Abstract
Protein biosynthesis on the ribosome requires accurate reading of the genetic code in mRNA. Two conformational rearrangements in the small ribosomal subunit, a closing of the head and body around the incoming tRNA and an RNA helical switch near the mRNA decoding site, have been proposed to select for complementary base-pairing between mRNA codons and tRNA anticodons. We determined x-ray crystal structures of the WT and a hyper-accurate variant of the Escherichia coli ribosome at resolutions of 10 and 9 Å, respectively, revealing that formation of the intact 70S ribosome from its two subunits closes the conformation of the head of the small subunit independent of mRNA decoding. Moreover, no change in the conformation of the switch helix is observed in two steps of tRNA discrimination. These 70S ribosome structures indicate that mRNA decoding is coupled primarily to movement of the small subunit body, consistent with previous proposals, whereas closing of the head and the helical switch may function in other steps of protein synthesis.
Selection of the correct aminoacyl-tRNA (aa-tRNA) during each cycle of polypeptide elongation on the ribosome requires binding of a ternary complex composed of elongation factor (EF)-Tu, aa-tRNA, and GTP to the ribosomal acceptor site (A site) (1, 2). When the small ribosomal subunit detects complementary base-pairing between the mRNA and cognate aa-tRNA in the A site, the large ribosomal subunit stimulates GTP hydrolysis by EF-Tu, which then releases the aa-tRNA. After accommodation of the aa-tRNA into the A site of the large subunit, peptide bond formation occurs. The initial selection of aa-tRNA and the subsequent proofreading (accommodation) step lead to a 1,000-fold preference for cognate tRNA over near-cognate tRNA in mRNA decoding (1). Although various components of the ribosome that promote translational fidelity have been identified (1, 3–13) (Fig. 1 a and b), many of which are conserved throughout evolution (14), the mechanism of mRNA decoding by the intact ribosome is not well understood.
Fig. 1.
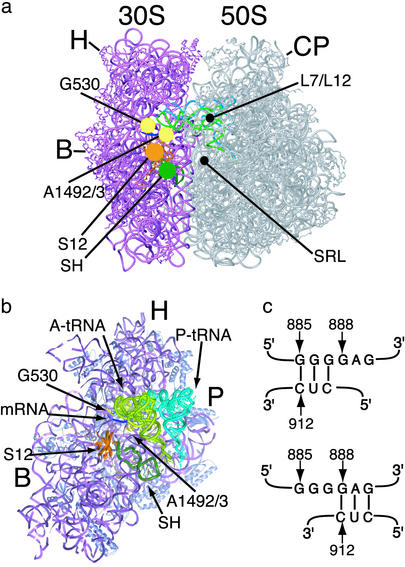
Components of the ribosome involved in mRNA decoding. (a) View of the ribosome from the A-site side. Elements of the small (30S) subunit and large (50S) subunit that contribute to binding of the anticodon of aa-tRNA and EF-Tu are marked: G530 and A1492/3, nucleotides in 16S rRNA; S12, protein S12; SH, switch helix; L7/L12, proteins L7/L12; SRL, sarcin-ricin loop in 23S rRNA. Other features of the ribosome are marked as follows: H, head of the small subunit; B, small subunit body; P, small subunit platform; CP, central protuberance of the large subunit. (b) View of the interface side of the small subunit, with the large subunit removed for clarity. Nucleotides G530, A1492, and A1493 (yellow) contact both the mRNA (blue) and A-site tRNA (light green). Protein S12 (orange) and the switch helix (dark green) are located adjacent to the decoding site. (c) Proposed switch in base-pairing in helix 27 of 16S rRNA. The 885 conformation (Upper) involves base-pairing between nucleotides 885–887 and 910–912. In the 888 conformation (Lower), nucleotides 888–890 base-pair with nucleotides 910–912.
Structures of the 30S subunit at atomic or near-atomic resolution have shown that the small subunit exists in “open” and “closed” conformations (10). In the closed conformation, the head and body of the 30S subunit enclose the A site-bound tRNA anticodon stem loop (Fig. 1a). This closure occurs only when cognate tRNA is bound to the A site or when the error-inducing antibiotic paromomycin is bound in the decoding site. Based on these structural observations, closure of the small subunit around the A-site tRNA has been proposed to stabilize cognate tRNA binding or drive the forward reactions of tRNA selection (2, 10). Mutations in protein S12 that lead to a hyper-accurate phenotype such as resistance to or dependence on the error-inducing antibiotic streptomycin (1) should disrupt contacts in the closed form of the small ribosomal subunit and may therefore favor the open conformation of the 30S subunit in the absence of the antibiotic (2, 10).
