Abstract
Recent studies indicated that the leucine zipper domain protein Par-4 induces apoptosis in certain cancer cells by activation of the Fas prodeath pathway and coparallel inhibition of NF-κB transcriptional activity. However, the intracellular localization or functional domains of Par-4 involved in apoptosis remained unknown. In the present study, structure-function analysis indicated that inhibition of NF-κB activity and apoptosis is dependent on Par-4 translocation to the nucleus via a bipartite nuclear localization sequence, NLS2. Cancer cells that were resistant to Par-4-induced apoptosis retained Par-4 in the cytoplasm. Interestingly, a 59-amino-acid core that included NLS2 but not the C-terminal leucine zipper domain was necessary and sufficient to induce Fas pathway activation, inhibition of NF-κB activity, and apoptosis. Most important, this core domain had an expanded target range for induction of apoptosis, extending to previously resistant cancer cells but not to normal cells. These findings have identified a unique death-inducing domain selective for apoptosis induction in cancer cells (SAC domain) which holds promise for identifying key differences between cancer and normal cells and for molecular therapy of cancer.
Development of cancer is a multistep process involving accumulation of multiple genetic aberrations. Most notable among such alterations is the loss of apoptotic responses that normally serve as built-in checkpoints against the emergence of cell populations with dysfunctional traits or the acquisition of prosurvival mechanisms that override the apoptotic signals (18). Loss of apoptotic mechanisms often results in abridged response to cancer therapy, and therefore, alternative or combinatorial approaches to kill the cancer cells and induce tumor regression are actively pursued (20). An essential feature of anticancer strategies is selective action against cancer cells, with little or no damage inflicted on normal cells. Most chemotherapeutic agents, ionizing radiation-based approaches, and combination therapies act within a confined dosage range to produce differential action in the cancer cell compartment with relatively fewer, albeit not completely absent, untoward effects in the normal tissue. Identification of molecules that can specifically target tumor cells therefore constitutes a significant area of cancer research. Molecules with selective action against tumor cells are valuable not only for their therapeutic potential but also for their potential applications as tools for dissection of the fundamental differences between normal and cancer cells.
The par-4 gene, first identified in prostate cancer cells undergoing apoptosis (32), encodes a proapoptotic protein that is remarkably effective in inducing cancer cell apoptosis and tumor regression in animal models. Our recent studies indicated that Par-4 induces apoptosis in androgen-independent prostate cancer cells, such as PC-3 and DU145, and in Ras-transformed mouse fibroblasts but not in androgen-dependent LNCaP prostate cancer cells, immortalized cells, or primary cultures of normal prostate epithelial and stromal cells (5, 26). The ability of Par-4 to directly induce apoptosis is distinct from the previously reported apoptosis-sensitization function of Par-4 (2, 31). Analysis of the latter function was always performed by overexpression of Par-4 in the presence of an additional apoptotic insult such as chemotherapeutic agents, tumor necrosis factor, serum deprivation, or ionizing radiation. Under these conditions, cells that are resistant to direct apoptosis by Par-4 become hypersensitive to apoptotic insults (16, 31, 32).
The par-4 gene maps to human chromosome 12q21 (22), which is often deleted in pancreatic cancer (23). This is also one of the regions that undergoes reorganization in Wilms' tumors (22). Par-4 expression is downregulated in renal cell carcinoma relative to the normal proximal tubular cell compartment (8). Moreover, oncogenes such as ras, raf, and src downregulate Par-4 expression, and restoration of the Par-4 level results in inhibition of oncogene-induced transformation of cells (1, 26). Par-4 expression is elevated in various neurodegenerative diseases such as ALS, Alzheimer's, Parkinson's, and Huntington's diseases and stroke, and inhibition of Par-4 with a dominant-negative mutant or an antisense oligodeoxynucleotide protects neuronal cells from apoptosis in cell culture and animal models of neurodegenerative diseases and stroke (9, 13, 14, 17, 28).
The transcriptional activity of NF-κB is an essential antiapoptotic target of Par-4 (26). NF-κB plays an essential role in oncogenesis and in the resistance of tumor cells to ionizing radiation and chemotherapy (30). The transcription potential of NF-κB, a heterodimer between subunits p65 (RelA) and p50, is fully activated by translocation to the nucleus and phosphorylation of RelA. Interestingly, Par-4 inhibits Ras- and Raf-induced NF-κB transcriptional activity (5, 26), and tumor necrosis factor alpha-induced NF-κB activation (11). Inhibition of NF-κB activity by superrepressor IκBα is sufficient to induce apoptosis in NIH 3T3 cells expressing oncogenic ras (25). As expected, overexpression of Par-4 is sufficient to induce apoptosis in these cells (26). However, in androgen-independent prostate cancer cells, inhibition of NF-κB transcriptional activity and activation of the Fas pathway are both required for Par-4-induced apoptosis (5). Fas (CD95) is a member of the tumor necrosis factor receptor family of death receptors that is activated by binding to Fas ligand (FasL), leading to the formation of death-inducing signaling complex (DISC) and the subsequent activation of caspase 8 and downstream caspases and apoptosis (24, 29). Par-4 activates the Fas pathway by promoting the Fas/FasL translocation to the cell membrane and by protecting the Fas apoptotic pathway from the inhibitory effects of ζ protein kinase C (5, 10).
Despite the advances made in understanding the mechanism of action of Par-4, the active domain(s) of Par-4 and its functional localization are largely unknown. The amino acid sequence of Par-4 protein predicts a leucine zipper domain at its C-terminal end (15, 16, 31, 32), between amino acids 292 and 332. This domain is involved in binding to all currently known partners of Par-4: WT1 (21), ζ protein kinase C (12), p62 (6), and Dlk (27). The leucine zipper domain is required for sensitization by Par-4 to apoptotic stimuli (16, 31). These findings led to the interpretation that the C-terminal leucine zipper domain is indispensable for the apoptosis-sensitizing function of Par-4 (12, 31).
The relationship between the intracellular localization of Par-4 and apoptotic function has not been adequately addressed. Par-4 contains two putative nuclear localization sequences, NLS1 at amino acids 20 to 25 and NLS2 at amino acids 137 to 153, both in the N-terminal half of Par-4. In normal tissues and cell types, endogenous Par-4 is localized in the cytoplasm (3, 17). Par-4 is cytoplasmic in the immortalized fibroblast cells NIH 3T3, and deletion of the first 68 amino acids, including NLS1, does not affect the apoptosis-sensitizing function of Par-4 (11). The role of NLS domains in direct apoptotic action of Par-4, however, has not been addressed. The following independent observations support a nuclear function for Par-4: (i) the presence of a bipartite nuclear localization sequence (NLS2); (ii) the ability of Par-4 to inhibit NF-κB transcriptional activity; (iii) direct binding to the nuclear proteins Dlk and WT1 (21, 27); and (iv) binding to WT1 and inhibition of the bcl-2 promoter (7).
