Abstract
The genetic code is thought to have developed from an early system of RNA-dependent peptide synthesis. To investigate one kind of template-like peptide synthesis that might emerge from an RNA world, we constructed highly reactive aminoacyl phosphate oligonucleotides as adaptors that bound to RNA guide sequences. The reactive aminoacyl groups mimic a chemistry found in modern protein biosynthesis. Guide sequence interactions with adaptors were borrowed in part from universal contacts seen between tRNAs and rRNA. With these constructions, di- and tripeptides formed in a single guide sequence-dependent reaction. The order of amino acids was not random but directional in a way consistent with substrate reactivities. No ribosomes or ribozymes were required. Thus, aminoacyl phosphate adaptors and RNA guides could, in principle, have been intermediates in the transition from the RNA world to modern template-dependent protein synthesis.
Development of modern peptide biosynthesis with RNA templates and aminoacyl-RNA esters in the form of tRNA adaptors was a critical step in the transition from the putative RNA world to the theater of proteins (1). To better understand this development, early work focused on self-condensation reactions with high concentrations of mononucleoside or oligonucleotide aminoacyl esters in reactions that ran for days (2–5). Here we introduced the use of highly reactive aminoacyl phosphate oligonucleotide adaptors and template-like RNA guide sequences. The use of aminoacyl phosphate oligonucleotides was inspired by contemporary systems that use aminoacyl phosphate (mononucleotide) adenylates as intermediates for aminoacyl adaptor (tRNA) synthesis.
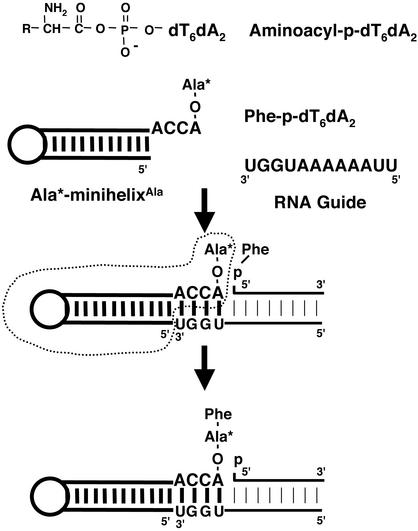
The free energy of hydrolysis of aminoacyl phosphates is ≈3 kcal·mol–1 (1 cal = 4.184 J) greater than that of the aminoacyl esters (aminoacyl-tRNAs) (6) that donate amino acids to growing polypeptide chains in modern template-dependent protein biosynthesis. The aminoacyl esters are formed by transfer of the aminoacyl moieties from aminoacyl phosphates (in the form of aminoacyl adenylates) to the 3′ ends of adaptors (tRNAs) in reactions catalyzed by aminoacyl-tRNA synthetases (7–9). With this transfer reaction in mind, we envisioned a scenario where the ester was eliminated and the high energy of the aminoacyl phosphate was captured by the adaptor itself. For this purpose, we designed a system of three components (Fig. 1). These were an aminoacyl minihelix, an aminoacyl phosphate oligonucleotide, and an RNA guide sequence. The idea was that, with the proper design of each of these three components, peptide synthesis might occur simply on mixing them together.
Fig. 1.
Structure of the aminoacyl-phosphate nucleotide used in this study and the reaction scheme for dipeptide synthesis. [14C]Ala-minihelix (enclosed by dotted lines), template oligonucleotides (guide), and 5′-Phe-p-dT6dA2 were mixed together.
Materials and Methods
Synthesis and Preparation of Substrates. MinihelixAla and the template oligoribonucleotides were synthesized on an Expedite 8909 synthesizer (PE Biosystems). Oligodeoxyribonucleotides with a phosphate group at the 5′ position were synthesized by Invitrogen. MinihelixAla and the template oligoribonucleotides were deprotected by using the methods of Wincott et al. (10). Escherichia coli tRNAPhe transcript was produced by a published method (11). All molecules were purified by denaturing PAGE. Aminoacylation of minihelixAla and tRNAPhe were performed by a fragment of E. coli alanyl-tRNA synthetase (AlaRS) (12) and E. coli crude aminoacyl-tRNA synthetases (Sigma), respectively. Aminoacyl-p-oligodeoxyribonucleotides were synthesized and purified by a published procedure with slight modifications (13, 14).
