Diabetes has been termed the epidemic of the 21st century and over the past 50 years in Western societies has been doubling in incidence every 15 years. It exacts a huge socioeconomic toll because of its devastating microvascular and macrovascular complications and the need of patients to maintain a lifetime daily therapeutic regimen. The childhood-onset form of diabetes, which accounts for 10% of all cases in humans [autoimmune or type 1 diabetes (IDDM)], is the product of a T lymphocyte-dependent autoimmune process that specifically destroys the insulin-secreting β cells of the pancreas without affecting contiguous endocrine cells or the surrounding exocrine tissue (1). Such tissue and cell specificity might logically be the product of an autoimmune reactivity directed at β-cell-specific molecular targets mediated by direct cellular contact between the target β cell and effector cytotoxic T lymphocytes. The article by Lieberman et al. (2) in a recent issue of PNAS supports such a notion and identifies the molecular target of a diabetogenic CD8 T cell in diabetes-susceptible nonobese diabetic (NOD) mice as the β-cell-specific protein islet glucose-6-phosphatase (G6Pase) catalytic subunit-related protein (IGRP). Moreover, it shows on the basis of T cell antigen receptor (TCR) Vα-chain usage that the TCR usage of the T cells studied in these experiments represents a dominant specificity in the NOD mouse, and that such cells are the major component of the CD8 T cell population infiltrating the islet up to the onset of diabetes (3).
Type 1 diabetes is a polygenic disorder both in humans and in the NOD mouse involving ≥20 loci (4) but with a major contribution to susceptibility from the MHC class II region (up to 50% in humans). The only other alleles un-equivocally identified lie within the variable number tandem repeat region of the insulin gene (IDDM2) and CTLA-4 (IDDM12) (5). The NOD mouse possesses a single MHC class II molecule, I-Ag7, that corresponds to the DQ8 locus in the human MHC that is associated with disease in humans (DQA1*0301-DQB1*0302 linked to DR4 and DQA1*0501-DQB1*0201 linked to DR3). A lesser contribution may be made by class I loci (notably A24 in humans and H2-Kd in the NOD mouse). Such associations presumably relate to the presentation of specific peptides to the T cell receptors that initiate or modulate the progress of disease. This could occur at any level from establishment of self-tolerance and development of the T cell repertoire to a response to foreign proteins that leads to molecular mimicry. Transgenic B6 mice carrying susceptible human MHC class II allele DQA1*0301-DQB1*0302 in the absence of endogenous mouse class II develop an immune-mediated diabetes if the costimulatory molecule B7 is also expressed on the pancreatic β cells (6). The human MHC class I A2.1 allele as a transgene in NOD mouse also accelerates the course of the disease, again supporting the conclusion that lessons learned from these animals are directly relevant to the human disease process (7). A key question, and one that is addressed in the article by Lieberman et al. (2), concerns the identity of the peptides these MHC molecules present to autoreactive T cells and how this relates to the specificity and diversity of CD4 and CD8 T cell responses that contribute to the disease. Such questions are difficult to address in the case of human disease, because the islets of Langerhans and draining lymph nodes where the disease occurs are inaccessible to biopsy or noninvasive imaging techniques.
The majority of diabetic autoantigens that are defined molecularly have been discovered either by a candidate-gene approach or by serological investigations in diabetic humans. Insulin, the 65-kDa form of glutamate decarboxylase, and the insulin granule membrane proteins ICA512 (IA2) and phogrin (IA2β) are the major targets of islet cell autoantibodies that are detected by immunofluorescence microscopy of islets. Longitudinal studies of at-risk neonates show that the number of autoantibody specificities rather than individual titers is a stronger indicator of risk of disease progression. There is no rigid hierarchy among the humoral autoantigens in terms of their order of appearance, although statistically insulin autoreactivity tends to appear earlier, at least in young individuals. Of the known autoantigens, only insulin seems β-cell-specific, whereas glutamic acid decarboxylase 65 (GAD65) and the IA2 members are broadly distributed among neuroendocrine tissues such as the brain, pituitary, and adrenal medulla. If there is any common link between these molecules it would seem to be an association with the regulated pathway of secretion where these proteins are localized intracellularly. Such a connection is also evident for some of the less prominent autoantigens, namely carboxypeptidase E and ICA69.