A second conformational change thought to modulate mRNA decoding involves a 3-bp helical switch in the small subunit that cycles between “hyper-accurate” and “error-prone” conformations (15). Mutations in 16S rRNA that preferentially stabilized one base-pairing configuration (888, or hyper-accurate, Fig. 1c) were shown to complement error-prone phenotypes such as those conferred by mutations in protein S5 in the small ribosomal subunit. In contrast, 16S rRNA mutations that favored the other base-pairing configuration (885, or error-prone, Fig. 1c) complemented mutations in protein S12 that result in a hyper-accurate phenotype (1, 15). Mutations in the ribosome that stabilize one or the other pairing in the switch helix induce large-scale conformational changes in the ribosome (16). However, mutations in the switch helix in yeast 18S rRNA lead to decoding phenotypes that may be inconsistent with the switching model (17). Furthermore, no change in the switch helix base-pairing has been observed in x-ray crystal structures of the 30S subunit or intact ribosome (8–10, 18).
The switching model predicts that the equilibrium between the two base-pairing conformations would be shifted by mutations in proteins S12 or S5 that lead to hyper-accurate or error-prone phenotypes, respectively (15). Mutations in protein S12 that confer the extremely hyper-accurate phenotype of streptomycin dependence should stabilize the hyper-accurate base-pairing of the switch helix in the absence of the antibiotic (15).
We determined x-ray crystal structures of the WT and hyper-accurate forms of the Escherichia coli 70S ribosome at resolutions of 10 and 9 Å, respectively, to test whether two conformational rearrangements in the small ribosomal (30S) subunit that are thought to play a role in tRNA selection (10, 15) occur in the intact ribosome as proposed. These 70S ribosome structures, along with a structure of the Thermus thermophilus ribosome (18) and chemical probing results, indicate that mRNA decoding is coupled primarily to movement of the small subunit body and not to closing of the small subunit head or to an RNA helical switch near the mRNA decoding center.
Materials and Methods
Ribosome Purification. Tight-coupled ribosomes from E. coli strain MRE600 and a strain of MRE600 that contains a mutation in protein S12 that confers streptomycin dependence (G92D) were purified as described (19, 20), except that 60 mM NH4Cl was used in the final sucrose gradient purification on a Beckman SW-28 rotor (20). Ribosomes from the streptomycin-dependent (smD) strain were extensively dialyzed during purification to remove streptomycin.
Ribosome Crystallization. Ribosomes and ribosome complexes with a short leaderless mRNA (5′-pAUGUUCAAACGUAAUAAU-3′) and deacylated E. coli tRNAfMet (Subriden RNA, Rolling Bay, WA) were crystallized at 4°C by the vapor diffusion method against 18–22% 2-methyl-2,4-pentanediol (MPD), 0–5% ethanol, 20–25 mM MgCl2, 200–320 mM NH4Cl, 1 mM spermine, 0–0.5 mM spermidine, pH 6.0–6.5. smD ribosomes were stabilized for cryo-cooling in the presence of higher concentrations of MPD, 30 μM leaderless mRNA, and 30 μM deacylated tRNAfMet. Diffraction data were measured at beam lines 19ID at the Advanced Photon Source at the Argonne National Laboratory (Argonne, IL) and at beam lines 8.3.1, 8.2.2, and 8.2.1 at the Advanced Light Source at the Lawrence Berkeley National Laboratory. Data reduction, molecular replacement, and rigid-body refinement were carried out with denzo/scalepack and cns (21, 22) (Table 1).
Table 1. Diffraction statistics for the E. coli 70S ribosome crystals.
|
Crystal form
|
||
|---|---|---|
| WT 70S | smD 70S | |
| Space group | 1422 | 1422 |
| Unit cell | a = b = 683.9 Å; | a = b = 682.4 Å; |
| c = 386.7 Å | c = 386.3 Å | |
| α = β = γ = 90° | α = β = γ = 90° | |
| Data resolution | 300-9.5 Å | 175-8.7 Å |
| (9.68-9.5 Å) | (8.9-8.7 Å) | |
| Completeness (%) | 92.1 (71.8) | 98.5 (99.2) |
| No. of reflections | 26,821 | 36,338 |
| Measurement redundancy | 25.1 (9.7) | 5.2 (5.2) |
| Mean signal to noise (l/σ) | 44.8 (2.5) | 9.0 (2.5) |
| Rsym (%) | 14.2 (54.9)* | 15.0 (63.5)* |
| Overall χ2 | 1.7 (1.2) | 1.0 (1.0) |
| Molecular replacement statistics | ||
| Resolution | 70-9.5 Å | 70-8.7 Å |
| R, % | 39.4 | 39.1 |
| Rfree, % | 41.1 | 41.2 |
Two data measurement passes were used: a high-resolution sweep followed by a low-resolution sweep. Thus the Rsym values in the low-resolution ranges and the overall Rsym value of the data are inflated.