Dipeptide Formation. Dipeptide formation reactions were performed under the following conditions. Reaction mixtures contained (i) 50 mM potassium phosphate (pH 7.5), 1,000 mM NaCl, 10 mM MgCl2, 10 μM [14C]Ala-minihelixAla, 10 μM template (5′U2A6UGGU3′ or 5′U2A6CCCC3′), and 10 μM 5′-Phe-p-dT6dA2; (ii) 50 mM potassium phosphate (pH 7.5), 1,000 mM NaCl, 10 mM MgCl2, 10 μM [14C]Ala-minihelixAla, 10 μM template (5′AUGGU3′), and 20 μM 5′-Phe-p-dT; and (iii) 50 mM potassium phosphate (pH 7.5), 1,000 mM NaCl, 10 mM MgCl2, 10 μM Phe-tRNAPhe, 10 μM template (5′U2A6UGGU3′), and 10 μM 5′-[14C]Ala-p-dT6dA2. After incubation at 0°C for 1 h, the reaction mixture was treated with 100 mM KOH at 37°C for 12 h to liberate dipeptide. After treatment, Phe-Ala (1 mM) and Ala-Phe (1 mM) were added to the reaction mixture and resolved by TLC using Silica Gel 60 F254 plates (EM Science). The products were eluted with chloroform/methanol/acetic acid/water (50:30:10:5) (vol/vol). After the visualization of the nonradioactive standard materials (Phe-Ala, Ala-Phe) with ninhydrin, the products were analyzed on a PhosphorImager screen (LE177-906, Molecular Dynamics). Dipeptide formation was also monitored by measuring the trichloroacetic acid-precipitable radioactivity generated by the generation of [14C]Ala-Phe-tRNAPhe (15, 16).
Tripeptide Formation. Tripeptide formation reactions were performed under the following conditions. The reaction mixture contained 50 mM potassium phosphate (pH 7.5), 1,000 mM NaCl, 10 mM MgCl2, 20 μM [14C]Phe-tRNAPhe, 200 μM template (5′U2A6UGGU3′), 100 μM 5′-Phe-p-dT6dA2, and 100 μM 5′-Phe-p-dT5dA2. After incubation at 0°C for 8 h, the reaction mixture was treated with 100 mM KOH at 37°C for 12 h to liberate dipeptide and any tripeptide that might be formed. After the treatment, 1 mM Phe-Phe and Phe-Phe-Phe were added to the reaction mixture and resolved by TLC using Silica Gel 60 F254 plates (EM Science). The products were eluted with 1-butanol/water/acetic acid (10:1:1) (vol/vol). After the visualization of the cold standard materials (Phe-Phe and Phe-Phe-Phe) with ninhydrin, the results were analyzed by phosphorimaging.
MS. In the experiments analyzed by MS, nonradioactive materials were used. Oasis HLB cartridges (Waters) were used to purify the product or remove salts in the reaction mixture. The final product was characterized by electron spray MS (API 100, MDS Sciex, Concord, Canada).
Results
Reaction Design. The minihelix was borrowed from the organization (in three dimensions) of the tRNA cloverleaf into an L-shaped structure with two domains (17–19). It is a 12-bp stem–loop domain that terminates at the 3′ end in the universal CCA trinucleotide sequence, with the terminal adenosine being the site of aminoacylation (17). [The minihelix portion of tRNA binds to the peptidyltransferase center of the 23S rRNA in the 50S ribosome particle (12, 20) and is a substrate for specific aminoacylation by many aminoacyl-tRNA synthetases (21–23).] [14C]Ala-minihelixAla was prepared by direct aminoacylation with E. coli alanyl-tRNA synthetase (AlaRS) (12). For the acyl phosphate oligonucleotide, phenylalanine was coupled (with 1,3-dicyclohexylcarbodiimide) through a mixed phosphoanhydride linkage to the 5′-phosphate of a dT6dA2 oligonucleotide to yield Phe-p-dT6dA2 (13, 14). [Deoxynucleotides were used for technical convenience, in particular to avoid byproducts in the synthesis. However, the RNA–DNA hybrid formed with the guide sequence is expected to have the geometry of an RNA A-form helix (24).] The RNA guide sequence was designed to be complementary to both the ACCA of the minihelix and the Phe-bearing aminoacyl phosphate oligonucleotide.
