strain (Fig. 2I to L for 30 min and Fig. 2M to P for 60 min after transcription arrest; compare Fig. 2M with 2O). After 60 min of transcription block, the majority of the poly(A) signal disappeared in the nuclei of nup116-Δ strain (Fig. 2M). On the other hand under the same condition, the poly(A) signal still remained much stronger in the nuclei of nup116-Δ cbc1-Δ strain (Fig. 2O). Thus, this result clearly suggests that the decay of poly(A) RNA occurs in the nucleus of nup116-Δ strains, and this degradation requires Cbc1p. Importantly, these findings indicate that this method will allow us to determine the fate of mRNAs retained in the nucleus and thereby establish details of the DRN system.
Existence of DRN. (i) Representative mRNAs retained in the nucleus are degraded more rapidly.
In order to substantiate the cytological observation described above, we first tested if intranuclear retention of specific mRNAs would affect their steady-state levels. Total RNAs from the normal NUP116 strain and the isogenic nup116-Δ mutant (Table 1) were subjected to Northern blot analysis after the strains were shifted from 25°C to 37°C for 1 h, a condition that results in the complete nuclear retention of mRNAs in nup116-Δ (82) (see above). After a 1-h temperature shift to 37°C, the steady-state levels of mRNAs of the three representative transcripts, CYC1, CYH2, and ACT1, were found to be lower in the nup116-Δ mutant compared to the normal NUP116 strain (Table 2; Fig. 3), indicating that the normal mRNAs retained in the nucleus of nup116-Δ strain might be unstable. Subsequent half-life measurements established that the lower abundance of all the three mRNAs in the nup116-Δ mutant was due to faster degradation (Tables 3 and 4; Fig. 4 and 6). Mock shifted control conditions, on the other hand, did not reveal any significant difference between the NUP116 and nup116-Δ strains in either the abundance (Table 2; Fig. 3) or half-lives of these transcripts (Table 3; Fig. 4 and 6) when the export of mRNAs in nup116-Δ strain is not blocked. The lower abundance in steady-state level and the enhanced instability of these representative mRNAs observed only with the nup116-Δ strain under the condition of complete retention of poly(A) RNA, i.e., after 1 h at 37°C, provides strong evidence for the existence of the DRN pathway, a conclusion that is also substantiated by the FISH analysis described above.
TABLE 2.
Relative steady-state levels of CYC1, ACT1, and CYH2 pre-mRNA and mRNA in various strainsa
| Pertinent genotype | Steady-state level
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
CYC1
|
CYH2
|
CYH2 pre-mRNA
|
ACT1
|
|||||
| 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | |
| NUP116 | 100 | 100 | 100 | 100 | 20 ± 0.5 | 23 ± 1.5 | 100 | 100 |
| nup116-Δ | 94 ± 2 | 27 ± 2.5 | 102 ± 2 | 38 ± 2.5 | 21 ± 1.5 | 10 ± 0.25 | 101 ± 3 | 53 ± 2.5 |
| nup116-Δ cbc1-Δ | 91 ± 4 | 76 ± 3 | 101 ± 2 | 84 ± 3 | 19 ± 2 | 29 ± 3 | 102 ± 1 | 81 ± 3 |
| nup116-Δ rrp6-Δ | 86 ± 3.5 | 66 ± 2 | 109 ± 8 | 85 ± 5 | 24 ± 3 | 26 ± 3 | 101 ± 3.5 | 75 ± 4 |
| nup116-Δ rail-Δ | 94 ± 4 | 55 ± 1.5 | 87 ± 8 | 71 ± 6 | 18 ± 0.5 | 20 ± 2 | 83 ± 4 | 70 ± 2 |
| nup116-Δ upf1-Δ | 92 ± 6 | 37 ± 2 | 83 ± 7 | 45 ± 2 | 102 ± 4 | 13 ± 1.5 | 96 ± 7 | 43 ± 2.5 |
| nup116-Δ cbc1-Δ rrp6-Δ | 105 ± 6 | 66 ± 2.5 | 92 ± 2 | 90 ± 6 | 23 ± 1 | 22 ± 2.5 | 93 ± 1 | 79 ± 6 |
The steady-state levels were determined with total RNA isolated from the indicated strains either from 25°C or after 1 h of temperature shift at 37°C as described in Materials and Methods. The level of each mRNA at each temperature in the NUP116 strain was arbitrarily chosen as 100%, and the relative levels at which mRNA in different strains were expressed as a percentage of that of strain NUP116 at respective temperatures. The level of CYH2 pre-mRNA in each strain was expressed as a percentage of the CYH2 mRNA level in strain NUP116 at each temperature. Where applicable, the mean values are presented (determined from three independent experiments) to nearest whole number. The number after the mean value represents the range of three independent experiments.
TABLE 3.
Half-lives of ACT1, CYH2, and CYC1 mRNAs in various strainsa
| Pertinent genotype | Half-life (min) of:
|
|||||
|---|---|---|---|---|---|---|
|
ACT1
|
CYH2
|
CYC1
|
||||
| 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | |
| NUP116 | 52 ± 4.5 | 32 ± 1.5 | 50 ± 3.5 | 26 ± 2.0 | 50 ± 3.5 | 31 ± 2.5 |
| nup116-Δ | 48 ± 3.25 | 18 ± 1.5 | 55 ± 4.0 | 11 ± 1.0 | 45 ± 2.0 | 18 ± 1.5 |
| nup116-Δ cbc1-Δ | 54 ± 4.25 | 60 ± 2.0 | 57 ± 3.75 | 37 ± 3.0 | 65 ± 3.0 | 61 ± 1.0 |
The half-lives were determined with total RNA from thiolutin-treated cells as described in Materials and Methods. The values of multiple determinations fell within 15% of the mean. Where applicable, the mean values are presented (determined from three independent experiments) to nearest whole number. The number after the mean value represents the range of three independent experiments.