The molecular replacement models consist of atomic-resolution structures of the bacterial 30S and 50S ribosomal subunits (8, 23), tRNAPhe as modeled in the T. thermophilus 70S ribosome structure (18), and homology modeling of additional components (8, 24, 25). In the rigid-body refinement of the resulting models, 5% of the measured diffraction amplitudes were set aside as a test set for calculation of Rfree values (22). The same 5% were set aside for refinement of the WT and smD ribosome structures. Details of the molecular replacement and refinement will be published elsewhere. Coordinates for the structures have been deposited in the Protein Data Bank.
Calculation of Difference Electron Density Maps. Difference electron density maps comparing the smD to the WT E. coli ribosome structures used experimental amplitudes from both crystal forms (Table 1) and filtered phases from the WT structure model (22, 26) and were calculated at a resolution of 9.5 Å. Difference electron density maps comparing the smD ribosome to ribosome models lacking mRNA and tRNA in the P site were calculated at a resolution of 8.7 Å by using observed diffraction amplitudes and calculated amplitudes and phases: (Fobs,smD – Fcalc,smD,unliganded)expiα,smD,unliganded. Difference electron density maps comparing the ribosome to models refined in the presence (+sm) or absence (–sm) of streptomycin (8) were calculated at a resolution of 8.7 Å by using observed diffraction amplitudes and calculated amplitudes and phases: (Fobs,smD – Fcalc,smD,+sm) expiα,smD,+sm, and (Fobs,smD – Fcalc,smD,–sm) expiα,smD,–sm. Models containing or lacking streptomycin were also refined against the WT E. coli ribosome diffraction amplitudes. Calculation of the corresponding difference electron density maps at a resolution of 9.5 Å yielded similar results to those with the smD diffraction amplitudes (data not shown).
Superposition of 30S Subunit Models. A subset of phosphorus atoms from the different 30S subunit models was chosen based on their limited conformational variability in the open and closed conformations of the 30S subunit structures, as described (10). These phosphorus atoms, 60 in number in comparisons to the E. coli ribosome and 52 in number in comparisons to the T. thermophilus ribosome, were superimposed by using the program o (27). The rms deviation in phosphorus positions between 30S models ranged from 0.7 Å (E. coli comparisons) to 1.6 Å (T. thermophilus comparisons). The T. thermophilus ribosome model was built manually (18) and contains some sequence register shifts, which might explain the higher rms deviation in phosphorus superpositions when using this model. The representative comparisons in Fig. 2 used structures of the small subunit from Protein Data Bank ID codes 1N34 (open 30S conformation), 1IBL (closed 30S conformation), and IGIX (T. thermophilus 70S ribosome). Sequence register shifts in Protein Data Bank model 1GIX were taken into account in Fig. 2. In all of the figures, phosphates in the backbone of the spur region were removed, because the spur is involved in significant crystal contacts in the 30S subunit structures (8). All figures were made by using the programs ribbons (28) and pymol (29).
Fig. 2.
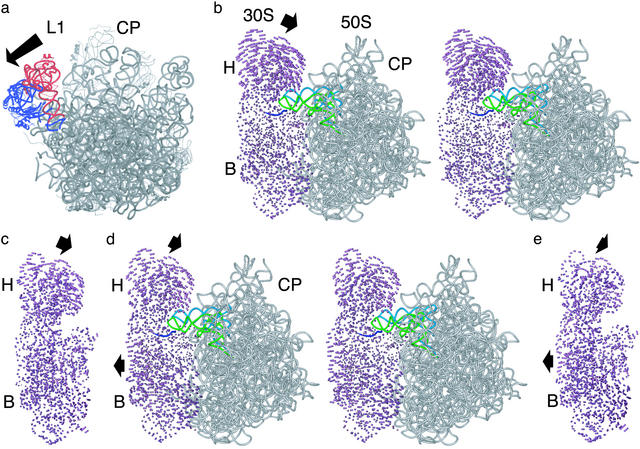
Superpositions of ribosomal subunit structures. (a) Position of the L1 stalk in the smD E. coli ribosome structure (blue) compared with that in the T. thermophilus 70S ribosome (red). The 50S subunit is shown from the 30S interface side. The arrow indicates the direction of motion required to move from the closed to open position of the L1 stalk. (b) Stereoview comparing the open 30S subunit conformation to the small subunit in the E. coli smD ribosome. Difference vectors between all conserved phosphorus atoms in the small subunit are shown. Arrows indicate the direction of movement in going from the open conformation to that in the intact ribosome. The large subunit (gray), mRNA (blue), A-site tRNA (light green), and P-site tRNA (light blue) bound to the E. coli smD ribosome are shown for reference. Other components of the ribosome are marked as in Fig. 1. (c) Comparison of the open 30S subunit conformation to the small subunit in the T. thermophilus 70S ribosome structure. Difference vectors are shown as described above. The orientation is the same as in b.(d) Stereoview comparing the closed 30S subunit conformation to the small subunit in the E. coli smD ribosome. (e) Comparison of the closed 30S subunit conformation to the small subunit in the T. thermophilus 70S ribosome structure. The orientation is the same as in d.
