Abstract
Sphingosine and sphingosine-1-phosphate (SPP) are interconvertible sphingolipid metabolites with opposing effects on cell growth and apoptosis. Based on sequence homology with LBP1, a lipid phosphohydrolase that regulates the levels of phosphorylated sphingoid bases in yeast, we report here the cloning, identification, and characterization of a mammalian SPP phosphatase (mSPP1). This hydrophobic enzyme, which contains the type 2 lipid phosphohydrolase conserved sequence motif, shows substrate specificity for SPP. Partially purified Myc-tagged mSPP1 was also highly active at dephosphorylating SPP. When expressed in yeast, mSPP1 can partially substitute for the function of LBP1. Membrane fractions from human embryonic kidney HEK293 cells transfected with mSPP1 markedly degraded SPP but not lysophosphatidic acid, phosphatidic acid, or ceramide-1-phosphate. Enforced expression of mSPP1 in NIH 3T3 fibroblasts not only decreased SPP and enhanced ceramide levels, it also markedly diminished survival and induced the characteristic traits of apoptosis. Collectively, our results suggest that SPP phosphohydrolase may regulate the dynamic balance between sphingolipid metabolite levels in mammalian cells and consequently influence cell fate.
Sphingosine-1-phosphate (SPP) is a bioactive sphingolipid metabolite that regulates diverse biological processes (reviewed in refs. 1 and 2). Many of its pleiotropic actions appear to be mediated by a family of specific cell surface G protein-coupled receptors (GPCR), known as EDG (endothelial differentiation genes) receptors. Binding of SPP to EDG-1 expressed on endothelial cells enhances survival (3), chemotaxis, and in vitro angiogenesis (4), and adherens junction assembly leading to morphogenetic differentiation (5), whereas binding of SPP to EDG-5 and EDG-3 induces neurite retraction and soma rounding (6, 7). SPP induces activation of Gi-gated inward rectifying K+-channels in atrial myocytes (8) and inhibits motility of melanoma cells (9) through as yet uncharacterized GPCRs.
SPP also plays important roles inside cells. In response to diverse external stimuli, sphingosine kinase, the enzyme that catalyzes the phosphorylation of sphingosine to SPP, is activated (10–15). Intracellular SPP, in turn, mobilizes calcium from internal stores independently of inositol triphosphate (15, 16), as well as eliciting diverse signaling pathways leading to proliferation (17, 18) and suppression of apoptosis (19–22).
Because of its dual function as a ligand and second messenger and its pivotal role in cell growth and survival, the synthesis and degradation of SPP should be tightly regulated in a spatial-temporal manner. Until recently, however, little was known of the enzymes involved in SPP metabolism. We have purified sphingosine kinase to apparent homogeneity from rat kidney (23) and subsequently cloned and characterized a mammalian sphingosine kinase (24), which belongs to a highly conserved gene family (24, 25). Enforced expression of sphingosine kinase markedly enhanced proliferation and survival, substantiating the importance of intracellularly generated SPP in cell fate decisions (26).
SPP can be metabolized by two distinct pathways: catabolism by a microsomal pyridoxal phosphate-dependent lyase to palmitaldehyde and phosphoethanolamine, which then can be used for the biosynthesis of glycerolipids, or dephosphorylation by specific phosphatases to sphingosine (27). Recently, the genes encoding these enzymes were identified in Saccharomyces cerevisiae (28–30), and genetic manipulation has demonstrated an important role for long-chain phosphorylated sphingoid bases in growth and survival of yeast after nutrient deprivation and heat stress (29, 31–33) in a manner that is reminiscent of their effects on mammalian cells. Although the mammalian counterpart of the yeast SPP lyase recently has been identified (34), a specific mammalian SPP phosphatase has not been previously cloned.
The yeast SPP phosphatases encoded by LBP1 and LBP2 are members of type 2 lipid phosphate phosphohydrolases, a family of magnesium-independent, membrane-bound enzymes that share sequence conservation within three domains that are predicted to be involved in the coordination and hydrolysis of the phosphate moiety (35). A search of the yeast genome for enzymes containing the three conserved domains revealed the presence of four genes encoding putative type 2 lipid phosphatases. Two of these, DPP1 and LPP1, were shown to encode phosphatases with activity against phosphatidic acid (PA), lysophosphatidic acid (LPA), and diacylglycerol pyrophosphate (36, 37). In contrast, LBP1 (also known as YSR2 or LCB3) and LBP2 (YSR3) encode phosphatases with remarkable specificity for phosphorylated sphingoid bases and without activity toward glycerolipid substrates (29, 30, 33).
In crude rat liver and cerebellum extracts, high-affinity SPP phosphatase activity has been previously described (38), with enzymatic properties similar to yeast SPP phosphatases. Although three isoforms of type 2 lipid phosphate phosphohydrolases, known as LPP1/PAP2a, LPP3/PAP2b, and LPP2/PAP2c, have been cloned from mammalian cells (reviewed in ref. 39), these gene products appear to have broad substrate specificity with similar efficiencies against PA, LPA, SPP, ceramide-1-P, and diacylglycerol pyrophosphate, when assayed in vitro in lipid/detergent micelles. We now report the cloning and characterization of a mammalian homolog of yeast SPP phosphatase, murine SPP phosphatase-1 (mSPP1). The mammalian enzyme differs from lipid phosphatase phosphatases (LPP) in its sequence, properties, and in its high specificity for SPP. Our results suggest that mSPP1, which regulates the dynamic balance between ceramide and SPP levels in mammalian cells, may play an important role in regulating cell survival.