TABLE 4.
Comparison of half-lives of ACT1, CYH2, and CYC1 mRNAs in different mutant strains under the restrictive condition of 37°C after temperature shifta
| Pertinent genotype | Half-life (min) of:
|
||
|---|---|---|---|
| ACT1 | CYH2 | CYC1 | |
| NUP116 | 32 ± 1.5 | 26 ± 2.0 | 31 ± 2.5 |
| nup116-Δ | 18 ± 1.5 | 11 ± 1.0 | 18 ± 1.5 |
| nup116-Δ cbc1-Δ | 60 ± 2.0 | 37 ± 3.0 | 61 ± 1.0 |
| nup116-Δ rrp6-Δ | 56 ± 6.5 | 28 ± 3.0 | 52 ± 5.5 |
| nup116-Δ rail-Δ | 38 ± 2.5 | 30 ± 2.5 | 38 ± 3.0 |
| nup116-Δ upf1-Δ | 16 ± 1.5 | 10 ± 1.0 | 24 ± 1.0 |
| nup116-Δ cbc1-Δ rrp6-Δ | 44 ± 3.0 | 26 ± 2.0 | 48 ± 3.5 |
| HPR1 | 34 ± 3.5 | NDb | ND |
| hpr1-Δ | 22 ± 1.5 | ND | ND |
The half-lives were determined with total RNA from thiolutin-treated cells as described in Materials and Methods. The values of multiple determinations fell within 15% of the mean. Where applicable, the mean values are presented (determined from three independent experiments) to nearest whole number. The number after the mean value represents the range of three independent experiments.
ND, not determined.
(ii) Instability of mRNAs retained in the nucleus is dependent on Cbc1p.
In subsequent experiments, we used Northern blot analysis of mRNA levels and decay rates to confirm the cytological observation that Cbc1p is responsible for decay of total poly(A) RNA in the nucleus (Fig. 2). Total RNA from a nup116-Δ strain and nup116-Δ cbc1-Δ strains shifted to 37°C for 1 h was isolated and subjected to Northern blot analysis, which revealed that the steady-state levels of the three representative transcripts, CYC1, CYH2, and ACT1, increased by approximately 1.5- to 3-fold compared to the levels in the nup116-Δ strain under the same condition (Table 2; Fig. 3). Also no significant difference in the levels of the mRNAs was observed in the control experiment where the nup116-Δ cbc1-Δ strain was maintained at 25°C for 1 h (Table 2; Fig. 3). Subsequent measurement of mRNA half-lives after inhibition of transcription of NUP116, nup116-Δ, and nup116-Δ cbc1-Δ strains (Table 1) by thiolutin revealed that the CYC1, CYH2, and ACT1 mRNAs are degraded more rapidly in nup116-Δ strain when the strains are shifted to 37°C, a condition that prevents export of RNAs from the nucleus (Table 3; Fig. 4 and 6). Furthermore, this rapid degradation is suppressed by cbc1-Δ, resulting in half-lives that were more-than-threefold longer than those obtained with nup116-Δ (Table 3; Fig. 4 and 6). In fact these half-lives are even higher than those of NUP116 mRNAs. It appears from the combined results of the FISH and Northern analyses that mRNAs are rapidly degraded if retained in the nucleus and that Cbc1p is a required component of this degradation system.
DRN is distinct from NMD.
We have investigated potential relationships between DRN and NMD by testing possible effects of upf and other mutations on the degradation of representative mRNAs in nup116-Δ strains. This issue was addressed by investigating the degradation of CYC1, CYH2, and ACT1 mRNAs in the nup116-Δ and nup116-Δ upf1-Δ strains and revealed that the upf1-Δ deletion did not stabilize any of the transcripts (Fig. 5; Table 4). These results indicate that the decay of transcripts in the nucleus of nup116-Δ strain is independent and distinct from the cytoplasmic NMD pathway. Furthermore, the steady-state levels of the intron-containing CYH2 pre-mRNA, one of the well characterized natural substrate of NMD pathway (23), was determined in the NUP116, nup116-Δ, nup116-Δ cbc1-Δ, and nup116-Δ upf1-Δ strains. As expected, the level of the CYH2 pre-mRNA remained the same in NUP116, nup116-Δ, and nup116-Δ cbc1-Δ strains at 25°C but accumulated by approximately 10-fold in the nup116-Δ upf1-Δ strain under the same condition (Fig. 3). Because the export of poly(A) RNA can occur in the nup116-Δ strain at 25°C, the majority of the CYH2 pre-mRNA is cytoplasmic and thus accumulates in the nup116-Δ upf1-Δ strain due to the lack of NMD. In contrast, CYH2 pre-mRNA did not accumulate when the nup116-Δ upf1-Δ strain was incubated at 37°C for 1 h, a condition that prevents the export of pre-mRNA and allows degradation in the nucleus. These findings confirm that transcripts in the nup116-Δ and nup116-Δ upf1-Δ strains were degraded in the nucleus and not in the cytoplasm. Thus, the stability analysis of different transcripts and the in situ localization study described above clearly identifies, DRN, a novel, Cbc1p-dependent nuclear pathway of mRNA decay that is distinct from the previously known cytoplasmic pathways.
We were unable to determine if the transcripts were stabilized in nup116-Δ xrn1-Δ strains, because the double deletion mutant was not recovered, suggesting that xrn1-Δ may be synthetically lethal with nup116-Δ.