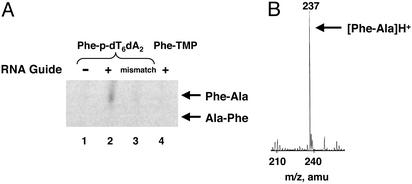
Dipeptide Formation. Incubation of 10 μM each of [14C]AlaminihelixAla, guide sequence, and 5′-Phe-p-oligonucleotide produced dipeptide after a 1-h reaction at pH 7.5, 0°C. [These concentrations are up to 2,000-fold lower than those used in the self-condensation studies with aminoacyl mononucleoside esters, where reaction times extended as long as 17 days (2, 3). A low temperature was used in the present studies to reduce hydrolysis of the aminoacyl phosphate (half-life of ≈1 h) and enhance base pairing.] By using the whole reaction mixture, which contained both oligonucleotides, the dipeptidyl residue was liberated by treatment with KOH and then was analyzed by TLC (Fig. 2A). The dipeptide was first identified by comparing the positions of ninhydrin-stained unlabeled standards (Phe-Ala and Ala-Phe) that were mixed into the reaction sample just before application to the TLC plate. Because the positions of Phe-Ala and Ala-Phe are different in the solvents used, the direction of dipeptide formation can be determined. Comparison of the phosphor image of the TLC plate (derived originally from [14C]Ala attached to the 3′-terminal adenosine of minihelixAla) with the ninhydrin-stained spots showed that the product was Phe-Ala, and not Ala-Phe. This result is consistent with an aminoacyl phosphate compound being higher in energy than the aminoacyl ester linkage, so that attack (on the activated carbonyl carbon of Phe) of the amino group of the aminoacyl moiety of Ala would strongly favor formation of Phe-Ala over Ala-Phe. (Thus, synthesis proceeds from the C to the N terminus because of the thermodynamics.) The yield of dipeptide was ≈7%. [This yield is considerably more than that seen with oligonucleotide ester-based reactions run at higher concentrations (5). In addition, in a preliminary experiment, we constructed a 5′-Phe-o-dT7ddT, 5′ ester-linked Phe-octamer, and complementary guide sequence with the alanyl minihelix. No dipeptide synthesis was seen under the same conditions used with the aminoacyl phosphate system described above (unpublished data).] MS of the product (with nonradioactive materials) gave a peak at 237 Da, the expected mass of protonated Phe-Ala (Fig. 2B).
Fig. 2.
Dipeptide formation. (A) TLC analysis of reaction products. Lanes 1 and 2, reaction of [14C]Ala-minihelix with 5′-Phe-p-dT6dA2 in the absence (lane 1) or presence (lane 2) of template oligonucleotide (5′U2A6UGGU3′). Lane 3, a “mismatched” template oligonucleotide (5′U2A6CCCC3′) was used. Lane 4, reaction of [14C]Ala-minihelix with 5′-Phe-p-dT in the presence of template oligonucleotide (5′AUGGU3′). (B) MS of reaction products produced from Ala-minihelix, template oligonucleotides, and 5′-Phe-p-oligonucleotide (A, lane 2). The peak at 237 mass units corresponds to Phe-Ala.
In contrast to earlier studies with glycyl mononucleoside esters (3), where the cyclic diketopiperazines were the predominant product, the noncyclic dipeptide formed readily in our system. Further work suggested that steric conflicts associated with bulkier side chains (e.g., Phe) and not associated with the glycyl residue inhibited diketopiperazine formation in our studies (unpublished data).
RNA Guide Sequence Dependence and Direction of Synthesis. Formation of dipeptide depended on the guide sequence: absence of the guide or the use of a guide with a noncomplementary sequence (CCCC replacing UGGU at the 3′ end of the guide sequence) yielded no dipeptide (Fig. 2 A). As a further control, we replaced Phe-p-dT6dA2 with Phe-p-dT and 5′U2A6UGGU3′ with 5′AUGGU3′, respectively, to evaluate the importance of the interaction between the aminoacyl phosphate adaptor and RNA guide (and to test for the possibility of the high energy aminoacyl phosphate reacting directly with the charged minihelix). However, no dipeptide was formed, consistent with a direct interaction between the guide and adaptor being needed for peptide synthesis at the low concentrations (10 μM) being used (Fig. 2 A).