In the present study, we performed structure-function analysis to address the relationship between the localization or the domains of Par-4 and its apoptotic function. Our studies indicated that for direct apoptosis by Par-4, entry into the nucleus is essential but the leucine zipper domain is not required. Most importantly, we identified a 59-amino-acid core domain of Par-4 that is essential for nuclear entry, Fas activation, inhibition of NF-κB activity, and induction of apoptosis in cancer cells. The spectrum of cells susceptible to apoptosis by this core domain covers both Par-4-sensitive and Par-4-resistant cancer cells but not normal cells.
MATERIALS AND METHODS
Plasmids, antibodies, and chemical reagents.
The pCB6+ vector, Par-4, ΔZip, and 163 (Par-4-ΔCTH) expression constructs in the pCB6+ background were described previously (21, 31). pSVβgal was from Promega. The RelA luciferase reporter system, composed of the Gal4-luciferase (Gal4-Luc) plasmid (which contains four Gal4 consensus DNA binding sites derived from Saccharomyces cerevisiae located upstream of the luciferase reporter gene) and the Gal4-RelA plasmid (containing the S. cerevisiae Gal4 DNA binding domain fused to the transactivation domain [TA1] of RelA), was from A. Baldwin (University of North Carolina at Chapel Hill, Chapel Hill, N.C.). The dominant-negative Fas-activated death domain (FADD), which prevents DISC formation, has been described (5). The green fluorescent protein (GFP) cloning plasmids pcDNA3.1/CT-GFP-TOPO and pcDNA3.1/NT-GFP-TOPO were from Invitrogen Life Technologies.
Par-4 deletion mutants (see Fig. 2 to 4) were constructed by PCR amplification with Par-4 as a template, followed by ligation in pcDNA3.1/CT-GFP-TOPO, and then left in the GFP plasmid or digested with XbaI and KpnI and subcloned into pCB6+ to prepare the untagged constructs.
FIG. 2.
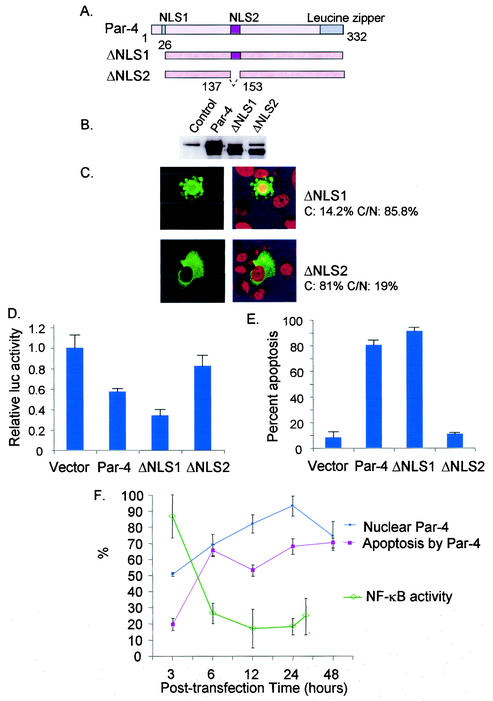
NLS2 is required for nuclear entry, inhibition of NF-κB activity, and induction of apoptosis. Schematic representation of full-length Par-4, ΔNLS1, and ΔNLS2 is shown in panel A. PC-3 cells were transiently transfected with the vector control, Par-4, ΔNLS1, and ΔNLS2 and their GFP-tagged derivatives. Expression of endogenous Par-4 (control), ectopic Par-4, ΔNLS1, and ΔNLS2 proteins was examined by Western blot analysis (B). Intracellular localization of ΔNLS1 and ΔNLS2 (C) and their ability to induce apoptosis (E) were examined as indicated in the legend to Fig. 1. The percentage of cytoplasmic and cytoplasmic and nuclear Par-4 and mutant protein is indicated in panel C. Also note the apoptotic morphology of the ΔNLS1-transfected cell in the same panel. To determine inhibition of NF-κB transcriptional activity by Par-4 and its mutants (D), cells were transfected for 48 h with the RelA reporter system and β-galactosidase expression plasmid together with vector, Par-4, and mutant constructs. Whole-cell lysates were subjected to luciferase assays, and luciferase activity was normalized to the corresponding β-galactosidase activity. Luciferase activity by Par-4 and its mutants is expressed relative to the activity noted with the vector. The relative time course of nuclear translocation of ectopic Par-4, inhibition of NF-κB transcriptional activity, and induction of apoptosis is shown in panel F.
FIG. 4.
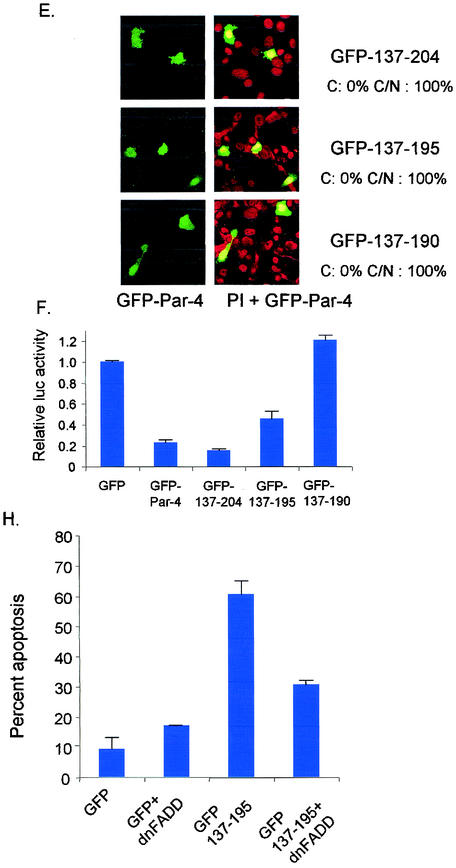
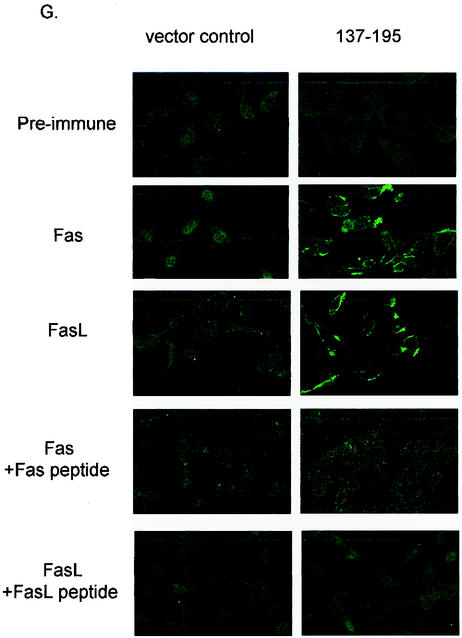
Identification of core domain of Par-4 that is sufficient for apoptosis. PC-3 cells were transiently transfected with GFP-tagged derivatives of Par-4 deletion mutants 137-332 and 148-332 (A) and examined for intracellular localization (B). Cells were transiently transfected with various deletion mutants of Par-4, 137-204, 137-195, and 137-190, and their GFP-tagged derivatives (C) and examined for expression by Western blot analysis (D), intracellular localization (E), inhibition of NF-κB activity (F), and apoptosis induction in the presence and absence of ectopic dominant-negative FADD (dnFADD) (H), as described in the legend to Fig. 1. The percentage of cells with strictly cytoplasmic and cytoplasmic and nuclear Par-4 mutant protein is indicated in panels B and E. To study Fas and FasL trafficking to the cell membrane, PC-3 cells were transfected with the 137-195 expression construct and the control vector for 48 h. The cells were then subjected to immunofluorescent staining with Fas and FasL antibody, preimmune normal rabbit antibody, Fas antibody preabsorbed with Fas peptide, and FasL antibody preabsorbed with FasL peptide as controls (G).