Materials and Methods
cDNA Cloning.
blast searches using the LBP1 sequence identified expressed sequence tag (EST) clones: 1247076 (GenBank accession no. AA574626) had homology through the phosphatase-conserved domains, and 1664615 (GenBank accession no. AI098466) contained 5′ sequences. PCR primers (5′-CGGAACTGGGCAACGAGCTCTTC and 5′-CTCGGAATACAGCATGCCCTCCACGC) were used in 3′ rapid amplification of cDNA ends (RACE) reactions using a Marathon cDNA Amplification kit and mouse brain cDNA (CLONTECH). A 2.6-kb PCR product was cloned into pCR2.1 (Invitrogen), and its sequence was found to match EST AA574626 with additional 3′ sequence including the putative stop codon. The full-length cDNA was constructed by ligating a 0.5-kb HindIII/BstYI fragment from AI098466 to a 1.0-kb BstYI/EcoRI fragment from the RACE PCR product into pcDNA3.1 (Invitrogen). NotI sites flanking the cDNA were used to subclone mSPP1 into pRS414-ADH for expression in yeast. To construct the N-terminal myc-tagged gene, PCR primers (5′-TCAGGATCCATGTCCCTGGGGCAGCGG, 5′-CTGCAGATATCCAGCACAGTGGCGGCC) were used to amplify mSPP1, which was digested with BamHI and EcoRI and ligated into myc-pcDNA3.1 (Invitrogen).
Northern Blot Analysis.
Poly(A)+ RNA blots containing 2 μg of RNA from mouse adult tissues (CLONTECH) were probed with a 590-bp EagI/EcoRV fragment from mSPP1 that was gel-purified and labeled with [32P]dCTP by random priming. Blots were hybridized in ExpressHyb Solution (CLONTECH) at 68°C for 1 h and washed following the manufacturer's protocol. Bands were quantified by using a Molecular Dynamics Storm 860.
Yeast Long-Chain Sphingoid Base Phosphate Phosphohydrolase Assay.
Yeast (lbp1Δsur2-2) were transformed with mSPP1pRS414-ADH or pRS414-ADH control DNA, and membranes were prepared as described (29). In brief, phosphatase activity was measured in 200 μl containing 50 mM KPO4, pH 7.2, 0.02% tergitol, 2 μM [3H]dihydrosphingosine-1-phosphate (40,000 cpm), 2 mM semicarbazide, and 0.3–5 μg of membrane protein. After a 30-min incubation at 37°C, the assay was terminated with 200 μl 7 M NH4OH. One milliliter of chloroform/methanol (3:2) was added, and 50 μl of the chloroform layer was counted by liquid scintillation. TLC was used to verify that the chloroform layer contained only the [3H]dihydrosphingosine product.
Yeast Growth Assay.
Growth inhibition was determined by using a microtiter broth dilution assay in synthetic complete-tryptophan yeast nitrogen base medium (Difco) containing 2% glucose and 0.078% tryptophan-free supplement mixture (Bio 101). Cells were inoculated at OD600 = 0.001 (≈1 × 104 yeast cells/ml), and serial 2-fold dilutions of australifungin were made from 5 μg/ml. Growth after 48 h at 30°C was measured by absorbance readings.
Mammalian Cell Culture.
NIH 3T3 fibroblasts (ATCC CRL-1658) and human embryonic kidney cells (HEK293, ATCC CRL-1573) were grown in high-glucose DMEM containing 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine supplemented with 10% calf serum or FBS, respectively (24).
Transfection.
pcDNA3.1 plasmids were transfected into cells using Lipofectamine Plus (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. In some experiments, cells were transfected with plasmids plus pCEFL GFP (5:1), which encodes green fluorescent protein. Transfection efficiencies were typically 30% and 40% for NIH 3T3 and HEK293 cells, respectively.
SPP Phosphatase Assay.
Forty-eight hours after transfection, cells were washed twice in PBS. Then, 200 μl of buffer A [100 mM Hepes (pH 7.5) containing 10 mM EDTA, 1 mM DTT, and 10 μg/ml each leupeptin, aprotinin, and soybean trypsin inhibitor] was added to each well, and cells were scraped on ice. Cells were subsequently freeze-thawed seven times, then centrifuged at 100,000 × g for 1 h. Supernatants were removed, and the membrane fractions were resuspended in 200 μl of buffer A and homogenized. Protein concentrations were determined with the Bradford assay. 32P-labeled SPP was prepared by phosphorylation of sphingosine with recombinant sphingosine kinase essentially as described (24). SPP phosphatase activity was measured by adding 32P-labeled SPP (2 nmol, 25,000 cpm, 0.3% BSA complex) to membrane or cytosol preparations (2 μg) in 200 μl of buffer A and incubated for 30 min at 37°C. Remaining 32P-labeled SPP was extracted and analyzed by TLC as described (29). To determine phosphatase activity of immunoprecipitated myc-mSPP1 fusion protein, HEK 293 cells were transfected with pcDNA3.1 containing c-myc-mSPP1 or empty vector, and membrane proteins prepared as described above were resuspended in buffer A containing 0.1% Tween-20 for 30 min at 4°C. Triton X-100 could not be used for this purpose because it strongly inhibited phosphatase activity. After centrifugation, proteins (500 μg) were immunoprecipitated for 2 h with 5 μg of mouse anti-c-myc (Zymed). Then, 30 μl of protein A/G agarose (Santa Cruz Biotechnology) were added for 1 h. The beads were washed five times with buffer A, resuspended in a total volume of 60 μl, and assayed for SPP phosphatase activity. In some experiments, dephosphorylation of 32P-labeled lysophosphatidic acid, phosphatidic acid, and ceramide-1-phosphate, prepared by phosphorylation using Escherichia coli diacylglycerol kinase and [γ-32P]ATP, was determined as described (40).