The Rrp6p and possibly Rat1p exoribonucleases are components of DRN.
We investigated the major question of which exoribonuclease is associated with DRN by determining if mutation of any of the known nuclear exoribonucleases could suppress cyc1-512, similar to suppression by cbc1-Δ. We considered the well-characterized 5′→3′ exoribonuclease, Rat1p (41), which is involved in 5′-end processing of snoRNAs and rRNAs and degradation of spacer fragments of pre-snRNA and pre-rRNAs (25, 61). The nuclear Rat1p is homologous to the cytosolic Xrn1p, which is engaged in 5′→3′ degradation of mRNAs. In spite of being homologous, Xrn1p is nonessential, whereas Rat1p is essential (39). Examination of the iso-1-cytochrome c level in a cyc1-512 rat1-1 strain, constructed with a conditional allele of RAT1, revealed that rat1-1 did not suppress cyc1-512, suggesting that Rat1p may not be involved in DRN. However, the results with conditional mutants can be ambiguous, as the lack of growth at the restrictive temperature may prevent manifestation of suppression.
The role of Rat1p was addressed further by examining the degradation rates of representative CYC1, CYH2, and ACT1 mRNAs in a nup116-Δ rai1-Δ strain. RAI1 is an unessential gene that binds to and modulates the activity of Rat1p both in vivo and in vitro. Furthermore, rai1-Δ strains are viable and lack Rat1p activity in vivo (84). However, rai1-Δ strains, including the nup116-Δ rai1-Δ strain, grow poorly (Fig. 1). Northern blots of total RNA from the nup116-Δ rai1-Δ strain were analyzed at 37°C and at different times after the temperature shift and transcription inhibition, according to the procedure used for the other strains. The results of the steady-state levels and half-life measurements at 37°C of the three representative CYC1, CYH2, and ACT1 mRNA transcripts in a nup116-Δ rai1-Δ strain (Fig. 3 and 5; Tables 2 and 4) indicated a twofold increase in both steady-state levels and stability compared to that of nup116-Δ under the same condition, suggesting that Rat1p possibly plays a minor role in DRN.
In addition, we have also considered the possibility of involvement of nuclear exoribonuclease Rrp6p, a protein involved in the 3′ processing of the 5.8S rRNA (7) and part of nuclear exosome (2). Burkard and Butler (8) provided direct evidence for the 3′→5′exonuclease activity of this protein in vitro (8). RRP6 is not essential for viability (7), and a strain carrying a precise deletion of RRP6 is impaired in growth at all temperatures and is nonviable at 37°C. We have established its role in DRN by demonstrating suppression of cyc1-512 by rrp6-Δ; the level of cytochrome c was increased from 10% of the normal level in the cyc1-512 strain to 30% in the cyc1-512 rrp6-Δ strain (data not shown). Consistent with this genetic evidence, the abundance of the CYC1, ACT1, and CYH2 mRNAs and CYH2 pre-mRNA (Fig. 3; Table 2) was found to increase by approximately threefold. Furthermore, degradation of the representative transcripts, CYC1, CYH2, and ACT1 mRNAs, was diminished approximately threefold in a nup116-Δ rrp6-Δ strain at 37°C under the condition of complete nuclear retention of poly(A) RNA, compared to the control nup116-Δ strain. This finding is similar to the effect of cbc1-Δ (Fig. 5 and 6; Table 4). These results indicate that Rrp6p is a component of DRN and that the degradation takes place at least in part in a 3′→5′ direction. Furthermore, because the rates of ACT1, CYH2, and CYC1 mRNA degradation in the nup116-Δ cbc1-Δ rrp6-Δ strain were similar to the rates of degradation in nup116-Δ cbc1-Δ and nup116-Δ rrp6-Δ strains (Fig. 5; Table 4), both Cbc1p and Rrp6p appear to be involved in the same pathway, not in separate parallel pathways. Taken together these results revealed a major role of Rrp6p in DRN.
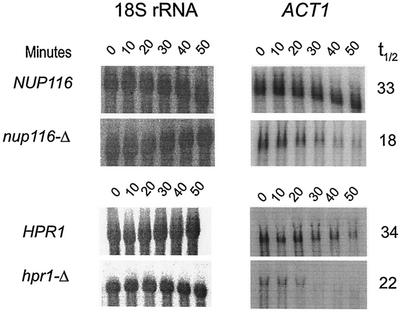
Another block in mRNA export enhances mRNA degradation.
We also tested the degradation of ACT1 mRNA in an additional conditionally lethal mutant, hpr1-Δ, defective in mRNA export. Similar to the results with the nup116-Δ (see above) and rat7-1 mutants (15), the steady-state levels (data not shown) and half-lives of the ACT1 mRNA were diminished in the hpr1-Δ mutant (Table 4; Fig. 7). Therefore it appears that this accelerated nuclear mRNA decay in nup116-Δ strains is not specific for this export defect. Rather, accelerated mRNA decay occurs as a consequence of nuclear retention of mRNAs, whether the deficiency is due to defective nucleoporins, such as that with rat7-1 and nup116-Δ, or due to a defect in another pathway, such as that with hpr1-Δ, which causes defects in elongation and metabolism of nascent mRNA and mRNA export proteins (38) and which results in RNA export defects at 37°C (69). In addition, increased degradation can also be due to the intrinsic defect in the sequence or the structure of mRNA that causes partial retention of a specific defective mRNA in the nucleus without any export or processing defect, such as in the cyc1-512 mutant (15).
FIG. 7.