Polyclonal antibodies for Par-4, NF-κB (p65/RelA), Fas, and FasL were from Santa Cruz Biotechnology Inc. The anti-GFP rabbit polyclonal was from Torrey Pine Biolabs. Terminal deoxynucleotide transferase-mediated dUTP-biotin nick end labeling (TUNEL) reagents were purchased from Roche Molecular Biochemicals. Propidium iodide was from Clonetech. Peptides representing Fas and FasL were from Oncogene Research Products.
Cell lines.
Androgen-independent PC-3 and DU145 prostate cancer cells, androgen-dependent LNCaP prostate cancer cells, normal PrE and PrS primary prostatic cells, NIH 3T3 fibroblast cells, and NIH 3T3 Ras-transformed fibroblast cells have been described previously by us (5). Androgen-dependent LAPC-4 prostate cancer cells were from Charles Sawyers (University of California at Los Angeles), and MDA 2b cells were from Nora Navone (M. D. Anderson Cancer Center). Androgen-independent LNCaP/IGFBP5 and LNCaP/IL-6 prostate cancer cells are isogenic derivatives of LNCaP cells. LNCaP/IGFBP5 cells were prepared by Martin Gleave (Vancouver General Hospital, Vancouver, British Columbia, Canada) by stable transfection with the IGFBP5 expression construct. LNCaP/IL-6 cells were prepared by Allen C. Gao (University of Pittsburgh Medical Center, Pittsburgh, Pa.) by stable transfection with interleukin-6 expression construct. Immortalized PZ-HPV-7 human prostate epithelial cells and SQ20B and SCC66 head and neck cancer cells were from Mansoor Ahmed (University of Kentucky). The human lung cancer cells A549, H157, H838, and H460 were from John Yannelli (Internal Medicine Department, University of Kentucky). The human breast cancer cells MCF-7 and MDA 231 were from Guo-Min Li (Pathology, University of Kentucky). The immortalized breast epithelial cells MCF10a were from M. Johnson (Georgetown University). Immortalized HMEC human endothelial cells were from Mariana Karakashian (University of Kentucky).
Transfection and RelA/NF-κB reporter assays.
Cells were transiently transfected with the indicated plasmid constructs with Lipofectamine Plus (from Invitrogen Life Technologies) following the manufacturer's protocol. Cells were harvested after 48 h, and whole-cell lysates were subjected to Western blot analysis with the Par-4 polyclonal antibody (from Santa Cruz Biotechnology, Inc.) or luciferase and β-galactosidase assays, as previously described (5), to quantify and normalize RelA activity.
Indirect immunofluorescence.
Cells were transfected with the appropriate plasmid construct, and after being fixed, they were subjected to indirect immunofluorescence with Fas, FasL, or preimmune antibody and a secondary antibody conjugated with the fluorescent dye Alexa Fluor 488 (green) or Alexa Fluor 594 (red) from Molecular Probes, Inc., as previously described by us (5). The specificity of staining was determined by preabsorbing the Fas or FasL primary antibodies with excess (1:10) Fas or FasL peptide for 2 h at room temperature before adding the antibody to fixed cells. Background (nonspecific) staining was determined with preimmune normal rabbit antibody. For localization and apoptosis studies, nuclei were stained with propidium iodide and 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI), respectively, for 20 min after cell fixation.
Apoptosis assays and indirect immunofluorescence.
Cells plated in chamber slides were transiently transfected with untagged or GFP-tagged Par-4 or its derivatives. Apoptosis was determined with the TUNEL assay and by DAPI staining. The cells expressing untagged protein were visualized by staining with anti-Par-4 antibody followed by DAPI staining. Apoptotic nuclei were determined among transfected cells as described previously (5).
RESULTS
Nuclear translocation of Par-4 correlates with susceptibility to apoptosis.
Previous immunohistochemical studies localized Par-4 to both the cytoplasm and nucleus of prostate cancer cells but primarily to the cytoplasm in normal tissues (3, 16). Toward the goal of determining the relationship between intracellular localization of Par-4 and apoptosis, we first tested the localization of endogenous Par-4 in PC-3 and LNCaP prostate cancer cell cells, which were shown previously (5) to be sensitive and resistant, respectively, to apoptosis by ectopic Par-4.
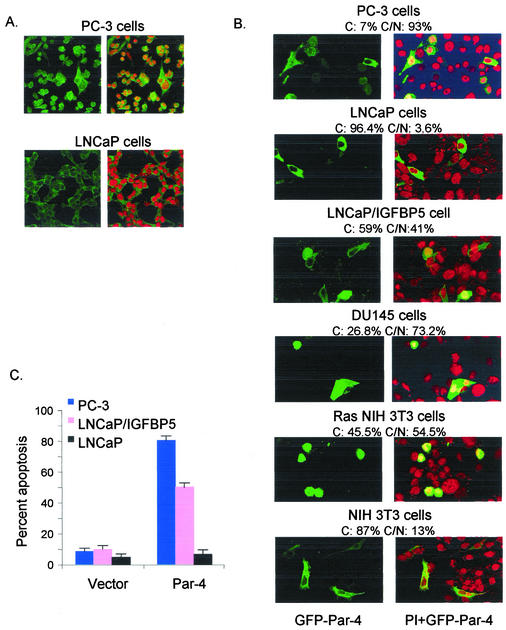
As shown in Fig. 1A, PC-3 cells showed endogenous Par-4 in both the cytoplasm and the nucleus; by contrast, LNCaP cells showed strictly cytoplasmic localization of endogenous Par-4. The findings of these initial experiments raised the possibility that Par-4 localization may correlate with its ability to induce apoptosis. To determine the precise relationship between Par-4 localization and apoptosis induction, a broad panel of transformed and nontransformed cells was transiently transfected with GFP-Par-4 and GFP vector for a control, and the transfectants were studied for intracellular localization of Par-4 and apoptosis by confocal microscopy. These experiments included NIH 3T3 Ras, PC-3, and DU145 cells that were previously shown to be sensitive to apoptosis induction by Par-4 (5, 26), LNCaP, PrE, PrS, and NIH 3T3 parental cells that were resistant to Par-4 (5, 26), and LNCaP/IGFBP5 and LNCaP/IL-6 (which are isogenic derivatives of LNCaP), MDA 2b, and LAPC-4 cells that had not been previously tested for susceptibility to Par-4 (Table 1).