Measurement of Mass Levels of SPP, Sphingosine, and Ceramide.
Mass levels of SPP, sphingosine, and ceramide were measured essentially as described in refs. 41, 42, and 43, respectively.
Determination of Apoptosis.
Apoptosis was assessed by staining cells with Hoechst in 30% glycerol/PBS for 10 min at room temperature as described (22). Forty-eight hours after transfection of NIH 3T3 fibroblasts, cells expressing GFP were examined with an inverted fluorescence microscope. Apoptotic cells were distinguished by condensed, fragmented nuclear regions. The percentage of intact and apoptotic nuclei in cells expressing GFP fluorescence was determined (7). A minimum of 500 cells were scored in a double-blinded manner to minimize subjective interpretations. In some experiments, apoptotic cells were determined from their rounded cell morphology.
Results
Cloning of mSPP1.
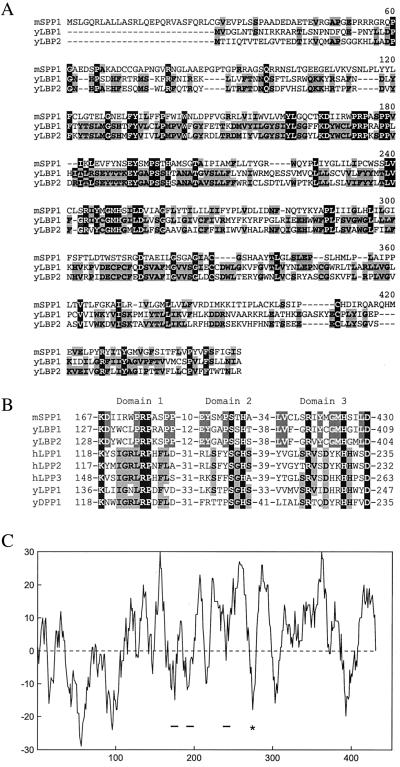
To identify a mammalian homolog of the yeast sphingoid base phosphate phosphatase, we searched the EST database. EST AA574626 contained all three domains predicted for a type 2 lipid phosphatase and had higher homology to the yeast LBP1 and LBP2 genes (29) than to the three isoforms of mammalian type 2 LPPs (39). The EST clone was partial, apparently truncated at the 5′ and 3′ ends of the putative SPP phosphatase. Therefore, RACE reactions on a mouse brain library were performed to retrieve the 3′ end, and ligation to the 5′ end obtained from the EST clone AI098466 provided the full-length cDNA, named murine SPP phosphohydrolase1 (mSPP1). The complete translational ORF encodes a 430-aa protein with 17% identity and 34% similarity at the amino acid level to yeast LBP1 and LBP2 (Fig. 1A), but with little overall homology to the LPP phosphatases, except at the conserved amino acids predicted to be in the active site (Fig. 1B). A hydropathy plot using the Kyte–Doolittle algorithm indicates 8–10 hydrophobic regions that are potential membrane-spanning helixes (Fig. 1C).
Figure 1.
(A) Alignment of amino acid sequences of mSPP1 with yeast Lbp1p and Lbp2p. Residues that are identical in two or three of the gene sequences are shaded in gray or black, respectively. (B) Alignments of the three highly conserved type 2 phosphohydrolase domains of LPPs and SPP phosphatases. (C) Hydropathy plot of mSPP1 using the Kyte–Doolittle algorithm and a window size of nine residues. The location of the three conserved domains are underlined, and the potential glycosylation site is marked with an *.
mSPP1 Is a Functional Homolog of LBP1.
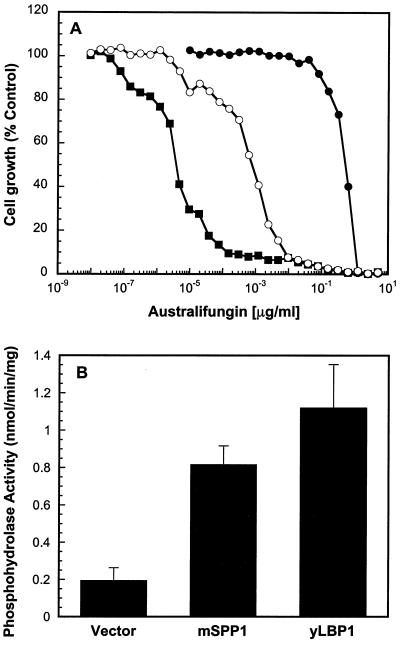
Yeast cells defective in LBP1 shunt sphingolipid metabolites into phosphatidylethanolamine and phosphatidylcholine synthesis and become extremely sensitive to ceramide synthase inhibition by the antifungal agent australifungin (29). If mSPP1 is a functional homolog of LBP1 and is appropriately expressed in yeast, it should convey the phenotypes associated with LBP1: reverse sensitivity to australifungin, and have in vitro long chain base phosphate phosphatase activity. Indeed, expression of mSPP1 under the control of a strong constitutive yeast promoter in lbp1Δ mutants partially attenuated the hypersensitivity to the ceramide synthase inhibitor (Fig. 2A), providing in vivo evidence for complementation of intracellular sphingoid base phosphatase activity. Moreover, membranes prepared from the lbp1Δ mutant transformed with mSPP1 had a 4-fold increase in specific activity of dihydrosphingosine-1-phosphate phosphatase, which brought the level of phosphohydrolase activity to 75% of cells expressing the yeast SPP phosphatase (Fig. 2B).