Northern blot analysis revealing an increased degradation of ACT1 mRNA (right panels), which is retained in the nucleus because of the export deficiency caused by nup116-Δ or hpr1-Δ mutations at the restrictive temperature of 37°C when compared to the corresponding isogenic normal strain. The NUP116 (normal), nup116-Δ, and HPR1 and hpr1-Δ strains were grown at 25°C to the mid-logarithmic phase of growth. Subsequently, one-half of each culture was transferred to the restrictive temperature of 37°C; the cultures were further incubated for one additional hour at that temperature; and transcription was inhibited by the addition of thiolutin (4 μg/ml), as described in Materials and Methods. Northern blots were prepared using total RNA extracted from cells after various times, 0 to 50 min, of thiolutin addition. The half-lives, presented in Table 4, were determined from these blots after normalization to the 18S rRNA signals shown at the left panels. The numbers beside each panel represents the half-lives in minutes.
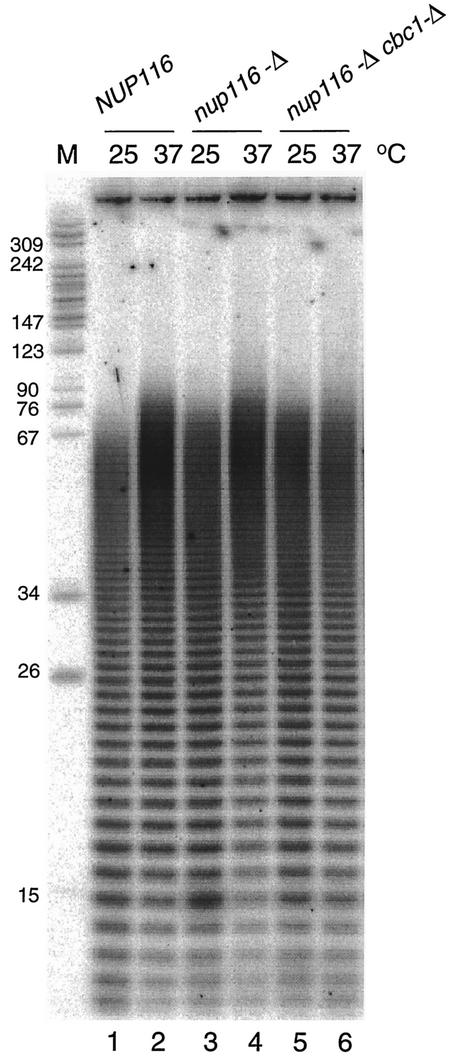
The nup116-Δ mutation does not cause hyperadenylation.
Mutations that block the export of mRNA from the nucleus often result in hyperadenylation of transcripts and are associated with mRNA retention in nuclear foci that may represent the site of transcription (37). We tested whether nuclear retention of mRNAs caused by the nup116-Δ mutation resulted in hyperadenylation of mRNAs by labeling their poly(A) tails and comparing the amounts and lengths in NUP116 and nup116-Δ strains. The results for the normal and mutant strains show an increase in the amount of the longest poly(A) tails, which is typical for yeast strains grown at 37°C (37, 63). Cells with the nup116-Δ mutation show a slight increase of ∼15 to 20 nucleotides (nt) in the poly(A) lengths at 25°C and after a shift to 37°C for 60 min (Fig. 8, lanes 1 to 4). This small increase in poly(A) tail length is not affected by the cbc1-Δ mutation (Fig. 8, lanes 5 to 6). These findings indicate that while the nup116-D mutation results in a small increase in the longest poly(A) tails, it does not result in the 50- to100-nt increase observed for other nuclear export mutations (37).
FIG. 8.
Poly(A) tail analysis of mRNA from strains carrying the nup116-Δ mutation. Poly(A) tails were analyzed by 3′ end labeling of 1 μg of total RNA with [32P]CP and RNA ligase, followed by hydrolysis with RNase A and RNase T1, electrophoretic separation on a 16% acrylamide-8 M urea gel, and storage phosphorimager analysis. Lane M indicates pBR322 MspI-cut length markers.
DISCUSSION
DRN is a general consequence of retention of mRNAs and pre-mRNAs in the nucleus.
This investigation clearly revealed that normal mRNAs are degraded when retained in the nucleus, and that this so-called DRN degradation is suppressed by the rrp6-Δ, rai1-Δ, and cbc1-Δ deletions. Thus, DRN involves the following: the 3′→5′ nuclear exonuclease, Rrp6p; the 5′→3′ nuclear exonuclease, Rat1p; and CBC, the nuclear cap binding complex, which may direct the mRNAs to the site of degradation. Degradation was investigated primarily by examining the stability of representative ACT1, CYH2, and CYC1 mRNAs after inhibition of transcription with thiolutin in nup116-Δ strains, which retain mRNAs in the nucleus under restrictive conditions.
The decay rates of specific eukaryotic mRNAs can vary by more than 100-fold (11, 64, 65) and can range from approximately 1 min to more than 90 min in S. cerevisiae (27, 51, 79). The diversity in turnover can be attributed in part to a wide variety of mechanisms that have been suggested to take place by-and-large in the cytosol (12, 56). Here we demonstrate that normal mRNAs are degraded also in the nucleus. These studies extend the findings of Das et al. (15), who reported that ACT1 and CYH2 mRNAs were only marginally stabilized, 20 to 25%, whereas the longer cyc1-512 transcripts were stabilized by approximately 200% by cbc1-Δ. Because the turnover of CYC1, as well as ACT1 and CYH2 mRNAs were not significantly affected by cbc1-Δ, Das et al. (15) suggested that long cyc1-512 mRNAs are partially retained in the nucleus, and the net destruction of a particular mRNA is determined in part by the length of time spent in the nucleus. It is reasonable to suggest that the turnover of certain mRNAs is also controlled in part by intrinsic properties of restricting their export from the nucleus. Therefore, it would be of considerable interest to determine if any wild-type mRNAs have a property similar to the mutant cyc1-512 mRNAs, a study that is in progress.