Insulin autoantibodies are present in the prediabetic phase in NOD animals but are not a prerequisite for subsequent emergence of disease (8). On the other hand, disease-related antibodies to GAD65 and the IA2 proteins cannot be detected by most investigators. Islet-specific CD4 and CD8 T cell clones have been isolated from spleen, lymph nodes, or islet infiltrates of pre- or newly diabetic NOD mice (9–11), and many accelerate disease in naive recipients or transfer diabetes susceptibility to NOD severe combined immunodeficient and NOD rag2 (–/–) mice, animals that have no endogenous T or B cells but are otherwise immune-competent. In terms of antigens, insulin and phogrin seem to be the target of spontaneous and/or primed T cell responses, and in both cases reactivity is directed at a small set of peptide epitopes (12, 13). Primed T cell responses to GAD65 can also be observed (14); however, their relevance to autoimmunity is questionable because the mouse islet expresses vanishingly small amounts of this GAD isoform. The spontaneous islet-specific diabetogenic CD4 or CD8 T cell clones that were not initially selected on candidate antigens are of particular interest, because these are likely to be most informative as to the molecular targets of spontaneous disease. The panel of clones produced by Wegmann et al. (12) (CD4), Haskins et al. (15) (CD4), Santamaria and coworkers (16) (CD4 and CD8), and Serreze and coworkers (17) (CD8) are of particular interest in this context. The restricted usage of Vα-chain in the TCRs of many of these clones (17–19) points to a restricted number of antigenic epitopes and antigens being responsible. Except in the case of insulin, none of these seem to correspond to the humoral antigens (12), and the majority have not been identified despite more than a decade of effort in some cases.
The article by Leiberman et al. (2) redraws the map in that regard in showing that the extensively studied CD8 clone NY8.3 developed by the Santamaria group (16) and the corresponding TCR transgenic animal recognize a hitherto undocumented antigen that is specific to the pancreatic β cell and of potential interest to the regulation of β cell energy metabolism. Similar to all the other diabetic autoantigens, it seems to be confined to membrane components of the secretory pathway.
The NY8.3 clone was isolated almost a decade ago from islet infiltrates of acutely diabetic NOD/Leiter mice from one of many H2-Kd-restricted T cell lines and clones with a common TCR restriction (9). NY8.3 and splenocytes from the CD8.3 TCR transgenic mouse greatly accelerate diabetes after adoptive transfer into young NOD recipients. In NOD severe combined immunodeficient mice it can produce disease by itself but is enhanced by cotransfer of CD4+ cells. T cells with the same Vα17 MR(D/E)Jα42 chain combination as NY8.3 appear in NOD insulitic lesions as early as 4–6 weeks of age. It is note-worthy that single-chain CD8.3β-chain transgenic animals combine with the same α-chain configuration, suggesting that it is a major determinant in antigen specificity and that the antigen targets are restricted in number. Other studies have documented avidity maturation of CD8.3-related T cells as insulitis progresses toward a more invasive form and disease onset (20). MHC class I tetramer bearing a mimotope peptide (NRP-V7) marks up to 40% of the CD8+ T cells present in NOD islet infiltrates and a significant number in the circulation (3). By comparison, MHC class II tetramers loaded with the BDC2.5 mimotope mark <1% of CD4 cells in the islet (K. Haskins and L. Teyton, unpublished observations).
The approach taken by the Serreze group (17) to identify the antigen was simple and direct, yet heroic. They took a CD8 cytotoxic T lymphocyte (CTL) bioassay that could detect endogenous peptides eluted from Kd receptors of the NIT1 cell line [an insulinoma cell line derived from NOD mice bearing an insulin promoter simian virus 40 T antigen transgene (21)]. After initial rounds of preparative liquid chromatographic fractionation, samples were directed to a high-end liquid chromatography tandem mass spectrometer for direct sequencing with the effluent split for CTL assay. The 9-mer sequence corresponded to a known pancreatic β cell protein that was subsequently shown to stimulate the CD 8.3 cells when transfected into engineered H2-Kd-presenting cells.