Figure 2.
(A) mSPP1 complements the hypersensitivity of Δlbp1 to australifungin. Δlbp1 mutant transformed with control vector (■), or mSPP1 in pRS414-ADH (○), or wild-type strain W3031a (●) were tested for their sensitivity to australifungin. Data are expressed as percent of cell growth in the absence of inhibitor. (B) Transformation of yeast with mSPP1 enhances long-chain base phosphate phosphohydrolase activity. Phosphohydrolase activity was measured in Δlbp1 mutants carrying vector, or mSPP1 on pRS414-ADH, or in wild-type cells as described in Materials and Methods. Results are means ± SD of quadruplicate determinations.
Tissue Distribution of mSPP1 mRNA Expression.
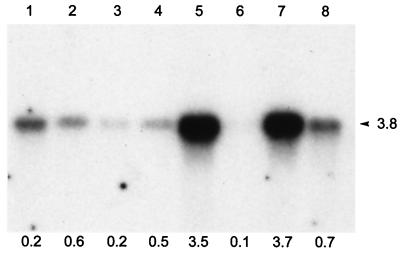
We next examined expression of mSPP1 mRNA in adult mouse tissues by Northern blotting. A single 3.8-kb transcript was detected in all tissues (Fig. 3). However, the level of expression was markedly variable among the different tissues. The mRNA levels were highest in mouse liver and kidney, and there were barely detectable levels in skeletal muscle. Interestingly, expression of mSPP1 in various mouse tissues does not closely correlate with the relative SPP phosphohydrolase activities measured in rat tissue extracts, as it was previously found that brain had about 3.5 times greater specificity activity than liver or kidney (38). A similar 3.8-kb transcript was detected in over 30 human tissues that were surveyed (data not shown), indicating that this gene is also ubiquitously expressed in humans.
Figure 3.
Tissue-specific expression of mSPP1. A 32P-labeled mSPP1 probe was hybridized to poly(A)+ RNA from the indicated mouse tissues (2 μg/lane). Lanes: 1, heart; 2, brain; 3, spleen; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, testis. The numbers underneath indicate relative levels of expression compared with actin as quantified with the Molecular Dynamics Storm 860.
mSPP1 Is a Highly Specific Phosphohydrolase.
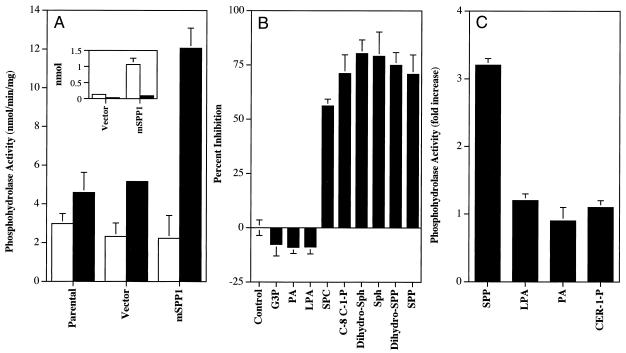
To investigate whether mSPP1 encodes a bona fide SPP-specific phosphohydrolase, HEK293 cells were transiently transfected with a pcDNA3.1 expression vector containing mSPP1 cDNA, and phosphohydrolase activity was measured. Modest levels of endogenous phosphatase activity were present in cytosolic and membrane fractions from control cells (either untransfected or transfected with an empty vector) (Fig. 4A). Cells transfected with mSPP1 exhibited 4-fold increased membrane SPP phosphatase activity 24 h after transfection that remained at this level for 2–3 days. There were no detectable increases in cytosolic SPP phosphatase activity, consistent with the hydropathy plot of mSPP1 predicting 8–10 potential membrane-spanning domains. Similar results also were found in NIH 3T3 fibroblasts (data not shown). It should be noted that transfection efficiency was quite high and similar in both cell lines.
Figure 4.
Characterization of mSPP1 activity in transfected cells. (A) Phosphohydrolase activity in cytosolic and membrane fractions. HEK293 cells were transfected with pcDNA3.1 or pcDNA3.1-mSPP1. Cytosolic (open bars) and membrane fractions (filled bars) were isolated, and SPP phosphohydrolase activity was determined by measuring hydrolysis of [32P]SPP as described in Materials and Methods. Results are means ± SD of triplicate determinations. Similar results were obtained in three additional experiments. (Inset) Phosphohydrolase activity of mSPP1 immunoprecipitated with anti-c-myc. Myc-tagged mSPP1 was partially purified from HEK293 cells transfected with empty vector or myc-tagged mSPP1 as described in Materials and Methods, and phosphohydrolase activity was measured with 32P-SPP (open bars) or 32P-LPA (filled bars). Data are expressed as nmol hydrolyzed per 30 min. (B) Inhibition of SPP phosphohydrolase activity by various lipids. Membrane fractions from HEK 293 fibroblasts transfected with pcDNA3.1-mSPP1 were incubated with [32P]SPP (10 μM) for 30 min at 37°C in the absence or presence of the indicated lipids (100 μM), and phosphohydrolase activity was measured. Data are expressed as percent inhibition and are means ± SD. Dihydro-Sph, sphinganine; Sph, sphingosine; Dihydro-SPP, sphinganine-1-phosphate; C-8 C-1-P, C8-ceramide-1-phosphate; SPC, sphingosylphosphocholine; LPA, lysophosphatidic acid; PA, phosphatidic acid; G3P, glycerol-3-phosphate. (C) mSPP1 specifically dephosphorylates SPP. Phosphohydrolase activity was measured with 32P-labeled lipids: SPP, LPA, PA, and ceramide-1-phosphate (CER-1-P). Results are expressed as fold stimulation of phosphohydrolase activity compared with vector-transfected HEK293 cells.