Our results showed that increased rates of ACT1 mRNA degradation were observed when they were retained in the nucleus by any of the mutations—nup116-Δ, rat7-1, and hpr1-Δ—which act by a variety of mechanisms. Furthermore, these results are consistent with the results of the FISH analysis, which indicate that total poly(A) RNA was similarly affected. However, most of the studies in this investigation were carried out with nup116-Δ strains, because the mRNA export defect in this strain is essentially complete at the restrictive condition and because the nup116-Δ defect was not apparently suppressed by any of the rrp6-Δ, rai1-Δ, and cbc1-Δ deletions, which were used to characterize DRN. In contrast, cbc1-Δ suppressed the temperature-sensitive growth of rat7-1 (15) and hpr1-Δ; consequently, these mutants were avoided for examining mRNA retention and decay in the nucleus. Uemura et al. (75) also reported that a cbc1 mutation suppressed the temperature-sensitive growth of hpr1 mutants, which are conditionally defective in the nuclear export of poly(A) RNA (69).
DRN involves both 3′→5′ and 5′→3′ degradation pathways.
The suppression of degradation of ACT1, CYH2, and CYC1 mRNAs in thiolutin treated nup116-Δ rrp6-Δ strains established that the 3′→5′ nuclear exonuclease, Rrp6p, is the major nuclease acting in DRN, adding to its other major function, nuclear pre-rRNA processing (8, 9). Rrp6p specifically associates with the nuclear form of the exosome (2, 8). Although RRP6 is not essential for viability (7) all other components of the exosome were found to be essential (1, 2, 53, 54). Like the other exosome mutants the rrp6-Δ strain is defective in the 3′ processing of the 5.8S rRNA, but differed from the others insofar as it accumulated a discrete species, 5.8S + 30, which was 3′ extended by ∼30 nt (7). These differences in phenotypes of rrp6− and other exosome mutants led Burkard and Butler (8) to speculate that Rrp6p may act independently of the exosome as a monomeric exonuclease or in conjunction with another set of proteins. Our results show a threefold increase in both the steady-state levels and in the stability of all the representative normal mRNAs in nup116-Δ rrp6-Δ strains thereby demonstrating that Rrp6p plays a major role in DRN. It is notable, however that although a 5- to 10-fold stabilization of various intron-containing pre-mRNAs in absence of Rrp6p was previously reported (6), we did not observe such a high degree of stabilization. This difference in the degree of stabilization may reflect the differences in the experimental systems employed in the previous work. Finally, we would like to speculate that DRN might be taking place in the nucleolus as Rrp6p is localized more densely in that region although it is present throughout the nucleoplasm (9).
Rat1p, the major nuclear 5′→3′exoribonuclease participates in a variety of functions such as the 5′-end processing of snoRNAs and rRNAs and degradation of spacer fragments of pre-snRNA and pre-rRNAs (25, 61) and requires Rai1p for both of its in vitro and in vivo activity (84). Because rat1-Δ deletion mutants are lethal, we examined rai1-Δ strains, which lack Rat1p activity in vivo (84). A relatively modest degree of suppression of degradation was observed in nup116-Δ rai1-Δ strains which, although are viable, exhibit greatly reduced growth, suggesting the elimination of certain critical functions described above. This observation suggests that Rat1p, might also act in DRN, but its effect is relatively modest; thus, we suggest that both the 5′→3′ and 3′→5′ pathways participate in DRN with 3′→5′ pathway being the major pathway. However, we have not determined the relative contribution of each.
DRN requires CBC, the nuclear cap binding complex.
CBC, the nuclear cap binding complex, is a critical component of DRN and defines this pathway (15). A major question is the mechanism by which CBC is required for DRN. CBC consists of a heterodimer of two proteins, denoted CBP80 and CBP20 in higher eukaryotes and Cbc1p and Cbc2p, respectively, in yeast (19, 34, 35, 36, 40, 41, 59). Mutant forms of the CBC components were recovered in numerous genetic screens with yeast, and CBC1 has been previously designated SUT1 (15), GCR3 (75), and STO1 (13), whereas CBC2 has also been designated MUD13 (13). Similar to higher eukaryotic CBC, yeast CBC is primarily located in the nucleus. Experiments performed in vivo and in vitro indicate that CBC plays a role in both pre-mRNA splicing and U snRNA export (34, 35, 49). CBC associates with the cap structures of pre-mRNA and nuclear mRNA in vivo and accompanies mRNA through nuclear pore complexes to the cytoplasm (78).