Chemical Probing. Chemical probing and primer extension were carried out essentially as described (4). WT or smD ribosomes were programmed with mRNA and deacylated tRNAfMet in the P site, and Phe-tRNAPhe was bound to the A site by means of EF-Tu ternary complexes containing GTP or the nonhydrolyzable analogue GMPPNP. The positioning of the A-site tRNA was confirmed by examination of protections from chemical probing (4, 30). Ribosome complexes were formed at 37°C in 6 mM MgCl2, 100 mM NH4Cl, and 20 mM Tris, pH 7.5, and chemical probing was carried out at the same temperature. Primers were labeled on the 5′ end with [33P]phosphate.
Results
Structural Comparison of 30S Subunits with 70S Ribosomes. To test whether smD mutations in the ribosome favor the open form of the small subunit, we examined the conformation of the 30S subunit in three x-ray crystal structures of the 70S bacterial ribosome: a 10-Å resolution structure of the WT E. coli ribosome, a 9-Å resolution structure of a smD form of the E. coli ribosome (Table 1), and the 5.5-Å resolution structure of the T. thermophilus ribosome (18). The WT ribosome from E. coli was crystallized in the absence of ligands. Crystals of smD ribosomes were obtained as a ribosome complex with a short mRNA and deacylated tRNAfMet bound to the P site. Crystals stabilized in the presence of 30 μM deacylated tRNAfMet gave a low level of noncognate tRNA binding to the A site. Binding of deacylated tRNAfMet to the A site of smD ribosomes was observed to be ≈50%. For comparison, WT T. thermophilus ribosomes bound deacylated tRNAfMet in the A site to a similar extent when stabilized in the presence of just 1 μM tRNAfMet (31), concentrations at which noncognate tRNA binding is not observed in the smD E. coli ribosome crystals (data not shown). Thus, the low level of noncognate tRNA bound in the A site is consistent with the hyper-accurate nature of the smD ribosomes. The positions of the A-site and P-site tRNAs in the smD E. coli ribosome structure are indistinguishable, at a resolution of 9 Å, from the positions of A-site and P-site tRNAs in the structure of the T. thermophilus ribosome (18) and the positions of tRNA fragments poised for the peptidyl transfer reaction in the atomic-resolution structure of the 50S subunit (32).
Taken together, these observations suggest that the smD E. coli ribosome structure contains a correctly positioned P-site tRNA and the accommodated state of A-site tRNA on the ribosome (1), similar to the 7-Å structure of the T. thermophilus 70S ribosome and the 30S ribosomal subunit structures (10, 18). In contrast to the T. thermophilus ribosome structures (18), the L1 stalk in the E. coli ribosome structures is rotated away from the central protuberance of the large subunit, similar to the conformation seen in the structure of the Deinococcus radiodurans 50S subunit (23) (Fig. 2a).
The small subunits from the intact ribosome structures were superimposed with either the open or closed conformations of the isolated 30S subunit by using a subset of phosphorus atoms previously shown to be constant in their relative position in the two states (10). In all three structures of the intact ribosome, the head of the small subunit tilts 5–8 Å toward the central protuberance of the large subunit when compared with the position of the head in the open subunit conformation (Fig. 2 b–e). The tilting is in the same direction as seen in the closing of the isolated 30S subunit structure (9, 10), yet is greater in magnitude. Thus in the intact ribosome, the small subunit adopts a more closed conformation than observed in any of the isolated 30S subunit structures (9, 10). In contrast, the body of the small subunit in all three 70S ribosome structures exists in an open conformation, similar to that observed in the open conformation of isolated 30S subunits (Fig. 2 b–e). Comparisons of the small subunits within the E. coli 70S ribosome structures do not reveal any significant differences at the present resolution (data not shown).