It was previously demonstrated that LPP1, LPP2, and LPP3 have activity toward a broad range of phospholipid substrates, including both glycerophospholipids and sphingolipids (reviewed in ref. 39). In contrast, yeast LBP1 and LBP2 are highly specific long-chain sphingoid base phosphate phosphohydrolases (29, 30). Thus, it was of interest to determine the substrate specificity of recombinant mSPP1. In competition studies using 32P-SPP as substrate for the phosphatase activity present in membrane preparations from cells overexpressing mSPP1, it appears to be highly specific for long-chain sphingoid bases because only lipids that contained sphingosine or dihydrosphingosine were able to inhibit mSPP1 activity (Fig. 4B). Other phospholipids, including phosphatidic acid, lysophosphatidic acid, and glycerol-3-phosphate, were not competitive substrates. Moreover, in agreement, there were no detectable increases in dephosphorylation of 32P-labeled phosphatidic acid, lysophosphatidic acid, or ceramide-1-phosphate in membranes from mSPP1-transfected cells (Fig. 4C). This phosphatase seems to be unique because, unlike the previously described LPP phosphatases, mSPP1 was inhibited by a low concentration of Triton X-100 (0.05%). To exclude the possibility that the increase in SPP phosphatase activity was caused by activation of a regulatory protein that indirectly affected SPP metabolism, we tagged mSPP1 with a c-myc epitope and partially purified this fusion protein by immunoprecipitation with a c-myc antibody (Fig. 4A, Inset). It should be noted that the myc-tagged mSPP1 did not compromise mSPP1 activity. After transfection with either mSPP1 or myc-tagged mSPP1, the phosphatase-specific activity was increased 4-fold to 12 nmol/min/mg in crude membrane fractions of HEK 293 cells. The specific activity of the fusion protein purified by immunoprecipitation was greater than 3,100 nmol/min/mg. Moreover, this partially purified fusion protein did not have significant activity toward lysophosphatidic acid (Fig. 4A, Inset). Collectively, these data demonstrate that the mSPP1 clone indeed encodes a specific SPP phosphatase.
mSPP1 Overexpression Enhances Programmed Cell Death.
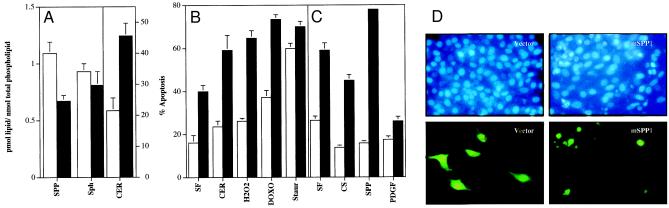
Next, we examined whether transfection of NIH 3T3 cells with mSPP1 resulted in changes in mass levels of sphingolipid metabolites. As expected, SPP levels were reduced. Although mass levels of sphingosine were not significantly altered, there was a marked increase in ceramide levels compared with cells transfected with vector alone (Fig. 5A). Thus, mSPP1 is active in intact cells, and overexpression alters the balance of sphingolipid metabolites within cells. If the dynamic balance of the sphingolipid metabolites, ceramide, and SPP is a critical factor that determines cell fate, then it is expected that overexpression of mSPP1 should enhance apoptosis. In agreement with previous studies (44), serum deprivation induced apoptosis in NIH 3T3 fibroblasts in a time-dependent manner (Fig. 5 B and C), where shrinkage and condensation of nuclei were clearly evident after 48 h (Fig. 5D). Transient expression of mSPP1 in NIH 3T3 fibroblasts enhanced the appearance of apoptotic nuclei induced by serum starvation (Fig. 5 B–D). Apoptosis induced by doxorubicin and oxidative stress, which are known to act at least in part through elevation of intracellular ceramide levels, or induced by cell permeable C2-ceramide, also was enhanced by expression of mSPP1 (Fig. 5B). In contrast, mSPP1 expression had almost no effect on apoptosis resulting from treatment with staurosporine (Fig. 5B), a broad-spectrum protein kinase inhibitor that induces apoptosis in many cell types (45). It should be pointed out that serum withdrawal markedly increases ceramide levels in many cell types, and it has been proposed that ceramide mediates, at least in part, serum deprivation-induced cell death (46). Even in the presence of serum or platelet-derived growth factor, where the degree of apoptosis was markedly reduced, mSPP1 overexpression still significantly enhanced apoptosis (Fig. 5C). In agreement with our previous results (19, 26), addition of 5 μM SPP to control (data not shown) or vector-transfected NIH 3T3 fibroblasts suppressed apoptosis to a similar extent as serum (Fig. 5C). However, the protection by SPP was completely blocked by expression of mSPP1 and even greater extent of apoptosis was detected in the presence of SPP (Fig. 5C), because of increased generation of ceramide.