CBC is not essential for growth in yeast, although the growth of cbc1-Δ strains is severely retarded on glucose medium but only mildly diminished on glycerol (74) and raffinose (15) media. Fortes et al. (19) relied on the nonessentiality of CBC to carry out a genetic screen for components that show synthetic lethality with a cbc1-Δ cbc2-Δ double deletion mutant strain. One group of synthetically lethal mutations was due to alterations that were complemented by components of U1 snRNP and the yeast splicing commitment complex. These interactions confirmed the role of CBC in commitment complex formation in yeast. Fortes et al. (19) also demonstrated the physical interaction of Cbc1p and Cbc2p with the commitment complex components Mud10p and Mud2p, which may directly mediate function. Most interestingly, Fortes et al. (19) identified five synthetically lethal mutations that were complemented by CBF5 and NOP58, which encode components of the two major classes of yeast snoRNPs functioning in the maturation of rRNA precursors. Cbf5p and Nop58p are essential nucleolar proteins that are core components of the box C+D and box H+ACA families of snoRNPs, respectively (42, 43). Both Nop58p and Cbf5p are required for the early pre-rRNA processing steps at sites A0, A1, and A2 in the pathway of 18S rRNA synthesis, and the synthetically lethal strains had defects in pre-rRNA processing at these steps (19). Most importantly, the cbc1-Δ cbc2-Δ strain by itself was defective in the cleaving at sites A0, A1, and A2, possibly explaining synergism and the synthetic lethality with CBF5 and NOP58 mutations. Although the role of CBC in nucleolar pre-rRNA processing has not been explained, it is tempting to speculate that the diminished cleavage of pre-rRNA and the diminished degradation of nuclear mRNAs have a common mode of action that involves the enhanced localization of capped RNAs, such as mRNAs and snoRNAs, to nucleoli. However, the cap may not be absolutely required for nucleolar localization of snoRNAs (45).
CBC and Rrp6p appear to act in the same pathway.
mRNA degradation in nup116-Δ revealed that CBC and Rrp6p both participate in the degradation of normal mRNAs in the nucleus. In order to determine whether the CBC and Rrp6p participate in the same or parallel pathways, the rates of ACT1, CYH2, and CYC1 mRNA degradation in the nup116-Δ cbc1-Δ rrp6-Δ strain were compared to the rates of degradation in nup116-Δ cbc1-Δ and nup116-Δ rrp6-Δ strains. The similar values and the lack of increased stability in the nup116-Δ cbc1-Δ rrp6-Δ strain (Table 4; Fig. 5) suggest that both CBC and Rrp6p are components of the same pathway and are in the same epistasis group.
NMD does not occur in the nucleus.
The extent of NMD of mRNA in the nucleus was assessed by determining the levels of CYH2 pre-mRNA in the nup116-Δ upf1-Δ strain at 25 and 37°C. As expected, the degradation of CYH2 pre-mRNA was clearly suppressed by upf1-Δ at the nonrestricted temperature of 25°C (Fig. 3; Table 2). In contrast, the CYH2 pre-mRNA levels at 37°C, a condition in which mRNA is retained in the nucleus, was approximately the same in both the nup116-Δ and nup116-Δ upf1-Δ strains. This finding clearly suggests that NMD does not act in the nucleus of S. cerevisiae. Consistent with this observation, Maderazo et al. (52) showed that NMD in S. cerevisiae most likely takes place in the cytoplasm.
Pathways for degrading abnormal mRNAs in the nucleus.
One major question is whether DRN, which acts on normal mRNAs and which is dependent on CBC, corresponds to any of the degradation pathways that act on abnormal and defective mRNAs. Recent studies have revealed nuclear surveillance systems that selectively degrade abnormal and defective RNA species, including the following: (i) pre-mRNAs that accumulate due to inefficient splicing, such as in splicing defective prp2-1 mutants (6); (ii) mRNAs produced after cessation of polyadenylation in a pap1-1 strain (8); (iii) aberrantly 3′-extended mRNAs due to lack of functional Rna14p and Rna15p, which are components of cleavage and polyadenylation factor CF1A (50, 72); (iv) mRNA that accumulates in the nucleus due to inefficient loading of essential mRNA export factors to the assembling mRNP (86); (v) hyperadenylated mRNAs, which appear as a consequence of nuclear retention, such as in export defective rat7-1 or rip1-Δ strains (37); and (vi) hypoadenylated mRNAs, which occurs in pap1-1 strains that lack functional poly(A) polymerase (28).
It should be emphasized that mRNAs in nup116-Δ strains are not hyperadenylated, as they are in other mutants defective in mRNA export, including rat7-1, rip1-Δ, gle1-4, mex67-5, rat8-2 (30, 37), and nab2-Δ (24). The marginal increases in poly(A) tail lengths of approximately 15 to 20 nt that is associated with the nup116-Δ mutation (Fig. 8) are similar to those seen in rna1-1 and prp20-1 mutants (18, 62), but contrast significantly with other nuclear export mutations, which resulted in poly(A) tail lengths up to 100 nt longer than normal (37). The reason for these differences remains unclear, but the results do indicate that retention of mRNAs in the nucleus does not necessarily lead to hyperadenylation. While the hyperadenylated mRNAs are clearly abnormal, the mRNAs in the nup116-Δ strains are considered to be normal.
Similar to the DRN pathway, Rrp6p is a component of the certain nuclear pathways acting on abnormal mRNAs. In the prp2-1 strain, Rrp6p acts as a 3′→5′ exonuclease degrading unspliced pre-mRNAs (6). In addition, Rrp6p appears to degrade unadenylated mRNAs in a pap1-1 strain (8). Furthermore, in the rna14-1 and rna15-2 strains, Rrp6p functions further to degrade the processively degraded pre-mRNA intermediates already acted on by the exosome and Dob1p (72). In this regard, pap1-1 is suppressed by rrp6-Δ (8). While DRN acts on CYH2 pre-RNA (Fig. 6), the results with nup116-Δ strains indicate that DRN action on pre-mRNA and mRNA may be similar.
However, the role of Rrp6p in degradation of unadenylated and hyperadenylated mRNAs in the intranuclear foci of the rat7-1, rip1-Δ, or pap1-1 strains is unclear (28). In fact, Hilleren et al. (28) demonstrated that PGK1 mRNA from pap1-1 strain becomes destabilized in absence of Rrp6p; they further demonstrated that unadenylated SSA4 mRNAs in pap1-1 strain accumulated at the specific intranuclear foci as a consequence of nuclear retention and that in pap1-1 rrp6-Δ strain they were released from these foci and exported. Thus, the degradation of unadenylated and hyperadenylated mRNAs at intranuclear foci differs significantly from the other nuclear pathways.