Conformation of the Switch Helix. The molecular replacement models used to solve the x-ray crystal structures of the WT and smD E. coli ribosomes presented here (Table 1) all contain the 30S subunit with the switch helix in its error-prone conformation, as observed in all of the previous x-ray crystal structures of the 30S subunit and 70S ribosome (8–10, 18). We therefore examined difference electron density maps to determine whether the switch helix adopts a different conformation in the smD ribosome, as predicted by the switch hypothesis.
Difference electron density maps comparing the WT and smD ribosome structures show positive electron density indicating the presence of tRNAs bound to the A and P sites of the smD ribosome (Fig. 3a) but do not reveal any differences between WT and smD ribosomes in the switch helix region (Fig. 3b). A difference electron density map calculated from a model of the smD ribosome refined without including mRNA and tRNAs reveals sharper features of the mRNA and P-site tRNA, including single-stranded stretches in the mRNA and P-site tRNA 3′ end, yet does not contain any differences in the switch helix region relative to the input model (Fig. 3 c and d). Thus, both the WT and smD E. coli ribosome x-ray crystal structures contain the switch helix in its error-prone base-pairing configuration.
Fig. 3.
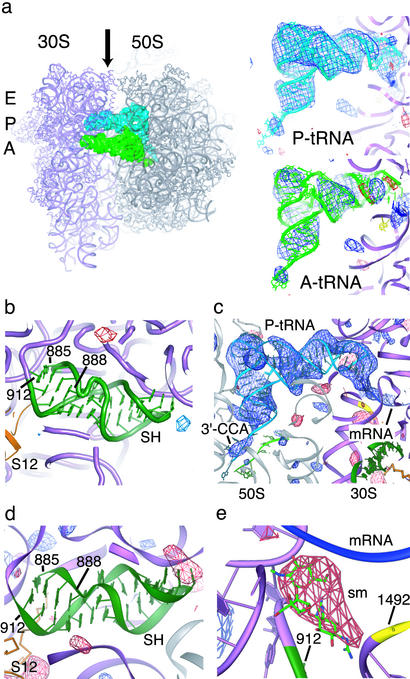
Conformation of the 70S ribosome in the switch helix region. (a) Difference electron density map comparing WT and smD 70S ribosomes from E. coli. The difference map was calculated by using observed diffraction amplitudes from both crystal forms, as described in Materials and Methods. The smD ribosomes contain mRNA, P-site tRNAfMet (P-tRNA, Upper Right) and noncognate A-site tRNAfMet at 50% occupancy (A-tRNA, Lower Right). (Left) A top view of the ribosome is shown with the A site, P site, and E site indicated to the left of the small subunit. The tRNAs are viewed from the perspective of the E site, as indicated by the arrow. The color scheme of the ribosome complex is the same as in Fig. 1. (b) The same difference electron density map in the switch helix region. (c) Difference electron density map comparing the smD 70S ribosome complex to a model lacking ligands throughout refinement. The positive electron density corresponds to P-site tRNA and mRNA bound to the ribosome. The perspective is the same as in a. (d) The same difference electron density map in the switch helix region. (e) Difference electron density map comparing the smD 70S ribosome complex to a ribosome model that contained streptomycin throughout refinement. The streptomycin binding pocket (sm) is shown. Positive electron density (blue, left) appeared after refinements of the model both in the absence and presence of the antibiotic, whereas negative density appeared only when streptomycin was included in the model. In all panels, difference electron density is contoured at 3.0 and –3.0 SDs from the mean (blue and red, respectively).
We confirmed that streptomycin is not bound to the smD ribosome in the crystal structure by adding or removing streptomycin from the molecular replacement model. Difference electron density maps generated by using a model that included streptomycin throughout the structure refinement show clear negative electron density in the streptomycin binding pocket (8). In contrast, models that did not include the antibiotic show no discernable difference electron density in the streptomycin binding site (Fig. 3e). The absence of streptomycin from the smD ribosome preparations was verified by chemical probing experiments that revealed changes in base protection patterns consistent with the smD mutation and the absence of the antibiotic (data not shown) (5, 33). P-site tRNA binding experiments confirmed the smD phenotype and showed decreased P-site tRNA binding in the presence of streptomycin (data not shown) (34). The fact that a small molecule like streptomycin can be distinguished in difference electron density maps of the ribosome at 9- to 10-Å resolution further supports the conclusion that the E. coli ribosome contains the switch helix in its error-prone base-pairing configuration in the x-ray crystal structures.