Figure 5.
Expression of mSPP1 increases ceramide levels and enhances apoptosis of NIH 3T3 fibroblasts. (A) NIH 3T3 fibroblasts were transfected with either pcDNA3.1 (open bars) or pcDNA3.1-mSPP1 (filled bars). Twenty-four hours after transfection, the medium was replaced with serum-free DMEM and sphingolipid metabolite levels were determined as described in Materials and Methods. (B and C) NIH 3T3 fibroblasts were transfected with pCEFL-GFP and either pcDNA3.1 (open bars) or pcDNA3.1-mSPP1 (filled bars). Twenty-four hours after transfection, the medium was replaced with serum-free DMEM (SF) in the absence or presence of C2-ceramide (CER, 7.5 μM), H2O2 (100 μM), doxorubicin (DOXO, 0.2 μg/ml), staurosporine (Staur, 25 nM), 10% calf serum (CS), SPP (5 μM), or platelet-derived growth factor (10 ng/ml). After an additional 24 h (B) or 48 h (C), total GFP-expressing cells and GFP-expressing cells displaying rounded morphology were counted and percent apoptotic cells expressing GFP was determined as described in Materials and Methods. A minimum of 500 cells in each field were scored. Data are mean ± SE of three independent experiments, each one done in triplicate. (D) Each set of panels shows the same field from vector-transfected and mSPP1-expressing cells cultured in serum-free medium examined by fluorescence microscopy to visualize GFP (green, Lower) and also stained with Hoechst dye (blue, Upper) to visualize fragmented nuclei indicative of apoptosis. Note the typical condensed fragmented nuclei of apoptotic cells in mSPP1-transfected but not in vector-transfected cells after serum deprivation for 48 h, which correlates with the rounded cell morphology revealed by green fluorescence.
Discussion
The type 2 LPPs are magnesium-independent, membrane-bound enzymes that are thought to regulate signaling pools of bioactive lipid mediators (35, 39, 40, 47, 48). Three mammalian isoforms, designated LPP1/PAP2a, LPP3/PAP2b, and LPP2/PAP2c, are the best-characterized examples; their sequences are approximately 50% identical and their hydrophobicity plots are almost superimposable with six well-defined transmembrane helices. Substrate specificity of the LPP enzymes as measured in vitro is broad, with phosphatase activity against glycerolipids and sphingolipids, but not phospholipase C-like activity. Characterization of type 2 phosphohydrolases in yeast revealed two isoforms of LPP-like enzymes encoded by LPP1 and DPP1 (36, 37, 47), and two other phosphatases encoded by LBP1 and LBP2 that specifically dephosphorylate sphingolipid substrates (29, 30, 33). The yeast SPP phosphatases diverge significantly in sequence from LPPs, except for the conserved residues predicted to be at the active site (35).
Here, we describe the cloning and characterization of a mammalian SPP phosphohydrolase, mSPP1, which appears similar to the previously described crude rat liver activity (38) and yeast SPP phosphatases (29) in its specificity for sphingolipid substrates and inhibition by Triton X-100. In addition to its substrate specificity, which distinguishes mSPP1 from the previously cloned type 2 phosphohydrolases, the sequence of mSPP1 is more similar to the yeast SPP phosphatases than to mammalian LPPs: SPP phosphatases are larger, consisting of 404–430 aa as compared with 281–312 aa; hydrophobicity plots predict 8–10 potential membrane-spanning domains in SPP phosphatases, in contrast to six well-defined transmembrane domains found in LPPs; and mSPP1 has one potential glycosylation site that is not conserved in the yeast orthologs and is predicted to be located on the opposite side of the membrane from the catalytic domains, unlike the LPPs, where the catalytic domains and glycosylation site are predicted to be in the first and second extracellular loops. Several experimental approaches have supported a topological model in which the N and C termini of LPPs are present at the cytosolic surface, and the active site and glycosylation site are exposed at the extracellular membrane surface (49). Consistent with a model of ecto-phosphatase activity is the finding that cells expressing LPP1 showed enhanced degradation of exogenous LPA and attenuated LPA-mediated activation of both the extracellular signal-regulated kinase pathway and DNA synthesis (48). Moreover, expression of PAP2a in insect cells resulted in cell surface phosphohydrolase activity (40). In contrast to a plasma membrane ectophosphatase activity ascribed to LPP1, the yeast SPP phosphatases have been localized to the endoplasmic reticulum, where they alter the balance of intracellular phosphorylated and nonphosphorylated sphingoid bases and ceramide, and consequently influence yeast cell growth and stress responses (29, 32, 33). Whereas lbp1 null mutants accumulated phosphorylated sphingoid bases, shunted sphingolipid metabolites into phospholipid synthesis, and became hypersensitive to ceramide synthase inhibition by australifungin (29, 30), overexpression of LBP1 increased ceramide levels and reduced the rate of cell growth (32). We observed that expression of mSPP1 in yeast resulted in functional dihydrosphingosine-1-phosphate phosphatase activity that was able to partially complement the defects associated with the lbp1Δ mutant. mSPP1 therefore can be regarded as the mammalian counterpart to LBP1, with a role in regulating intracellular sphingolipid metabolites. Future studies directed at the localization and topology of mSPP1 will be important to determine whether this enzyme also participates in dephosphorylation of extracellular SPP.