The relationship of DRN to the RNA surveillance pathways acting on defective forms of pre-mRNA as described by Bousquet-Antonelli et al. (6) and Torchet et al. (72) has yet to be defined. While all of these degradation pathways require Rrp6p and presumably the nuclear exosome, it is unknown whether the RNA surveillance pathways require CBC. Experiments with strains containing cbc1-Δ and other appropriate mutations should reveal the role of CBC in the RNA surveillance pathways.
DRN may play physiological roles in degrading aberrant mRNAs and in regulating the abundance of specific normal mRNAs.
Another major question is whether DRN is restricted to mRNAs that are retained in the nucleus by abnormal physiological conditions caused by the mutations affecting mRNA export. As stressed above, the degradation of certain cyc1-512 transcripts are suppressed by cbc1-Δ, cbc2-Δ and rrp6-Δ in strains having the normal mRNA export apparatus. In fact, the initial proposal of DRN was based on the on the assumption that long cyc1-512 transcripts are partially retained in the nucleus due to an intrinsic property of these mRNAs (15). We believe that DRN plays a positive role in identifying and eliminating defective mRNAs in the nucleus in wild type cells where export proceeds normally. Finding certain wild-type mRNAs that are particularly protected from degradation by cbc1-Δ and rrp6-Δ will provide evidence consistent with the view that DRN is a normal pathway acting at a high rate on a special class of normal mRNAs.
Acknowledgments
We thank Patricia Hinkle and John Puskas (Department of Pharmacology and Physiology, University of Rochester) for assistance in the use of the fluorescence microscope, Letian Kuai (Department of Biochemistry, University of Rochester) for assistance with processing of the fluorescence image, and Jay Greenberg (Department of Biochemistry, University of Rochester) for useful discussions. The Cy3-labeled oligo(dT) probe was kindly supplied by Pascal Chartrand (Department of Biochemistry, University of Montreal).
This work was supported by National Institutes of Health grants RO1 GM12702 (to F.S.) and RO1 GM59898 (to J.S.B.).
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 13:2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailer, S. M., C. Balduf, J. Katahira, A. Podtelejnikov, C. Rollenhagen, M. Mann, N. Pante, and E. Hurt. 2000. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275:23540-23548. [DOI] [PubMed] [Google Scholar]
- 4.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 5.Boeck, R., B. Lapeyre, C. E. Brown, and A. B. Sachs. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 7.Briggs, M. W., K. T. Burkard, and J. S. Butler. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273:13255-13263. [DOI] [PubMed] [Google Scholar]
- 8.Burkard, K. T., and J. S. Butler. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20:604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12:90-96. [DOI] [PubMed] [Google Scholar]
- 10.Butler, J. S., M. W. B. Briggs, and A. Proweller. 1997. Analysis of polyadenylation phenotypes in S. cerevisiae, p. 111-124. In J. Richter (ed.), mRNA formation and function. Academic Press, New York, N. Y.
- 11.Cabrera, C. V., J. J. Lee, J. W. Ellison, R. J. Britten, and E. H. Davidson. 1984. Regulation of cytoplasmic mRNA prevalence in sea urchin embryos. Rates of appearance and turnover for specific sequences. J. Mol. Biol. 174:85-111. [DOI] [PubMed] [Google Scholar]
- 12.Caponigro, G., and R. Parker. 1996. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60:233-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colot, H. V., F. Stutzand, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed] [Google Scholar]
- 14.Cui, Y., K. W. Hagan, S. Zhang, and S. W. Peltz. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9:423-436. [DOI] [PubMed] [Google Scholar]
- 15.Das, B., Z. Guo, P. Russo, P. Chartrand, and F. Sherman. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell Biol. 20:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daugeron, M. C., F. Mauxion, and B. Seraphin. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for requirement of deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 18.Forrester, W., F. Stutz, M. Rosbash, and M. Wickens. 1992. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 6:1914-1926. [DOI] [PubMed] [Google Scholar]
- 19.Fortes, P., J. Kufel, M. Fornerod, M. Polycarpou-Schwarz, D. Lafontaine, D. Tollervey, and I. W. Mattaj. 1999. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 19:6543-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frischmeyer, P. A., A. van Hoof, K. O'Donnell, A. L. Guerrerio, R. Parker, and H. C. Dietz. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258-2261. [DOI] [PubMed] [Google Scholar]
- 21.Gorsch, L. C., T. C. Dockerdorff, and C. N. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.He, F., S. W. Peltz, J. L. Donahue, M. Rosbash, and A. Jacobson. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc. Natl. Acad. Sci. USA 90:7034-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hector, R. E., K. R. Nykamp, S. Dheur, J. T. Anderson, P. J. Non, C. R. Urbinati, S. M. Wilson, L. Minvielle-Sebastia, and M. S. Swanson. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21:1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry, Y., H. Wood, J. P. Morrissey, E. Petfalski, S. Kearsey, and D. Tollervey. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentze, M. W., and A. E. Kulozik. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307-310. [DOI] [PubMed] [Google Scholar]
- 27.Herrick, D., R. Parker, and A. Jacobson. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 29.Hilleren, P., and R. Parker. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33:229-260. [DOI] [PubMed] [Google Scholar]
- 30.Hilleren, P., and R. Parker. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho, A. K., T. X. Shen, K. J. Ryan, E. Kiseleva, M. A. Levy, T. D. Allen, and S. R. Wente. 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20:5736-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izaurralde, E., J. Stepinski, E. Darzynkiewicz, and I. W. Mattaj. 1992. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 118:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657-668. [DOI] [PubMed] [Google Scholar]