The cognate peptide was derived from IGRP, a protein that was first identified from a subtractive hybridization screen that was performed to identify β-cell-specific proteins that could be potential diabetes autoantigens or regulators of insulin stimulus–secretion coupling (22). IGRP was initially investigated as a possible islet-specific G6Pase that had long been postulated as a key component of a glucose substrate cycle and control of energy metabolism in the β cell (23). IGRP has 50% sequence identity with liver G6Pase, spans the membrane in the same way, and has the conserved acid phosphatase signature but no discernible catalytic activity. Similar to insulin and islet amyloid polypeptide, it is expressed in a highly pancreatic β-cell manner (24) yet seems to be controlled by a different set of transcription factors (25). IGRP and mRNA are abundant in mouse and human islets but not expressed in the rat, where the single-copy gene has an altered transcriptional start site and a series of ORF frame-shift mutations and deletions (26). The human IGRP gene is located on chromosome 2q24-31 in humans, a short distance from glucagon and GAD67 genes in a region where IDDM7, NIDDM, and the Bardet–Biedl genes map. CTLA-4, a chromosome 2 candidate for IDDM12, maps to 2q33 (5).
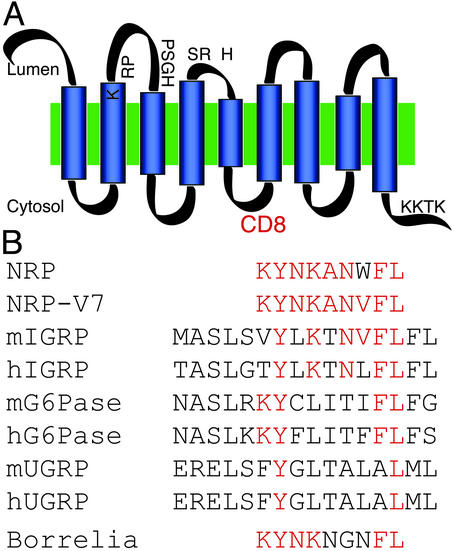
There are few clues from what we know of IGRP that single it out as a candidate for a diabetes autoantigen apart from its tissue specificity and an association with the secretory pathway. The COOH-terminal KKTK sequence of IGRP is an endoplasmic reticulum (ER) retention motif for transmembrane resident proteins, and its localization by immunofluorescence microscopy is consistent with the ER or secretory granule compartment (Fig. 1). It is conceivable that IGRP gains access to post-Golgi vesicular compartments such as the secretory granule particularly under conditions of ER stress, which can also be associated with surface delivery of proteins such as HSP70 that trigger an innate immune response. The epitope identified by DiLorenzo et al. (17) is predicted to reside on the cytoplasmic face of the molecule and in this regard is similar to the dominant T cell and humoral epitopes of GAD65- and IA2-related autoantigens. Similar to these molecules, IGRP is also membrane-associated, although in its case most of its sequence appears buried in the membrane with only short cytoplasmic and luminal peptide loops (Fig. 2 and ref. 23).
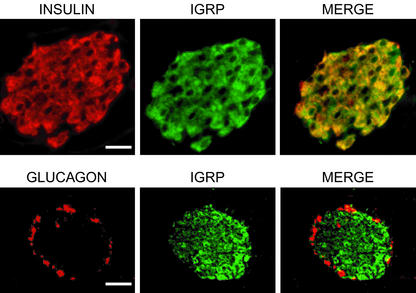
Fig. 1.
Immunolocalization of IGRP in BALB/c mouse pancreas with antibody raised to the full-length IGRP recombinant protein. IGRP immunoreactivity is localized to insulin-containing β cells and excluded from glucagon-containing α cells. In the β cell, IGRP staining overlaps with insulin except in areas close to the nuclear membrane, which is continuous with the ER. (Scale bars: Upper, 20 μm; Lower, 40 μm.)
Fig. 2.
(A) Structural relationship between NRP epitope peptides and G6Pase family members and mimotopes. The CD8 epitope that was identified by Lieberman et al. (2) appears to be located in the third 23-aa cytoplasmic loop of IGRP. The putative active-site residues in the second and third luminal domains are indicated. (B) Alignment of the NRP mimotope, the superagonist NRP-V7 variant, members of the G6Pase family, and Borrelia agonist of the CD8.3 T cell. The Kd anchor residues at positions 2 and 8 in the NRP molecule are conserved in each case. Six of the nine residues of NRP-V7 are conserved in murine IGRP (mIGRP) and the Borrelia peptide. Much weaker conservation is seen in G6Pase and ubiquitously expressed G6Pase catalytic subunit-related protein (UGRP).