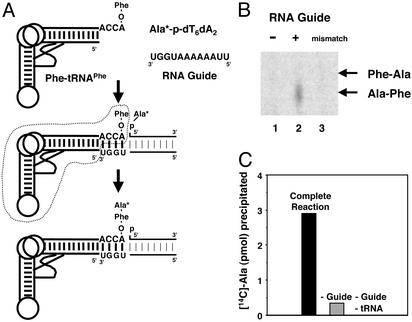
The selective formation of Phe-Ala over Ala-Phe in the experiment shown above might be caused by steric clashes of the bulky side chain of Phe. To clarify whether sequence-dependent steric effects govern formation of the dipeptide, the amino acids were reversed on the two reacting aminoacyl species. For this purpose, Phe was esterified to tRNAPhe by using phenylalanyl-tRNA synthetase (PheRS) in a standard aminoacylation reaction. (We used tRNAPhe instead of a minihelix because of the poor efficiency of aminoacylation of the latter with PheRS.) [14C]Ala was chemically joined to the 5′ phosphate of the dT6dA2 oligonucleotide to give [14C]Ala-p-dT6dA2 (Fig. 3A). TLC analysis of the reaction mixture (after the liberation of the peptide from tRNAPhe) produced Ala-Phe and not Phe-Ala. As expected, this dipeptide has the sequence opposite to the one obtained when the positions of the amino acids on the reacting substrates were reversed (Fig. 3B). (Thus, the reaction was governed by straightforward thermodynamic principles, as expected.) Here again, appearance of dipeptide depends on the RNA guide sequence and does not occur when the guide is mismatched.
Fig. 3.
Dipeptide formation with a “reversed” combination of two reacting aminoacyl species. (A and B) Reaction scheme for dipeptide synthesis using Phe-tRNAPhe, template oligonucleotides, and 5′-[14C]Ala-p-oligonucleotide (A) and TLC analysis of reaction products produced from mixing Phe-tRNAPhe and 5′-[14C]Ala-p-oligonucleotide without (lane 1) and with (lane 2) the template oligonucleotide (5′U2A6UGGU3′) (B). (B) A mismatched template oligonucleotide (5′U2A6CCCC3′) was used for lane 3. (C) Trichloroacetic acid precipitation of nascently formed dipeptide. After a 2-h incubation, three different reaction mixtures were precipitated with trichloroacetic acid. Complete reaction: [14C]Ala-p-dT6dA2, Phe-tRNAPhe, and template oligonucleotide (5′U2A6UGGU3′). –Guide: [14C]-Ala-p-dT6dA2 and Phe-tRNAPhe. –Guide, –tRNA: [14C]Ala-p-dT6dA2. Background (≈1 pmol) from the partial precipitation of [14C]Ala-p-dT6dA2 alone was subtracted in each case.
For confirmatory evidence that the dipeptide was formed on tRNAPhe, as expected from the directionality of synthesis, we took advantage of the ability to precipitate tRNA with trichloroacetic acid quantitatively (15, 16). By using this assay we could measure the incorporation of [14C]Ala from [14C]Ala-p-dT6dA2 into Phe-tRNAPhe. Indeed, [14C]Ala was only precipitated after 2 h when all three components ([14C]Ala-p-dT6dA2, Phe-tRNAPhe, and guide sequence) were present simultaneously. Omission of Phe-tRNAPhe and the guide sequence, or of the guide sequence alone, eliminated the formation of precipitable [14C]Ala (Fig. 3C). The yield of product by this method was similar to that determined from TLC analysis of product (see above).
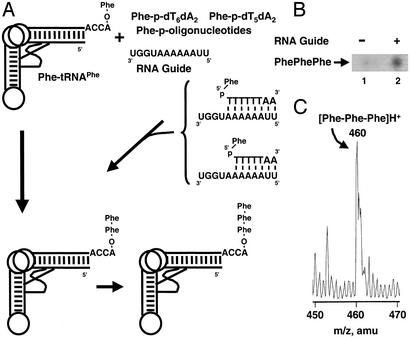
Guide Sequence-Dependent Tripeptide Synthesis. Because Watson–Crick base pairing between the guide sequence and the two reacting aminoacyl species is a dynamic equilibrium, a continuous reaction should in principle generate dipeptidyl-tRNA that could be extended to tripeptidyl-tRNA and even higher-order oligomers by further reactions with the aminoacyl phosphate oligonucleotide. To test this possibility, we raised the concentrations (20 μM Phe-tRNAPhe, 200 μM RNA guide sequence, 100 μM 5′-Phe-p-dT6dA2, and 100 μM 5′-Phe-p-dT5dA2). [We used not only the 5′-Phe-p-dT6dA2 octamer but also a 5′-Phe-p-dT5dA2 heptamer. Here the rationale was that reactivity might be enhanced by deleting one nucleotide so that a single-stranded base (on the guide RNA) creates a flexible “joint” at the junction of the two reacting substrates. Also, we used only Phe because of the high chromatographic resolution of (Phe)x oligomers (Fig. 4A).] The phosphor image of the TLC plate (derived originally from [14C]Phe attached to the 3′ terminal adenosine of tRNAPhe) suggested formation of not only Phe-Phe but also Phe-Phe-Phe (Fig. 4B). [As anticipated, the amount of tripeptide was less (<1%) than that of dipeptide. The formation of putative Phe-Phe-Phe depended on the presence of the guide sequence (Fig. 4B).] MS of the product (with nonradioactive materials) gave a peak at 460 Da, which is the expected mass of the protonated form of Phe-Phe-Phe (Fig. 4C).