- 36.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen, T. H., K. Patricio, T. McCarty, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 38.Jimeno, S., A. G. Rondon, R. Luna, and A. Aguilera. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21:3526-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, A. W. 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17:6122-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kataoka, N., M. Ohno, I. Moda, and Y. Shimura. 1995. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 23:3638-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kataoka, N., M. Ohno, K. Kangawa, Y. Tokoro, and Y. Shimura. 1994. Cloning of a complementary DNA encoding an 80 kilodalton nuclear cap binding protein. Nucleic Acids Res. 22:3861-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafontaine, D. L. J., and D. Tollervey. 1998. Birth of the snoRNPs: the evolution of the modification guide snoRNAs. Trends Biochem. Sci. 23:383-388. [DOI] [PubMed] [Google Scholar]
- 44.LaGrandeur, T. E., and R. Parker. 1998. Isolation and Characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange, T. S., A. V. Borovjagin, and S. A. Gerbi. 1998. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA 4:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larimer, F. W., and A. Stevens. 1990. Disruption of the gene XRN1, coding for a 5′→3′ exoribonuclease, restricts yeast cell growth. Gene 95:85-90. [DOI] [PubMed] [Google Scholar]
- 47.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 48.Lelivelt, M. J., and M. R. Culbertson. 1999. Yeast UPF proteins required for mRNA surveillance affect global gene expression of the yeast transcriptome. Mol. Cell. Biol. 19:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 50.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Losson, R., R. P. Fuchs, and F. Lacroute. 1983. In vivo transcription of a eukaryotic regulatory gene. EMBO J. 2:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madezaro, A., B., J. P. Belk, F. He, and A. Jacobson. 2003. Nonsense containing mRNAs that accumulate in the absence of a functional nonsense mediated decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 23:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell, P., E. Petfalski, and D. Tollervey. 1996. The 3′-end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 10:502-513. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonuclease activities. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10:193-198. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell, P., and D. Tollervey. 2001. mRNA turnover. Curr. Opin. Cell Biol. 13:320-325. [DOI] [PubMed] [Google Scholar]
- 57.Muhlrad, D., C. J. Decker, and R. Parker. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15:2145-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muhlrad, D., and R. Parker. 1994. Premature translation termination triggers mRNA decapping. Nature 340:578-581. [DOI] [PubMed] [Google Scholar]
- 59.Ohno, M., N. Kataoka, and Y. Shimura. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18:6989-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peltz, S. W., A. H. Brown, and A. Jacobson. 1993. mRNA destabilization triggered by premature translational termination depends on three mRNA sequence elements and at least one trans-acting factor. Genes Dev. 7:1737-1754. [DOI] [PubMed] [Google Scholar]
- 61.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piper, P. W., and J. L. Aamand. 1989. Yeast mutation thought to arrest mRNA transport markedly increases the length of the 3′ poly(A) on polyadenylated RNA. J. Mol. Biol. 208:697-700. [DOI] [PubMed] [Google Scholar]
- 63.Proweller, A., and S. Butler. 1996. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. J. Biol. Chem. 271:10859-10865. [DOI] [PubMed] [Google Scholar]
- 64.Ross, J. 1995. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 12:171-175. [DOI] [PubMed] [Google Scholar]
- 65.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothstein, R. J. 1983. One step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 67.Russo, P., W.-Z. Li, D. M. Hampsey, K. S. Zaret, and F. Sherman. 1991. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 10:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 69.Schneiter, R., C. E. Guerra, M. Lamp, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3-41. [DOI] [PubMed] [Google Scholar]
- 71.Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075-1083. [DOI] [PubMed] [Google Scholar]
- 72.Torchet, C., C. Bousquet-Antonelli, L. Milligan, E. Thompson, J. Kufel, and D. Tollervey. 2002. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9:1285-1296. [DOI] [PubMed] [Google Scholar]
- 73.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 74.Uemura, H., and Y. Jigmi. 1992. GCR3 encodes an acidic protein that is required for expression of glycolytic genes in Saccharomyces cerevisiae. J. Bacteriol. 174:5526-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uemura, H., S Pandit, Y. Jigmi, and R. Sternglanz. 1996. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces cerevisiae, suppress the temperature growth of hpr1 mutants. Genetics 142:1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Hoof, A., P. A. Frischmeyer, H. C. Dietz, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262-2264. [DOI] [PubMed] [Google Scholar]
- 77.Vilela, C., B. Linz, C. Rodrigues-Pousada, and J. E. McCarthy. 1998. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 26:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Visa, N., E. Izaurralde, J. Ferreira, B. Daneholt, and I. W. Mattaj. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 133:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Welch, E. M., and A. Jacobson. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18:6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wente, S. R., M. P. Rout, and G. Blobel. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119:705-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wente, S. R., and G. Blobel. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 84.Xue, Y, X. Bai, I. Lee, G. Kallstrom, J. Ho, J. Brown, A. Stevens, and A. W. Johnson. 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 20:4006-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaret, K. S., and F. Sherman. 1982. DNA sequence required for efficient transcription termination in yeast. Cell 28:563-573. [DOI] [PubMed] [Google Scholar]
- 86.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factorsYra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang, S., M. J. Ruiz-Echevarría, Y. Quan, and S. W. Peltz. 1995. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15:2231-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]