FIG. 1.
Nuclear localization of Par-4 correlates with apoptosis induction. PC-3 and LNCaP cells were fixed and probed with Par-4 antibody to detect expression of endogenous Par-4. Nuclei were visualized by staining with propidium iodide. Intracellular localization of endogenous Par-4 was recorded by confocal microscopy. Par-4 images are shown in the left panels, and overlay of Par-4 and propidium iodide images are shown in the right panels (A). Cells were transfected with GFP-Par-4 and treated with propidium iodide to detect nuclei. Intracellular localization of GFP-Par-4 was recorded by confocal microscopy (B). GFP-Par-4 images are shown in the left panels, and overlay of GFP-Par-4 and propidium iodide images are shown in the right panels. Note yellow color resulting from colocalization of GFP green fluorescence with reddish-orange propidium iodide staining (right panels), indicating nuclear expression of Par-4. The percentage of cells with strictly cytoplasmic and with cytoplasmic and nuclear Par-4 is indicated above each panel as C and C/N, respectively. To determine percent apoptosis, cells were transfected with GFP-Par-4 and the GFP vector as a control and subjected to DAPI staining. Apoptotic cells were quantified and expressed as a percentage of the total number of transfected cells (C).
TABLE 1.
Correlation between nuclear translocation and susceptibility to apoptosis by Par-4
| Cell type | Cell linea | Nuclear Par-4 | Cytoplasmic Par-4 | Apoptosis by Par-4b |
|---|---|---|---|---|
| Androgen-independent cancer cells | PC-3 | + | + | Sensitive |
| DU145 | + | + | Sensitive | |
| LNCaP/IGFBP5 | + | + | Sensitive | |
| LNCaP/IL-6 | + | + | Sensitive | |
| Androgen-dependent cancer cells | LNCaP | − | + | Resistant |
| MDA 2b | − | + | Resistant | |
| LAPC-4 | − | + | Resistant | |
| Immortalized epithelial cells | PZ-HPV-7 | − | + | Resistant |
| Primary epithelial cells | PrE | − | + | Resistant |
| Primary stromal cells | PrS | − | + | Resistant |
| Immortalized fibroblast cells | NIH 3T3 | − | + | Resistant |
| Transformed fibroblast cells | Ras NIH 3T3 | + | + | Sensitive |
LNCaP/IGFBP5 and LNCaP/IL-6 are isogenic derivatives of LNCaP cells.
Sensitive, 50 to 75% of the transfected cells underwent apoptosis; resistant, <10% of the transfected cells underwent apoptosis.
The efficiency of transfection and the expression level of ectopic Par-4 were comparable in normal and cancer cell lines and similar to those reported previously (5). Representative examples of nuclear versus cytoplasmic localization and susceptibility to apoptosis by Par-4 are shown in Fig. 1B and C. It was noted that Par-4 nuclear presence correlated with its ability to induce apoptosis (Fig. 1 and Table 1). Ectopic Par-4 was detected in the nucleus and cytoplasm in androgen-independent prostate cancer cells PC-3, DU145, LNCaP-derived cells LNCaP/IGFBP5 and LNCaP/IL-6, and Ras-transformed NIH 3T3 cells, all of which were sensitive to apoptosis induction by Par-4 (Table 1 and Fig. 1). On the other hand, ectopic Par-4 was strictly cytoplasmic in the apoptosis-resistant androgen-dependent prostate cancer cells LNCaP, LAPC-4, and MDA 2b, immortalized mouse fibroblast NIH 3T3 cells, and PrE primary prostate epithelial cells and PrS primary prostate stromal cells (Table 1 and Fig. 1). These findings suggested a correlation between translocation of ectopic Par-4 to the nucleus and apoptosis induction.
Moreover, immunocytochemical analysis indicated that the localization of endogenous Par-4 was similar to that of ectopic Par-4; i.e., in cells sensitive to apoptosis by ectopic Par-4, endogenous and ectopic Par-4 were present in both the cytoplasm and the nucleus, but in cells that were resistant to apoptosis by ectopic Par-4, both endogenous and ectopic Par-4 was strictly cytoplasmic (the localization data for endogenous Par-4 were similar to those for ectopic Par-4 shown in Fig. 1 and Table 1 and are therefore not shown). Translocation of Par-4 to the nucleus was not prevented by the general caspase inhibitor zVAD-fmk, by inhibition of the Fas prodeath pathway by coexpression of dominant-negative FADD, and by coexpression of RelA, all of which inhibit Par-4-induced apoptosis (data not shown), suggesting that nuclear entry preceded apoptosis by Par-4. Together, these findings suggested that nuclear entry of ectopic Par-4 was essential for apoptosis.
NLS2 is essential for nuclear entry.
The 332-amino-acid protein Par-4 has two putative nuclear localization sequences, NLS1 (amino acids 20 to 25) and NLS2 (amino acids 137 to 153), which are conserved in human, rat, and mouse Par-4 (16). To delineate the relationship between Par-4 entry into the nucleus and induction of apoptosis, we generated untagged and GFP-tagged derivatives of Par-4, ΔNLS1 and ΔNLS2, that lacked the NLS sequences (Fig. 2A). PC-3 cells were transiently transfected with these constructs and examined for expression (Fig. 2B), nuclear entry (Fig. 2C), inhibition of NF-κB transcriptional activity (Fig. 2D), and apoptosis (Fig. 2E). ΔNLS1 showed nuclear entry, whereas ΔNLS2, which lacked an intact NLS2 domain, failed to translocate to the nucleus, suggesting that NLS2 is a functional nuclear localization sequence in Par-4 (Fig. 2C). Moreover, loss of NLS2 but not NLS1 abrogated the ability of Par-4 to inhibit NF-κB transcriptional activity and induce apoptosis in PC-3 cells (Fig. 2D and 2E). These findings indicated that an intact NLS2 sequence was essential for nuclear entry of Par-4 and that the ability to inhibit NF-κB transcriptional activity and to induce apoptosis correlated with nuclear entry of Par-4.