Fig. 4.
Tripeptide formation. (A) Reaction for tripeptide synthesis. Phe-tRNAPhe, template oligonucleotide, and a mixture of 5′-Phe-p-oligonucleotides (5′-Phe-p-dT6dA2 and 5′-Phe-p-dT5dA2) were incubated. (B) Chromatogram showing the guide sequence-dependent appearance of tripeptide. (C) MS analysis of reaction products produced in A. The peak at 460 mass units corresponds to the protonated form of Phe-Phe-Phe.
Discussion
The results demonstrate the efficacy of aminoacyl phosphate oligonucleotide adaptors in template-like synthesis of di- and tripeptides. [An earlier attempt used aminoacyl phosphate mononucleotides in the absence of a template, with concentrations on the order of 100- to 1,000-fold higher than those used here (25).] Relatively low concentrations of the adaptors and minihelices and tRNAs were used here, with reaction times as short as 1 h. In principle, the reactions could be developed for even higher-order peptides, especially if sequestered in environments (such as those that reduce the effective concentration of water) where hydrolysis of the aminoacyl phosphate is attenuated. Indeed, because the half-lives of the aminoacyl phosphate used here are on the order of an hour, sequestration of the aminoacyl phosphate would be critical for development of high yields and longer chains. In contemporary systems, the same (as used here) high-energy 5′-aminoacyl phosphate linkage is formed in the first step of protein synthesis as an aminoacyl adenylate (7–9). The adenylate is shielded from the solvent by a tight binding pocket on the aminoacyl-tRNA synthetase (26). Plausibly, aminoacyl phosphate oligonucleotides were intermediates in the development of protein synthesis and later were replaced by the aminoacyl mononucleotide. [A ribozyme that catalyzes formation of an aminoacyl phosphate intermediate has been described (27).] Thus, in making the transition to the theater of proteins (1), a 5′-aminoacyl phosphate oligonucleotide could later have been sequestered as a 5′-aminoacyl mononucleotide (e.g., aminoacyl adenylate).
Modern protein biosynthesis reiteratively uses the attack of the amino group (of aminoacyl-tRNA adaptor) on the acylcarbon of the growing peptidyl-tRNA. In contrast, in the system described here, to grow long chains by reiterative synthesis the amino group of the peptidyl-tRNA must attack the acyl-carbon of the aminoacyl adaptor (phosphate oligonucleotide). With both systems, however, the polypeptide grows on the tRNA as peptidyl-tRNA.
The success of these experiments was based in part on the presence of a guide sequence. Without the guide sequence, no synthesis was observed even in the presence of an aminoacyl phosphate oligonucleotide. Thus, the guide sequence enabled the reactions to proceed at far lower concentrations than otherwise possible. The guide sequence used here interacts with the universal CCA end of tRNA. This same principle is used in the modern ribosome, where the CCA end interacts with specific nucleotides in 23S rRNA (28–30).
Acknowledgments
We thank A. Eschenmoser, L. Orgel, M. Sprinzl, D. Usher, T. Hasegawa, S. Akashi, K. Kamaike, and T. Kushiro for helpful discussions, H. Asahara for the kind gift of the E. coli tRNAPhe gene, and The Scripps Center for Mass Spectrometry for electron spray MS measurements. We also thank L. Orgel for specific comments on the manuscript. This work was supported by National Institutes of Health Grant GM15539 and a fellowship from the National Foundation for Cancer Research. K.T. was supported by a research fellowship from the Uehara Memorial Foundation.
References
- 1.Rich, A. (1962) Horizons in Biochemistry (Academic, New York), pp. 103–126.