To rule out the possibility that crystal packing influenced the conformation of the switch helix, we used chemical probing with kethoxal and dimethyl sulfate (DMS) to determine the base-pairing of the switch helix in various steps of mRNA decoding. In addition to unliganded ribosomes and ribosomes in a complex with mRNA and P-site tRNA, we probed ribosome complexes stopped at the codon recognition step of decoding and ribosomes with accommodated A-site tRNA (1, 4, 30). In all of these states, the switch helix was observed to be in the error-prone base-pairing configuration (Fig. 4). Although it is possible that helical switching might occur transiently after GTP hydrolysis by EF-Tu, but before aa-tRNA accommodation into the A site, the helix would then have to switch twice during the process, from error-prone to hyper-accurate and then back to error-prone, to be consistent with the chemical probing results.
Fig. 4.
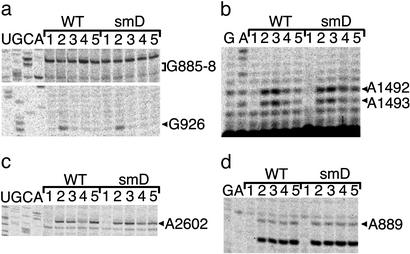
Chemical probing of E. coli ribosome decoding complexes. (a) Kethoxal modification of 16S rRNA showing P-site tRNA dependent protection of G926 (4) and the absence of reactivity in the 885–887 stretch in the switch helix as expected for the error-prone conformation (15). Sequencing lanes are marked U, G, C, and A. Lanes for both WT and smD ribosomes were as follows: 1, ribosomes with no kethoxal treatment; 2, ribosomes with kethoxal treatment; 3, ribosomes plus mRNA and tRNAfMet with kethoxal; 4, ribosomes plus mRNA, tRNAfMet, EF-Tu/GTP/Phe-tRNAPhe with kethoxal; 5, ribosomes plus mRNA, tRNAfMet, EF-Tu/GMP-PNP/Phe-tRNAPhe with kethoxal. (b) Protections from DMS modification at nucleotides A1492–A1493 of 16S rRNA dependent on A-site tRNA binding (4). Lanes were loaded as in a, replacing kethoxal with DMS. (c) Protection of A2602 in 23S rRNA from DMS dependent on release of Phe-tRNAPhe after GTP hydrolysis by EF-Tu (EFTu/GTP/Phe-tRNAPhe lanes), and modification of A2602 in codon recognition complexes (EFTu/GMPPNP/Phe-tRNAPhe lanes) (28). Lanes were loaded as in b. (d) Reactivity of A889 in 16S rRNA to DMS, in agreement with the error-prone conformation (15). Lanes were loaded as in b.
Discussion
Based on the x-ray crystal structures of the E. coli ribosome presented here and structures of the T. thermophilus 70S ribosome (18) and 30S subunit (10), conformational changes in the small subunit responsible for mRNA decoding might be limited to the body of the small subunit. According to this model (10), closing of the body of the small subunit is coupled energetically to the positioning of G530 and A1492-A1493, nucleotides that directly interact with the A-site mRNA codon and tRNA anticodon during tRNA discrimination (10) (Fig. 1). On the other hand, closing of the head of the small subunit accompanies formation of the intact ribosome from the two subunits, rather than mRNA decoding.
It is possible that rearrangements in the head that are coupled to mRNA decoding may be too subtle to detect at a resolution of 9 Å. Alternatively, the head of the small subunit may close even further than observed in the 70S ribosome crystal structures when cognate tRNA binds to the A site. However, several aspects of the structures presented here support the hypothesis that closing of the small subunit head depends solely on the formation of the 70S ribosome. First, tilting of the head toward the 50S subunit is independent of mRNA and P-site tRNA binding, because the structure of the unliganded WT E. coli ribosome adopts a similar conformation to those of the smD E. coli ribosome and T. thermophilus ribosome, both of which contain mRNA and P-site tRNA (not shown). The closing is also independent of tRNA binding to the E site and the position of the L1 stalk. In the E. coli ribosome structures, no tRNA is bound in the E site and the L1 stalk is in an open conformation (Fig. 2a), similar to that observed in the structure of the 50S subunit from D. radiodurans (23). In the T. thermophilus 70S ribosome structures, tRNA is bound in the E site and the L1 stalk is rotated toward the central protuberance (18) (Fig. 2a). The orientation of the head is also independent of crystal packing, given that E. coli ribosomes and the T. thermophilus ribosome crystallized in different packing arrangements.
Finally, the tilting of the small subunit head occurs independently of cognate tRNA binding in the A site, because the three 70S structures either contain noncognate tRNA bound to the A site or have an empty A site. It is worth noting that during protein synthesis binding of tRNA to the A site occurs only in the context of the intact 70S ribosome, because initiation protein IF1 blocks access to the A site during initiation (35). It is therefore possible that closure of the head observed in the isolated 30S subunit structures upon cognate tRNA binding (10) makes these structures more similar to that expected for the 70S ribosome. Interestingly, all three structures of the 70S ribosome have an open conformation of the small subunit body (Fig. 2 d and e). Thus, the structures are consistent with the model that opening and closing of the small subunit body contributes to mRNA decoding (10). Structures of the intact ribosome with cognate tRNA bound in the A site may shed more light on movements within the small subunit that are coupled to mRNA decoding.