To further confirm this relationship between nuclear Par-4 and apoptosis, the percentage of PC-3 cells with nuclear ectopic Par-4 and apoptosis induction was determined at various posttransfection time points (Fig. 2F). At 3 h posttransfection, about 50% of the cells expressed nuclear Par-4 and 20% were undergoing apoptosis. At the later time points, a coparallel increase in nuclear translocation of Par-4 and the percentage of apoptotic cells was noted (Fig. 2F). Moreover, when the kinetics of inhibition of NF-κB transcriptional activity by ectopic Par-4 was determined by luciferase reporter assays, a similar relationship indicating inhibition of NF-κB transcriptional activity following nuclear entry of ectopic Par-4 was noted (Fig. 2F).
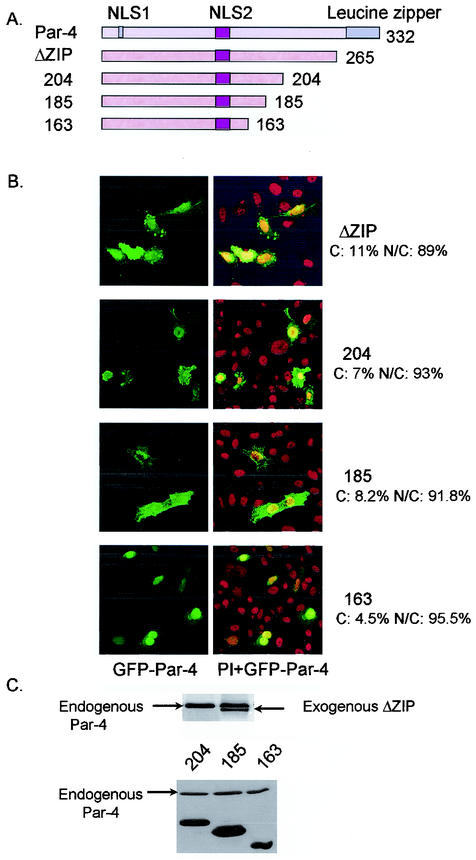
Leucine zipper domain of Par-4 is not essential for apoptosis.
We next sought to identify other domains of Par-4 that are essential for apoptosis. The C-terminal leucine zipper domain of Par-4 is required for sensitization of cells to exogenous apoptotic insults and for binding to all currently known partners of Par-4, including WT1, ζ protein kinase C, p62, and Dlk (6, 12, 21, 27). Transient transfection of a mutant lacking the leucine zipper domain (ΔZip) in PC-3 cells resulted in direct induction of apoptosis (Fig. 3E). This suggested that the mechanism by which Par-4 sensitizes cells to apoptosis is different from the mechanism required for direct induction of apoptosis.
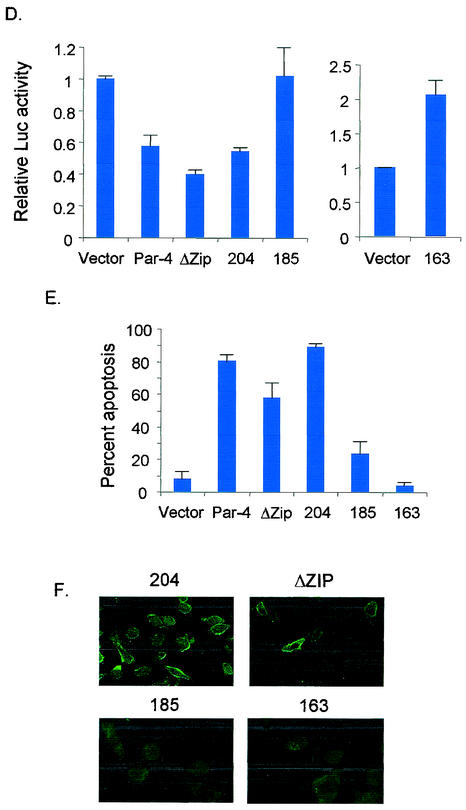
FIG. 3.
Leucine zipper domain is not essential for apoptosis. PC-3 cells were transiently transfected with untagged and GFP-tagged derivatives of various C-terminal deletion mutants of Par-4 (A). After 48 h, the cells were examined for intracellular localization of the mutants (B), expression of the mutant proteins by Western blot analysis (C), inhibition of NF-κB transcription activity (D), and apoptosis (E), as described in the legend to Fig. 1. The percentage of cells with strictly cytoplasmic and cytoplasmic and nuclear Par-4 mutant protein is indicated in panel B. To examine Fas translocation (F), PC-3 cells were transiently transfected with untagged C-terminal deletion mutants; after 48 h they were stained with anti-Fas antibody, and Alexa Fluor 488 green fluorescence was visualized by confocal microscopy.
To further characterize the C terminus of Par-4, untagged and GFP-tagged derivatives of various C-terminal deletion mutants were prepared and studied for nuclear localization, inhibition of NF-κB transcriptional activity and apoptosis in PC-3 cells. Similar to full-length Par-4 localization in PC-3 cells (Fig. 1A), all C-terminal mutants of Par-4 had a tendency to localize to the nucleus (Fig. 3B). However, NF-κB transcriptional activity and apoptosis were differentially affected by the extent of the C-terminal deletions (Fig. 3D and 3E). Par-4 mutants ΔZip and 1-204 inhibited NF-κB transcriptional activity and induced apoptosis; on the other hand, mutants 1-185 and 1-163 neither inhibited NF-κB transcriptional activity nor induced apoptosis (Fig. 3D and 3E). The slight reduction in apoptosis induction by the ΔZip mutant (Fig. 3E) may have resulted from the decreased stability of this mutant compared to full-length Par-4 and other mutants, as seen in the Western blot analysis in Fig. 2B and 3C. Moreover, the ability to induce Fas trafficking to the cell membrane by ΔZip and 1-204 correlated with their ability to induce apoptosis (Fig. 3F).
These experiments indicated that the amino acids downstream of 1 to 204, including the leucine zipper domain, were dispensable for apoptosis by Par-4 and that binding to the partner proteins via the leucine zipper domain involved in sensitization to various apoptotic stimuli (16) was not essential for inhibition of NF-κB transcription activity and direct apoptosis induction by Par-4.
Identification of the core domain of Par-4 required for apoptosis induction.