- 2.Chung, S. K., Copsey, D. B. & Scott, A. I. (1978) Bioorg. Chem. 7, 303–312. [Google Scholar]
- 3.Weber, A. L. & Orgel, L. E. (1980) J. Mol. Evol. 16, 1–10. [DOI] [PubMed] [Google Scholar]
- 4.Usher, D. A., Profy, A. T., Walstrum, S. A., Needels, M. C., Bulack, S. C. & Lo, K. M. (1984) Origins Life 14, 621–628. [DOI] [PubMed] [Google Scholar]
- 5.Tamura, K. & Schimmel, P. (2001) Proc. Natl. Acad. Sci. USA 98, 1393–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter, F. H. (1960) J. Am. Chem. Soc. 82, 1111–1122. [Google Scholar]
- 7.Berg, P. (1961) Annu. Rev. Biochem. 30, 293–324. [Google Scholar]
- 8.Schimmel, P. (1987) Annu. Rev. Biochem. 56, 125–158. [DOI] [PubMed] [Google Scholar]
- 9.Giegé, R., Puglisi, J. D. & Florentz, C. (1993) Prog. Nucleic Acid Res. Mol. Biol. 45, 129–206. [DOI] [PubMed] [Google Scholar]
- 10.Wincott, F., DiRenzo, A., Shaffer, C., Grimm, S., Tracz, D., Workman, C., Sweedler, D., Gonzalez, C., Scaringe, S. & Usman, N. (1995) Nucleic Acids Res. 23, 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson, J. R. & Uhlenbeck, O. C. (1988) Proc. Natl. Acad. Sci. USA 85, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sardesai, N. Y., Green, R. & Schimmel, P. (1999) Biochemistry 38, 12080–12088. [DOI] [PubMed] [Google Scholar]
- 13.Berg, P. (1958) J. Biol. Chem. 233, 608–611. [PubMed] [Google Scholar]
- 14.Illangasekare, M., Sanchez, G., Nickles, T. & Yarus, M. (1995) Science 267, 643–647. [DOI] [PubMed] [Google Scholar]
- 15.Barnett, W. E. & Jacobson, K. B. (1964) Proc. Natl. Acad. Sci. USA 51, 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura, S. & Novelli, G. D. (1964) Biochim. Biophys. Acta 80, 574–586. [DOI] [PubMed] [Google Scholar]
- 17.Schimmel, P., Giegé, R., Moras, D. & Yokoyama, S. (1993) Proc. Natl. Acad. Sci. USA 90, 8763–8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertus, J. D., Ladner, J. E., Finch, J. T., Rhodes, D., Brown, R. S., Clark, B. F. & Klug, A. (1974) Nature 250, 546–551. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. H., Suddath, F. L., Quigley, G. J., McPherson, A., Sussman, J. L., Wang, A. H., Seeman, N. C. & Rich, A. (1974) Science 185, 435–440. [DOI] [PubMed] [Google Scholar]
- 20.Noller, H. F. (1993) The RNA World (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 137–156.
- 21.Frugier, M., Florentz, C. & Giegé, R. (1994) EMBO J. 13, 2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinis, S. A. & Schimmel, P. (1997) tRNA: Structure, Biosynthesis, and Function (Am. Soc. Microbiol., Washington, DC), pp. 349–370.
- 23.Musier-Forsyth, K. & Schimmel, P. (1999) Acc. Chem. Res. 32, 368–375. [Google Scholar]
- 24.Arnott, S., Chandrasekaran, R., Millane, R. P. & Park, H.-S. (1986) J. Mol. Biol. 188, 631–640. [DOI] [PubMed] [Google Scholar]
- 25.Paecht-Horowitz, M., Berger, J. & Katchalsky, A. (1970) Nature 228, 636–639. [DOI] [PubMed] [Google Scholar]
- 26.Schimmel, P. R. & Söll, D. (1979) Annu. Rev. Biochem. 48, 601–648. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, R. K. & Yarus, M. (2001) Biochemistry 40, 6998–7004. [DOI] [PubMed] [Google Scholar]
- 28.Samaha, R. R., Green, R. & Noller, H. F. (1995) Nature 377, 309–314. [DOI] [PubMed] [Google Scholar]
- 29.Green, R., Switzer, C. & Noller, H. F. (1998) Science 280, 286–289. [DOI] [PubMed] [Google Scholar]
- 30.Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920–930. [DOI] [PubMed] [Google Scholar]