Santamaria and coworkers (27), in identifying mimotopes for the CD8.3 clone, showed that many naturally occurring peptides could stimulate the clone as efficiently as NRP-A7. They included an agonist peptide from Borrelia burgdorferi, an organism that has been implicated in the autoimmune-like reaction that results in arthritis accompanying Lyme disease (28). From the present study, the cognate IGRP peptide epitope would be classed as an agonist for the CD8.3 clone with the peptide producing a weaker response than NRP-A7 in CTL assays. An important question for the future is whether anti-genicity is related to molecular mimicry or whether it is some structural or cell biological feature of the molecule itself that is important. It will be of interest to see what other antigenic epitopes may reside in IGRP and if it is presented by IAg7 and DQ8 to CD4 cells in mice and humans, respectively. Whether it is an important target for CD8 T cells in humans may depend on what IGRP peptide epitopes can be presented on human MHC class I molecules, because humans have no precise equivalent of Kd. Given that 40% of the CD8 T cells in the insulitic lesion in NOD mice are directed at the specific Kd-binding sequence identified by these authors and that MHC class 1 tetramers targeted at insulin B chain 15–23 see a similar proportion of CD8 T cells in inflamed islets, there is not a lot of room for other CD8 epitope specificities in these animals. Given the general observation that both CD4 and CD8 responses in NOD mice act synergistically to produce disease (9), it will be of interest to know in this case whether they are linked to the same autoantigen or to different β-cell-specific proteins. An IGRP gene knockout animal should be quite informative in such studies, and, if the rat data are anything to go by, knockout animals should be viable. It could be argued that the very existence of the Biobreeding (BB) rat model of autoimmune diabetes indicates that IGRP is not essential for diabetes autoimmunity; however, the BB autoimmunity is linked to a Ian gene lymphopenia, and initiation of autoimmunity is probably not mechanistically the same as in the NOD mouse or humans (29). If IGRP is as important as these studies suggest and an animal can live without it, then a way to prevent autoimmune attack against IGRP may be to simply suppress its expression. This is an area in which ongoing studies on the transcriptional regulation of this gene may ultimately prove to be highly significant for diabetes therapy. This is clearly not an option for other β-cell-specific autoantigens such as insulin, although even here there is evidence that escape of central tolerance relating to the promoter activity in thymic medullary epithelium may be key to its subsequent recognition as an autoantigen (30).
See companion article on page 8384 in issue 14 of volume 100.
References
- 1.Castano, L. & Eisenbarth, G. S. (1990) Annu. Rev. Immunol. 8, 647–679. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman, S. M., Evans, A. M., Han, B., Takaki, T., Vinnitskaya, Y., Caldwell, J. A., Serreze, D. V., Shabanowitz, J., Hunt, D. F., Nathenson, S. G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8384–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trudeau, J. D., Kelly-Smith, C., Verchere, C. B., Elliott, J. F., Dutz, J. P., Finegood, D. T., Santamaria, P. & Tan, R. (2003) J. Clin. Invest. 111, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wicker, L. S., Todd, J. A. & Peterson, L. B. (1995) Annu. Rev. Immunol. 13, 179–200. [DOI] [PubMed] [Google Scholar]
- 5.Ueda, H., Howson, J. M., Esposito, L., Heward, J., Snook, H., Chamberlain, G., Rainbow, D. B., Hunter, K. M., Smith, A. N., Di Genova, G., et al. (2003) Nature 423, 506–511. [DOI] [PubMed] [Google Scholar]