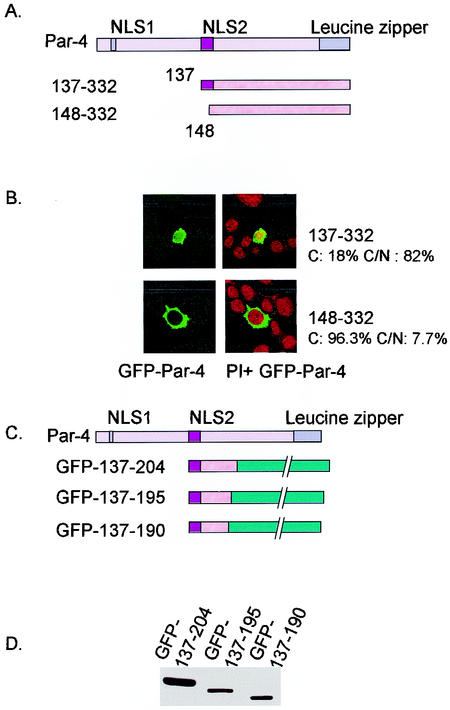
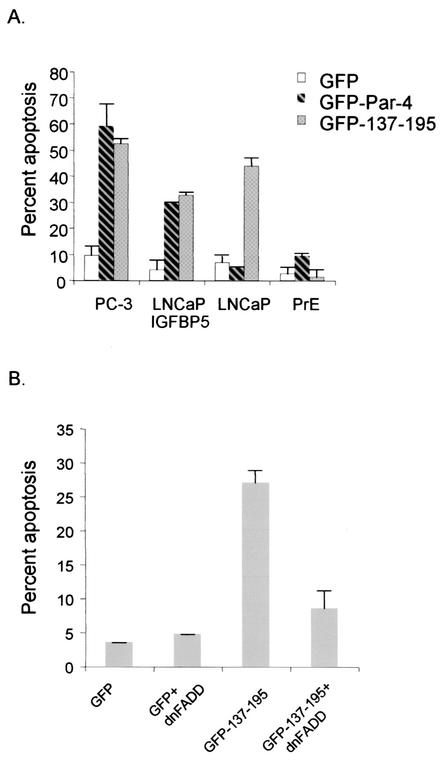
Up to this stage in our analysis, NLS2 was identified as the most critical sequence for nuclear localization and apoptosis induction. To further confirm the importance of NLS2, we constructed two N-terminal deletion constructs, 137-332 and 148-332, with an intact NLS2 and a disrupted NLS2, respectively (Fig. 4A). The construct with the intact NLS2, 137-332, translocated to the nucleus in PC-3 cells (Fig. 4B) and was able to induce apoptosis (note the apoptotic morphology of condensed nucleus in the 137-332-transfected cells). On the other hand, 148-332 did not induce apoptosis and localized strictly to the cytoplasm in PC-3-transfected cells (Fig. 4B). This suggested that NLS2 was critical for Par-4 function and that the N terminus of Par-4 was not required for Par-4 nuclear entry or apoptotic functions.
To define the minimal domain of Par-4 that is essential for apoptosis, we made additional GFP-tagged constructs that began with the intact NLS2 domain at the amino terminus and with various deletions upstream of amino acid 204 (Fig. 4C). Transient transfection of PC-3 cells indicated that 137-204 and 137-195 but not 137-190 inhibited NF-κB transcriptional activity and induced apoptosis (Fig. 4E and F and Table 2); note nuclear expression of all three deletion mutants (in Fig. 4E), reiterating that nuclear entry is not secondary to apoptosis. Moreover, the 137-195 mutant caused Fas and FasL translocation to the cell membrane (Fig. 4G), and cotransfection of 137-195 with dominant-negative FADD inhibited apoptosis (Fig. 4H), indicating that similar to full-length Par-4 (5), the 137-195 core domain induced apoptosis via the Fas prodeath pathway and inhibition of NF-κB transcriptional activity. Thus, these findings identified 137-195 as the minimal core domain of Par-4 that was essential and sufficient to induce apoptosis in PC-3 cells.
TABLE 2.
The Par-4 deletion mutant 137-195 induces apoptosis selectively in cancer cells
| Cell type | Par-4 reaction | Cell line | Apoptosis by Par-4 | Apoptosis by 137-195 | Apoptosis by 137-190 |
|---|---|---|---|---|---|
| Cancer cells | Sensitivea | PC-3 | + | + | − |
| DU145 | + | + | − | ||
| LNCaP/IGFBP5 | + | + | − | ||
| LNCaP/IL-6 | + | + | − | ||
| MDA MB231 | + | + | − | ||
| Resistantb | LNCaP | − | + | − | |
| LAPC-4 | − | + | − | ||
| MDA 2b | − | + | − | ||
| MCF-7 | − | + | − | ||
| Immortalized/normal cells | Resistantc | PZ-HPV-7 | − | − | − |
| PrS | − | − | − | ||
| PrE | − | − | − | ||
| MCF10a | − | − | − | ||
| HMEC | − | − | − |
+, apoptosis in 50 to 75% of the transfected cells.
+, apoptosis in 25 to 35% of the transfected cells.
−, apoptosis in <10% of the transfected cells for all the constructs, regardless of the cell type.
Core domain of Par-4 induces apoptosis specifically in cancer cells but not in normal cells.
To determine whether, similar to Par-4, the core domain 137-195 induced apoptosis exclusively in androgen-independent prostate cancer cells, we tested androgen-dependent and -independent prostate cancer cells and primary normal cells in apoptosis assays. Interestingly, 137-195 induced apoptosis in both androgen-dependent and -independent prostate cancer cells but not in normal cells (Fig. 5A). In view of this expanded target range of 137-195, we studied the susceptibility of a broad panel of cancer cells and primary normal and immortalized cells to apoptosis by Par-4 and 137-195, with 137-190 as a control. These experiments indicated that both Par-4-susceptible and Par-4-resistant cancer cells, regardless of whether or not they were of prostatic origin, were induced to undergo apoptosis by 137-195 but not by 137-190 (Table 2). Moreover, in addition to the cancer cell lines shown in Table 2, SQ20B and SCC66 head and neck cancer cells and A549, H157, H838 and H460 human lung cancer cells were all susceptible to apoptosis by Par-4 and its proapoptotic deletion mutant 137-195 (data not shown). Most importantly, neither Par-4 nor 137-195 (which, unlike Par-4, entered the nucleus in both cancer and normal cells; data not shown) induced apoptosis in the primary normal and immortalized cells (Table 2). The 137-195 mutant acquired the ability to induce apoptosis in cancer cell lines that were resistant to full-length Par-4, suggesting that an inhibitory domain(s) regulating the ability of Par-4 to induce apoptosis of certain cells is present at the C terminus. A more extensive deletion extending upstream of amino acid 195 disrupts the functional domains of Par-4, and therefore, such mutants (137-190, for example, for which data are shown in Table 2) are inactive in apoptosis induction.