Biochemical and genetic evidence has implicated components of the head of the small subunit in formation of the 70S ribosome (36) and in mRNA decoding (6, 7, 14). Chemical modifications in the head that inhibit subunit association (36) are likely caused by indirect effects, because there are no direct contacts between the small and large subunits in the modified region in the x-ray crystal structures of the intact ribosome (ref. 18 and this work). However, it is unclear at present how these observations correlate to movement of the head during the decoding process (2). Cryo-electron microscopy (EM) reconstructions of different steps of mRNA and tRNA translocation have revealed lateral movements of the head relative to the large subunit. However, because of the limitations in the resolution of the reconstructions (37), it is not clear whether the head is tilted toward the large subunit to a lesser or greater extent than in the x-ray crystal structures. Cryo-EM reconstructions of initiation complexes (38) implicate closing of the head of the small subunit in the correct positioning the start codon. Further structural characterization of competent initiation complexes (39) will be required to verify the importance of the observed closing during initiation.
The switch helix does not seem to change base-pairing during mRNA decoding, suggesting that the proposed 3-bp switch (Fig. 1c) does not occur during protein synthesis. On the other hand, the helical switch might in other steps of translation. One possibility is that the switch helix functions in the initiation of protein synthesis. The conformations of the 30S and 40S small ribosomal subunits have been shown to be heterogeneous before the proper positioning of the mRNA start codon (38, 40), which might be caused in part by rearrangements in the switch helix. Alternatively, the switch helix might play a role in mRNA and tRNA translocation after peptide bond formation. Consistent with this hypothesis, some of the rRNA mutations that stabilize the 888 conformation of the switch helix lead to increased frameshifting and higher sensitivity to spectinomycin (15). In addition, some streptomycin resistance mutations in protein S12 enhance EF-G independent translocation (41). This enhancement may be related to a shift in the equilibrium between the two pairings of the switch helix or reflect the lowering of an activation barrier between the two base-pairings that prevents uncatalyzed translocation.
Acknowledgments
We thank Barbara Schuwirth and James Holton for help with diffraction data measurement; Jennifer Doudna, Christina Shenvi, Venki Ramakrishnan, James Ogle, and Harry Noller for comments on the manuscript; and Farnoosh Seifoddini for help in figure preparation. A.V.-S. acknowledges Fidedigna Vila for balance and coherence. We thank the Whitehead Institute for Biomedical Research and the Massachusetts Institute of Technology for supporting the initial stages of this work. This work was supported by funding from the Searle Scholars Program and the National Institutes of Health.
Abbreviations: aa-tRNA, aminoacyl-tRNA; EF, elongation factor; DMS, dimethyl sulfate; smD, streptomycin-dependent.
Data deposition: Structural coordinates and diffraction amplitudes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1PNS, 1PNU, 1PNX, and 1PNY).
References
- 1.Rodnina, M. V. & Wintermeyer, W. (2001) Annu. Rev. Biochem. 70, 415–435. [DOI] [PubMed] [Google Scholar]
- 2.Ogle, J. M., Carter, A. P. & Ramakrishnan, V. (2003) Trends Biochem. Sci. 28, 259–266. [DOI] [PubMed] [Google Scholar]
- 3.Pettersson, I. & Kurland, C. G. (1980) Proc. Natl. Acad. Sci. USA 77, 4007–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moazed, D. & Noller, H. F. (1986) Cell 47, 985–994. [DOI] [PubMed] [Google Scholar]
- 5.Moazed, D. & Noller, H. F. (1987) Nature 327, 389–394. [DOI] [PubMed] [Google Scholar]
- 6.Moine, H. & Dahlberg, A. E. (1994) J. Mol. Biol. 243, 402–412. [DOI] [PubMed] [Google Scholar]