- 6.Wen, L., Wong, F. S., Tang, J., Chen, N. Y., Altieri, M., David, C., Flavell, R. & Sherwin, R. (2000) J. Exp. Med. 191, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marron, M. P., Graser, R. T., Chapman, H. D. & Serreze, D. V. (2002) Proc. Natl. Acad. Sci. USA 99, 13753–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abiru, N., Yu, L., Miao, D., Maniatis, A. K., Liu, E., Moriyama, H. & Eisenbarth, G. S. (2001) J. Autoimmun. 17, 1–6. [DOI] [PubMed] [Google Scholar]
- 9.Nagata, M., Santamaria, P., Kawamura, T., Utsugi, T. & Yoon, J. W. (1994) J. Immunol. 152, 2042–2050. [PubMed] [Google Scholar]
- 10.Haskins, K. & Wegmann, D. (1996) Diabetes 45, 1299–1305. [DOI] [PubMed] [Google Scholar]
- 11.Bowie, L., Tite, J. & Cooke, A. (1999) J. Immunol. Methods 228, 87–95. [DOI] [PubMed] [Google Scholar]
- 12.Wegmann, D. R., Gill, R. G., Norbury-Glaser, M., Schloot, N. & Daniel, D. (1994) J. Autoimmun. 7, 833–843. [DOI] [PubMed] [Google Scholar]
- 13.Kelemen, K., Wegmann, D. R. & Hutton, J. C. (2001) Diabetes 50, 1729–1734. [DOI] [PubMed] [Google Scholar]
- 14.Zekzer, D., Wong, F. S., Ayalon, O., Millet, I., Altieri, M., Shintani, S., Solimena, M. & Sherwin, R. S. (1998) J. Clin. Invest. 101, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haskins, K., Portas, M., Bradley, B., Wegmann, D. & Lafferty, K. (1988) Diabetes 37, 1444–1448. [DOI] [PubMed] [Google Scholar]
- 16.Verdaguer, J., Schmidt, D., Amrani, A., Anderson, B., Averill, N. & Santamaria, P. (1997) J. Exp. Med. 186, 1663–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiLorenzo, T. P., Graser, R. T., Ono, T., Christianson, G. J., Chapman, H. D., Roopenian, D. C., Nathenson, S. G. & Serreze, D. V. (1998) Proc. Natl. Acad. Sci. USA 95, 12538–12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simone, E., Daniel, D., Schloot, N., Gottlieb, P., Babu, S., Kawasaki, E., Wegmann, D. & Eisenbarth, G. S. (1997) Proc. Natl. Acad. Sci. USA 94, 2518–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson, B., Park, B. J., Verdaguer, J., Amrani, A. & Santamaria, P. (1999) Proc. Natl. Acad. Sci. USA 96, 9311–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amrani, A., Verdaguer, J., Serra, P., Tafuro, S., Tan, R. & Santamaria, P. (2000) Nature 406, 739–742. [DOI] [PubMed] [Google Scholar]
- 21.Hamaguchi, K., Gaskins, H. R. & Leiter, E. H. (1991) Diabetes 40, 842–849. [DOI] [PubMed] [Google Scholar]
- 22.Neophytou, P. I., Muir, E. M. & Hutton, J. C. (1996) Diabetes 45, 127–133. [DOI] [PubMed] [Google Scholar]
- 23.Arden, S. D., Zahn, T., Steegers, S., Webb, S., Bergman, B., O'Brien, R. M. & Hutton, J. C. (1999) Diabetes 48, 531–542. [DOI] [PubMed] [Google Scholar]
- 24.Ebert, D. H., Bischof, L. J., Streeper, R. S., Chapman, S. C., Svitek, C. A., Goldman, J. K., Mathews, C. E., Leiter, E. H., Hutton, J. C. & O'Brien, R. M. (1999) Diabetes 48, 543–551. [DOI] [PubMed] [Google Scholar]
- 25.Bischof, L. J., Martin, C. C., Svitek, C. A., Stadelmaier, B. T., Hornbuckle, L. A., Goldman, J. K., Oeser, J. K., Hutton, J. C. & O'Brien, R. M. (2001) Diabetes 50, 502–514. [DOI] [PubMed] [Google Scholar]
- 26.Martin, C. C., Bischof, L. J., Bergman, B., Hornbuckle, L. A., Hilliker, C., Frigeri, C., Wahl, D., Svitek, C. A., Wong, R., Goldman, J. K., et al. (2001) J. Biol. Chem. 276, 25197–25207. [DOI] [PubMed] [Google Scholar]
- 27.Amrani, A., Serra, P., Yamanouchi, J., Trudeau, J. D., Tan, R., Elliott, J. F. & Santamaria, P. (2001) J. Immunol. 167, 655–666. [DOI] [PubMed] [Google Scholar]
- 28.Guerau-de-Arellano, M. & Huber, B. T. (2002) Curr. Opin. Rheumatol. 14, 388–393. [DOI] [PubMed] [Google Scholar]
- 29.MacMurray, A. J., Moralejo, D. H., Kwitek, A. E., Rutledge, E. A., Van Yserloo, B., Gohlke, P., Speros, S. J., Snyder, B., Schaefer, J., Bieg, S., et al. (2002) Genome Res. 12, 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halbout, P., Briand, J. P., Becourt, C., Muller, S. & Boitard, C. (2002) J. Immunol. 169, 2436–2443. [DOI] [PubMed] [Google Scholar]