FIG. 5.
Core domain of Par-4 has an expanded but cancer-specific apoptotic ability. Various cell lines were transiently transfected with GFP vector, GFP-Par-4, or GFP-137-195 and examined for apoptosis in androgen-dependent and -independent prostate cancer cells and primary cells (A). LNCaP cells were transiently transfected with GFP vector or GFP-137-195, with or without dominant-negative FADD (dnFADD), and examined for apoptosis induction (B), as described in the legend to Fig. 1.
DISCUSSION
The findings of the present study indicate that Par-4 translocation into the nucleus is essential for induction of apoptosis. The NLS2 domain of Par-4 was essential for nuclear translocation, and resistance to apoptosis was noted in cells that did not translocate Par-4 to the nucleus and with Par-4 constructs that lacked an intact NLS2 domain. Moreover, nuclear translocation was essential for inhibition of NF-κB transcriptional activity by Par-4. Because apoptosis inhibitors such as zVAD-fmk, dominant-negative FADD, and RelA did not block nuclear entry, and because several deletion mutants of Par-4 (for example, amino acids 1 to 163, amino acids 1 to 185, and amino acids 137 to 190) translocated to the nucleus but failed to induce apoptosis, nuclear localization was not secondary to apoptosis. Because inhibition of NF-κB activity is critical for apoptosis by Par-4 (5, 26), these observations suggest that Par-4 has a nuclear function that includes NF-κB transcriptional inhibition and induction of apoptosis.
Most importantly, the deletion analysis identified a 59-amino-acid core domain of Par-4 that constitutes a minimal region of Par-4 that is sufficient for apoptosis. This core domain translocated to the nucleus in both cancer and normal or immortalized cells, but is selective in inducing apoptosis of the cancer cells, regardless of whether they are sensitive to wild type Par-4. Importantly, similar to Par-4, the core domain induces apoptosis by a mechanism involving the Fas prodeath pathway and inhibition of NF-κB transcription activity, indicating that the mechanism of apoptosis by Par-4 is encoded in the core domain. This core domain is 100% conserved in rat, mouse, and human Par-4 (16). Because this domain does not induce apoptosis in normal cells but induces apoptosis in diverse cancer cells, we designated it the selective for apoptosis induction cancer cells (SAC) domain. This domain resembles neither the death domains (19, 33) nor the death effector domains (4) of other proapoptotic proteins, nor does it show significant homology with other known proteins in GenBank, and unlike the previously characterized death domains and death effector domains, the SAC domain specifically induces apoptosis in cancer cells and not normal cells.
Studies to elucidate the precise mechanism of resistance of normal cells to the SAC domain are currently under way. Because Par-4 causes remarkable regression of solid tumors by apoptosis via the Fas prodeath pathway and inhibition of NF-κB transcription activity (5), the SAC domain, which utilizes a similar mechanism of apoptosis, is also expected to cause tumor regression by apoptosis. The cancer-specific apoptotic potential of the SAC domain renders it a promising candidate for directed molecular therapeutics of cancer. Moreover, the SAC domain has potential applications in elucidation of the fundamental cellular and molecular differences between normal and cancer cells.
The findings of the present study suggest that the leucine zipper domain of Par-4 is not essential for apoptosis induced by Par-4 acting alone. Cells that are resistant to apoptosis following overexpression of Par-4 become sensitized to the action of diverse apoptotic insults (16), and the leucine zipper domain of Par-4 is essential for the apoptosis sensitization function of Par-4 (31). The apoptosis-sensitizing action is regulated by the interaction of Par-4 via its leucine zipper domain, with WT1, Dlk, ζ protein kinase C, and p62 (reviewed in reference 16). Depending on the partner and the cellular context, the apoptosis-sensitizing function of Par-4 is implemented in the cytoplasm when regulated by ζ protein kinase C and p62 and in the nucleus when regulated by WT1 (16). Together, these findings imply that the domain and mechanism for apoptosis by Par-4 acting alone are distinct from those essential for apoptosis sensitization by Par-4 acting in conjunction with other apoptotic insults.
In summary, our studies indicate that nuclear entry is essential for apoptosis by Par-4. By deletion analysis, we have uncovered a unique core domain of Par-4 that is essential and sufficient for nuclear entry, Fas/FasL translocation to the cell membrane, activation of the Fas prodeath pathway, inhibition of NF-κB activity, and induction of apoptosis. This domain extends the susceptibility spectrum of Par-4 to diverse cancer cells, regardless of whether they are susceptible or resistant to Par-4. Because this domain shows selective action against cancer cells but not normal cells, it has both academic and therapeutic applications.
Acknowledgments
We thank Anthony Sinai (University of Kentucky) for thoughtful discussions.
This study was supported by NIH/NCI grants CA60872 and CA84511 (to V.M.R.).