- 7.O'Connor, M., Thomas, C. L., Zimmermann, R. A. & Dahlberg, A. E. (1997) Nucleic Acids Res. 25, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, A. P., Clemons, W. M., Jr., Brodersen, D. E., Morgan-Warren, R. J., Wimberly, B. T. & Ramakrishnan, V. (2000) Nature 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 9.Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F. & Yonath, A. (2000) Cell 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 10.Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. (2002) Cell 111, 721–732. [DOI] [PubMed] [Google Scholar]
- 11.Valle, M., Sengupta, J., Swami, N. K., Grassucci, R. A., Burkhardt, N., Nierhaus, K. H., Agrawal, R. K. & Frank, J. (2002) EMBO J. 21, 3557–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark, H., Rodnina, M. V., Wieden, H. J., Zemlin, F., Wintermeyer, W. & van Heel, M. (2002) Nat. Struct. Biol 9, 849–854. [DOI] [PubMed] [Google Scholar]
- 13.O'Connor, M. & Dahlberg, A. E. (1995) J. Mol. Biol. 254, 838–847. [DOI] [PubMed] [Google Scholar]
- 14.Liebman, S. W., Chernoff, Y. O. & Liu, R. (1995) Biochem. Cell. Biol. 73, 1141–1149. [DOI] [PubMed] [Google Scholar]
- 15.Lodmell, J. S. & Dahlberg, A. E. (1997) Science 277, 1262–1267. [DOI] [PubMed] [Google Scholar]
- 16.Gabashvili, I. S., Agrawal, R. K., Grassucci, R., Squires, C. L., Dahlberg, A. E. & Frank, J. (1999) EMBO J. 18, 6501–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velichutina, I. V., Dresios, J., Hong, J. Y., Li, C., Mankin, A., Synetos, D. & Liebman, S. W. (2000) RNA 6, 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 19.Moazed, D., Van Stolk, B. J., Douthwaite, S. & Noller, H. F. (1986) J. Mol. Biol. 191, 483–493. [DOI] [PubMed] [Google Scholar]
- 20.Blaha, G., Stelzl, U., Spahn, C. M., Agrawal, R. K., Frank, J. & Nierhaus, K. H. (2000) Methods Enzymol. 317, 292–309. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 24.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 25.Nikulin, A., Eliseikina, I., Tishchenko, S., Nevskaya, N., Davydova, N., Platonova, O., Piendl, W., Selmer, M., Liljas, A., Drygin, D., et al. (2003) Nat. Struct. Biol. 10, 104–108. [DOI] [PubMed] [Google Scholar]
- 26.Abrahams, J. P. & De Graaff, R. A. (1998) Curr. Opin. Struct. Biol. 8, 601–605. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt, G. J., Zou, J. Y., Kjeldgaard, M. & Jones, T. A. (2001) in International Tables for Crystallography, eds. Rossmann, M. G. & Arnold, E. (Kluwer, Dordrecht, The Netherlands), Vol. F, pp. 353–367. [Google Scholar]
- 28.Carson, M. (1997) Methods Enzymol. 277, 493–505. [PubMed] [Google Scholar]
- 29.Delano, W. L. (2002) The PyMOL User's Manual (Delano Scientific, San Carlos, CA).
- 30.Moazed, D. & Noller, H. F. (1989) Cell 57, 585–597. [DOI] [PubMed] [Google Scholar]
- 31.Cate, J. H., Yusupov, M. M., Yusupova, G. Z., Earnest, T. N. & Noller, H. N. (1999) Science 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 32.Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 33.Allen, P. N. & Noller, H. F. (1989) J. Mol. Biol. 208, 457–468. [DOI] [PubMed] [Google Scholar]
- 34.Karimi, R. & Ehrenberg, M. (1996) EMBO J. 15, 1149–1154. [PMC free article] [PubMed] [Google Scholar]
- 35.Carter, A. P., Clemons, W. M., Jr., Brodersen, D. E., Morgan-Warren, R. J., Hartsch, T., Wimberly, B. T. & Ramakrishnan, V. (2001) Science 291, 498–501. [DOI] [PubMed] [Google Scholar]
- 36.Herr, W., Chapman, N. M. & Noller, H. F. (1979) J. Mol. Biol. 130, 433–449. [DOI] [PubMed] [Google Scholar]
- 37.Frank, J. & Agrawal, R. K. (2000) Nature 406, 318–322. [DOI] [PubMed] [Google Scholar]
- 38.Spahn, C. M., Kieft, J. S., Grassucci, R. A., Penczek, P. A., Zhou, K., Doudna, J. A. & Frank, J. (2001) Science 291, 1959–1962. [DOI] [PubMed] [Google Scholar]
- 39.Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I. & Hellen, C. U. (2001) Proc. Natl. Acad. Sci. USA 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabashvili, I. S., Agrawal, R. K., Grassucci, R. & Frank, J. (1999) J. Mol. Biol. 286, 1285–1291. [DOI] [PubMed] [Google Scholar]
- 41.Asatryan, L. S. & Spirin, A. S. (1975) Mol. Gen. Genet. 138, 315–321. [DOI] [PubMed] [Google Scholar]