Analogous to its role in yeast (29, 32), mSPP1 expression also altered the balance of sphingolipid metabolites in mammalian cells; levels of ceramide were increased, whereas SPP decreased in NIH 3T3 cells transfected with mSPP1. Moreover, apoptosis of mSPP1-transfected cells induced by diverse stimuli was dramatically enhanced. Of interest, overexpression of sphingosine kinase not only increased levels of SPP and decreased ceramide levels, but also suppressed apoptosis because of ceramide elevation (26). Hence, the dynamic balance between the sphingolipid metabolites SPP and ceramide is an important factor in cell survival. Collectively, the data in this report suggest that mSPP1 and its yeast orthologs belong to an evolutionarily conserved family of enzymes that regulate the levels of bioactive sphingolipid intermediates and thereby influence cell fate.
Acknowledgments
This study was supported in part by National Institutes of Health Grant GM43880 (to S.S.) and a postdoctoral fellowship from the Spanish Ministry of Education and Science (to I.G.R.).
Abbreviations
- SPP
sphingosine-1-phosphate
- PA
phosphatidic acid
- LPA
lysophosphatidic acid
- LPP
lipid phosphate phosphohydrolase
- mSPP1
murine SPP phosphatase-1
- RACE
rapid amplification of cDNA ends
- GFP
green fluorescent protein
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF247177).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120146897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120146897
References
- 1.Goetzl E J, An S. FASEB J. 1998;12:1589–1598. [PubMed] [Google Scholar]
- 2.Spiegel S. J Leukocyte Biol. 1999;65:341–344. doi: 10.1002/jlb.65.3.341. [DOI] [PubMed] [Google Scholar]
- 3.Hisano N, Yatomi Y, Satoh K, Akimoto S, Mitsumata M, Fujino M A, Ozaki Y. Blood. 1999;93:4293–4299. [PubMed] [Google Scholar]
- 4.Wang, F., Van Brocklyn, J. R., Movafagh, S., Zukowska-Grojec, Z., Milstien, S. & Spiegel, S. (1999) J. Biol. Chem., in press. [DOI] [PubMed]
- 5.Lee M J, Thangada S, Claffey K P, Ancellin N, Liu C H, Kluk M, Volpi M, Sha'afi R I, Hla T. Cell. 1999;99:301–312. doi: 10.1016/s0092-8674(00)81661-x. [DOI] [PubMed] [Google Scholar]
- 6.Postma F R, Jalink K, Hengeveld T, Moolenaar W H. EMBO J. 1996;15:2388–2392. [PMC free article] [PubMed] [Google Scholar]
- 7.Van Brocklyn J R, Tu Z, Edsall L, Schmidt R R, Spiegel S. J Biol Chem. 1999;274:4626–4632. doi: 10.1074/jbc.274.8.4626. [DOI] [PubMed] [Google Scholar]
- 8.van Koppen C J, Meyer zu Heringdorf D, Laser K T, Zhang C, Jakobs K H, Bünnemann M, Pott L. J Biol Chem. 1996;271:2082–2087. doi: 10.1074/jbc.271.4.2082. [DOI] [PubMed] [Google Scholar]
- 9.Yamamura S, Yatomi Y, Ruan F, Sweeney E A, Hakomori S, Igarashi Y. Biochemistry. 1997;36:10751–10759. doi: 10.1021/bi970926s. [DOI] [PubMed] [Google Scholar]
- 10.Olivera A, Spiegel S. Nature (London) 1993;365:557–560. doi: 10.1038/365557a0. [DOI] [PubMed] [Google Scholar]
- 11.Choi O H, Kim J-H, Kinet J-P. Nature (London) 1996;380:634–636. doi: 10.1038/380634a0. [DOI] [PubMed] [Google Scholar]
- 12.Melendez A, Floto R A, Gillooly D J, Harnett M M, Allen J M. J Biol Chem. 1998;273:9393–9402. doi: 10.1074/jbc.273.16.9393. [DOI] [PubMed] [Google Scholar]
- 13.Xia P, Gamble J R, Rye K A, Wang L, Hii C S T, Cockerill P, Khew-Goodall Y, Bert A G, Barter P J, Vadas M A. Proc Natl Acad Sci USA. 1998;95:14196–14201. doi: 10.1073/pnas.95.24.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleuser B, Cuvillier O, Spiegel S. Cancer Res. 1998;58:1817–1824. [PubMed] [Google Scholar]
- 15.Meyer zu Heringdorf D, Lass H, Alemany R, Laser K T, Neumann E, Zhang C, Schmidt M, Rauen U, Jakobs K H, van Koppen C J. EMBO J. 1998;17:2830–2837. doi: 10.1093/emboj/17.10.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattie M, Brooker G, Spiegel S. J Biol Chem. 1994;269:3181–3188. [PubMed] [Google Scholar]
- 17.Rani C S, Berger A, Wu J, Sturgill T W, Beitner-Johnson D, LeRoith D, Varticovski L, Spiegel S. J Biol Chem. 1997;272:10777–10783. doi: 10.1074/jbc.272.16.10777. [DOI] [PubMed] [Google Scholar]
- 18.Van Brocklyn J R, Lee M J, Menzeleev R, Olivera A, Edsall L, Cuvillier O, Thomas D M, Coopman P J P, Thangada S, Hla T, Spiegel S. J Cell Biol. 1998;142:229–240. doi: 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuvillier O, Pirianov G, Kleuser B, Vanek P G, Coso O A, Gutkind S, Spiegel S. Nature (London) 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 20.Perez G I, Knudson C M, Leykin L, Korsmeyer S J, Tilly J L. Nat Med. 1997;3:1228–1232. doi: 10.1038/nm1197-1228. [DOI] [PubMed] [Google Scholar]