REFERENCES
- 1.Barradas, M., A. Monjas, M. T. Diaz-Meco, M. Serrano, and J. Moscat. 1999. The downregulation of the proapoptotic protein Par-4 is critical for Ras-induced survival and tumor progression. EMBO J. 18:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berra, E., M. M. Municio, L. Sanz, S. Frutos, M. T. Diaz-Meco, and J. Moscat. 1997. Positioning atypical protein kinase C isoforms in the UV-induced apoptotic signaling cascade. Mol. Cell. Biol. 17:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boghaert, E. R., S. F. Sells, A. J. Walid, P. Malone, N. M. Williams, M. H. Weinstein, R. Strange, and V. M. Rangnekar. 1997. Immunohistochemical analysis of the proapoptotic protein Par-4 in normal rat tissues. Cell Growth Differ. 8:881-890. [PubMed] [Google Scholar]
- 4.Boldin, M. P., E. E. Varfolomeev, Z. Pancer, I. L. Mett, J. H. Camonis, and D. Wallach. 1995. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 270:7795-7798. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, M., S. G. Qiu, K. M. Vasudevan, and V. M. Rangnekar. 2001. Par-4 drives trafficking and activation of Fas and Fasl to induce prostate cancer cell apoptosis and tumor regression. Cancer Res. 61:7255-7263. [PubMed] [Google Scholar]
- 6.Chang, S., J. H. Kim, and J. Shin. 2002. p62 forms a ternary complex with protein kinase Czeta and PAR-4 and antagonizes PAR- 4-induced protein kinase Czeta inhibition. FEBS Lett. 510:57-61. [DOI] [PubMed] [Google Scholar]
- 7.Cheema, S. K., S. K. Mishra, V. M. Rangnekar, A. M. Tari, R. Kumar, and G. Berestein. 2003. PAR-4 transcriptionally regulates Bcl-2 through a WT1 binding site on the bcl-2 promoter. J. Biol. Chem. 278:19995-20005. [DOI] [PubMed] [Google Scholar]
- 8.Cook, J., S. Krishnan, S. Ananth, S. F. Sells, Y. Shi, M. M. Walther, W. M. Linehan, V. P. Sukhatme, M. H. Weinstein, and V. M. Rangnekar. 1999. Decreased expression of the proapoptotic protein Par-4 in renal cell carcinoma. Oncogene 18:1205-1208. [DOI] [PubMed] [Google Scholar]
- 9.Culmsee, C., Y. Zhu, J. Krieglstein, and M. P. Mattson. 2001. Evidence for the involvement of Par-4 in ischemic neuron cell death. J. Cereb. Blood Flow Metab. 21:334-343. [DOI] [PubMed] [Google Scholar]
- 10.de Thonel, A., A. Bettaieb, C. Jean, G. Laurent, and A. Quillet-Mary. 2001. Role of protein kinase C zeta isoform in Fas resistance of immature myeloid KG1a leukemic cells. Blood 98:3770-3777. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Meco, M. T., M. J. Lallena, A. Monjas, S. Frutos, and J. Moscat. 1999. Inactivation of the inhibitory kappaB protein kinase/nuclear factor kappaB pathway by Par-4 expression potentiates tumor necrosis factor alpha-induced apoptosis. J. Biol. Chem. 274:19606-19612. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Meco, M. T., M. M. Municio, S. Frutos, P. Sanchez, J. Lozano, L. Sanz, and J. Moscat. 1996. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell 86:777-786. [DOI] [PubMed] [Google Scholar]
- 13.Duan, W., Z. Guo, and M. P. Mattson. 2000. Participation of par-4 in the degeneration of striatal neurons induced by metabolic compromise with 3-nitropropionic acid. Exp. Neurol. 165:1-11. [DOI] [PubMed] [Google Scholar]
- 14.Duan, W., V. M. Rangnekar, and M. P. Mattson. 1999. Prostate apoptosis response-4 production in synaptic compartments following apoptotic and excitotoxic insults: evidence for a pivotal role in mitochondrial dysfunction and neuronal degeneration. J. Neurochem. 72:2312-2322. [DOI] [PubMed] [Google Scholar]
- 15.Dutta, K., A. Alexandrov, H. Huang, and S. M. Pascal. 2001. pH-induced folding of an apoptotic coiled coil. Protein Sci. 10:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Guendy, N., and V. M. Rangnekar. 2003. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Exp. Cell Res. 283:51-66. [DOI] [PubMed] [Google Scholar]
- 17.Guo, Q., W. Fu, J. Xie, H. Luo, S. F. Sells, J. W. Geddes, V. Bondada, V. M. Rangnekar, and M. P. Mattson. 1998. Par-4 is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer disease. Nat. Med. 4:957-962. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, N., and S. Nagata. 1993. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 268:10932-10937. [PubMed] [Google Scholar]
- 20.Johnstone, R. W., A. A. Ruefli, and S. W. Lowe. 2002. Apoptosis: a link between cancer genetics and chemotherapy. Cell 108:153-164. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone, R. W., R. H. See, S. F. Sells, J. Wang, S. Muthukkumar, C. Englert, D. A. Haber, J. D. Licht, S. P. Sugrue, T. Roberts, V. M. Rangnekar, and Y. Shi. 1996. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms' tumor suppressor WT1. Mol. Cell. Biol. 16:6945-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnstone, R. W., N. Tommerup, C. Hansen, H. Vissing, and Y. Shi. 1998. Mapping of the human PAWR (par-4) gene to chromosome 12q21. Genomics 53:241-243. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, M., T. Furukawa, T. Abe, T. Yatsuoka, E. M. Youssef, T. Yokoyama, H. Ouyang, Y. Ohnishi, M. Sunamura, M. Kobari, S. Matsuno, and A. Horii. 1998. Identification of two common regions of allelic loss in chromosome arm 12q in human pancreatic cancer. Cancer Res. 58:2456-2460. [PubMed] [Google Scholar]
- 24.Kischkel, F. C., S. Hellbardt, I. Behrmann, M. Germer, M. Pawlita, P. H. Krammer, and M. E. Peter. 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayo, M. W., C. Y. Wang, P. C. Cogswell, K. S. Rogers-Graham, S. W. Lowe, C. J. Der, and A. S. Baldwin, Jr. 1997. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science 278:1812-1815. [DOI] [PubMed] [Google Scholar]
- 26.Nalca, A., S. G. Qiu, N. El-Guendy, S. Krishnan, and V. M. Rangnekar. 1999. Oncogenic Ras sensitizes cells to apoptosis by Par-4. J. Biol. Chem. 274:29976-29983. [DOI] [PubMed] [Google Scholar]
- 27.Page, G., D. Kogel, V. Rangnekar, and K. H. Scheidtmann. 1999. Interaction partners of Dlk/ZIP kinase: coexpression of Dlk/ZIP kinase and Par-4 results in cytoplasmic retention and apoptosis. Oncogene 18:7265-7273. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen, W. A., H. Luo, I. Kruman, E. Kasarskis, and M. P. Mattson. 2000. The prostate apoptosis response-4 protein participates in motor neuron degeneration in amyotrophic lateral sclerosis. FASEB J. 14:913-924. [DOI] [PubMed] [Google Scholar]
- 29.Peter, M. E., and P. H. Krammer. 1998. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr. Opin. Immunol. 10:545-551. [DOI] [PubMed] [Google Scholar]
- 30.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 18:6938-6947. [DOI] [PubMed] [Google Scholar]
- 31.Sells, S. F., S. S. Han, S. Muthukkumar, N. Maddiwar, R. Johnstone, E. Boghaert, D. Gillis, G. Liu, P. Nair, S. Monnig, P. Collini, M. P. Mattson, V. P. Sukhatme, S. G. Zimmer, D. P. Wood, Jr., J. W. McRoberts, Y. Shi, and V. M. Rangnekar. 1997. Expression and function of the leucine zipper protein Par-4 in apoptosis. Mol. Cell. Biol. 17:3823-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sells, S. F., D. P. Wood, Jr., S. S. Joshi-Barve, S. Muthukumar, R. J. Jacob, S. A. Crist, S. Humphreys, and V. M. Rangnekar. 1994. Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Differ. 5:457-466. [PubMed] [Google Scholar]
- 33.Tartaglia, L. A., T. M. Ayres, G. H. Wong, and D. V. Goeddel. 1993. A novel domain within the 55 kd tumor necrosis factor receptor signals cell death. Cell 74:845-853. [DOI] [PubMed] [Google Scholar]