- 21.Edsall L C, Pirianov G G, Spiegel S. J Neurosci. 1997;17:6952–6960. doi: 10.1523/JNEUROSCI.17-18-06952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuvillier O, Rosenthal D S, Smulson M E, Spiegel S. J Biol Chem. 1998;273:2910–2916. doi: 10.1074/jbc.273.5.2910. [DOI] [PubMed] [Google Scholar]
- 23.Olivera A, Kohama T, Tu Z, Milstien S, Spiegel S. J Biol Chem. 1998;273:12576–12583. doi: 10.1074/jbc.273.20.12576. [DOI] [PubMed] [Google Scholar]
- 24.Kohama T, Olivera A, Edsall L, Nagiec M M, Dickson R, Spiegel S. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 25.Nagiec M M, Skrzypek M, Nagiec E E, Lester R L, Dickson R C. J Biol Chem. 1998;273:19437–19442. doi: 10.1074/jbc.273.31.19437. [DOI] [PubMed] [Google Scholar]
- 26.Olivera A, Kohama T, Edsall L C, Nava V, Cuvillier O, Poulton S, Spiegel S. J Cell Biol. 1999;147:545–548. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiegel S, Merrill A H., Jr FASEB J. 1996;10:1388–1397. doi: 10.1096/fasebj.10.12.8903509. [DOI] [PubMed] [Google Scholar]
- 28.Saba J D, Nara F, Bielawska A, Garrett S, Hanun Y A. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 29.Mandala S, Thornton R, Tu Z, Kurtz M, Nickels J, Broach J, Menzeleev R, Spiegel S. Proc Natl Acad Sci USA. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao C, Wadleigh M, Jenkins G M, Hannun Y A, Obeid L M. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 31.Gottlieb D, Heideman W, Saba J D. Mol Cell Biol Res Commun. 1999;1:66–71. doi: 10.1006/mcbr.1999.0109. [DOI] [PubMed] [Google Scholar]
- 32.Mao C, Saba J D, Obeid L M. Biochem J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- 33.Skrzypek M S, Nagiec M M, Lester R L, Dickson R C. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Saba J D. Biochem Biophys Res Commun. 1998;242:502–507. doi: 10.1006/bbrc.1997.7993. [DOI] [PubMed] [Google Scholar]
- 35.Stukey J, Carman G M. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toke D A, Bennett W L, Oshiro J, Wu W I, Voelker D R, Carman G M. J Biol Chem. 1998;273:14331–14338. doi: 10.1074/jbc.273.23.14331. [DOI] [PubMed] [Google Scholar]
- 37.Toke D A, Bennett W L, Dillon D A, Wu W I, Chen X, Ostrander D B, Oshiro J, Cremesti A, Voelker D R, Fischl A S, Carman G M. J Biol Chem. 1998;273:3278–3284. doi: 10.1074/jbc.273.6.3278. [DOI] [PubMed] [Google Scholar]
- 38.De Ceuster P, Mannaerts G P, Van Veldhoven P P. Biochem J. 1995;311:139–146. doi: 10.1042/bj3110139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brindley D N, Waggoner D W. J Biol Chem. 1998;273:24281–24284. doi: 10.1074/jbc.273.38.24281. [DOI] [PubMed] [Google Scholar]
- 40.Roberts R, Sciorra V A, Morris A J. J Biol Chem. 1998;273:22059–22067. doi: 10.1074/jbc.273.34.22059. [DOI] [PubMed] [Google Scholar]
- 41.Edsall L C, Spiegel S. Anal Biochem. 1999;272:80–86. doi: 10.1006/abio.1999.4157. [DOI] [PubMed] [Google Scholar]
- 42.Olivera A, Rosenthal J, Spiegel S. Anal Biochem. 1994;223:306–312. doi: 10.1006/abio.1994.1589. [DOI] [PubMed] [Google Scholar]
- 43.Olivera A, Buckley N E, Spiegel S. J Biol Chem. 1992;267:26121–26127. [PubMed] [Google Scholar]
- 44.Brancolini C, Lazarevic D, Rodriguez J, Schneider C. J Cell Biol. 1997;139:759–771. doi: 10.1083/jcb.139.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobsen M D, Weil M, Raff M C. J Cell Biol. 1996;133:1041–1051. doi: 10.1083/jcb.133.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannun Y. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 47.Dillon D A, Chen X, Zeimetz G M, Wu W I, Waggoner D W, Dewald J, Brindley D N, Carman G M. J Biol Chem. 1997;272:10361–10366. doi: 10.1074/jbc.272.16.10361. [DOI] [PubMed] [Google Scholar]
- 48.Jasinska R, Zhang Q X, Pilquil C, Singh I, Xu J, Dewald J, Dillon D A, Berthiaume L G, Carman G M, Waggoner D W, Brindley D N. Biochem J. 1999;340:677–686. [PMC free article] [PubMed] [Google Scholar]
- 49.Waggoner D W, Xu J, Singh I, Jasinska R, Zhang Q X, Brindley D N. Biochim Biophys Acta. 1999;1439:299–316. doi: 10.1016/s1388-1981(99)00102-x. [DOI] [PubMed] [Google Scholar]