Abstract
Site-specific recombination typically occurs only between DNA sequences that have co-evolved with a natural recombinase enzyme to optimize sequence recognition, catalytic efficiency, and regulation. Here, we show that the sequence recognition and the catalysis functions of a recombinase can be specified by unrelated protein domains. We describe chimeric recombinases with a catalytic domain from an activated multiple mutant of the bacterial enzyme Tn3 resolvase, fused to a DNA recognition domain from the mouse transcription factor Zif268. These proteins catalyze efficient recombination specifically at synthetic target sites recognized by two Zif268 domains. Our results demonstrate the functional autonomy of the resolvase catalytic domain and open the way to creating “custom-built” recombinases that act at chosen natural target sequences.
Site-specific recombinases bring about programmed rearrangements of DNA sequences, by catalyzing cleavage and rejoining of DNA strands at specific sites to which they bind (ref. 1; Fig. 1A). Recently, site-specific recombination has found widespread application in experimental biology, biotechnology, and potential gene therapies (2, 3). However, appropriately positioned sites for the chosen recombinase must first be introduced into the DNA to be rearranged. The use of site-specific recombination in unmodified genomes will become practicable only when recombinases can be systematically designed to recognize endogenous target sequences with high specificity.
Fig. 1.
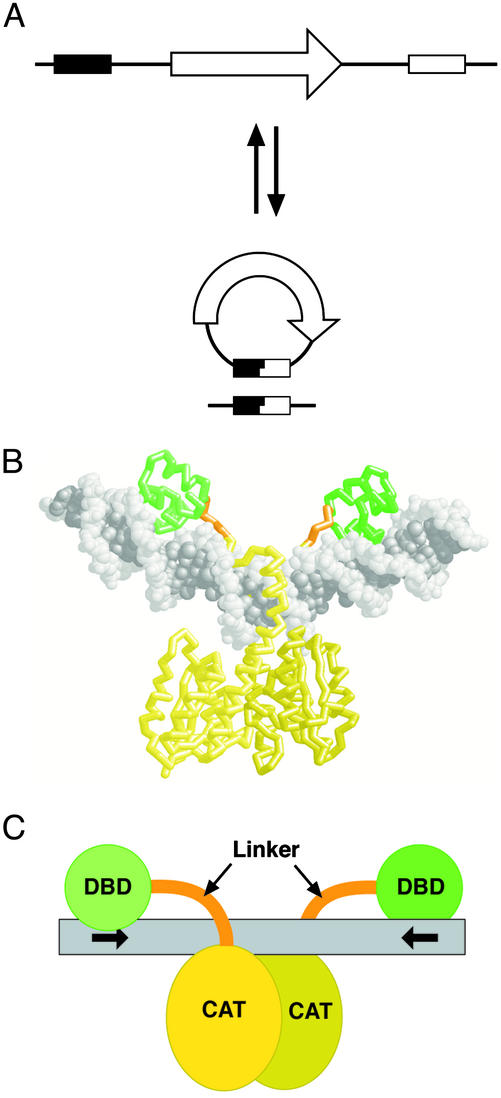
Site-specific recombination mediated by Tn3 resolvase. (A) Site-specific recombination reactions. A DNA segment containing a sequence of interest (arrow) may be excised by breaking and rejoining the DNA within two sites (boxes) recognized by a recombinase. (If the substrate is circular, this reaction is sometimes called resolution.) Similarly, the segment can be inserted by the intermolecular, reverse reaction. Another alternative, inversion of the orientation of the DNA segment between the sites, is not shown. (B) Crystal structure ofγδ resolvase dimer bound to a 34-bp DNA fragment containing res site I (10). The protein backbone is shown, with the N-terminal, catalytic domains in yellow, the C-terminal, DNA-binding domains in green, and the interdomain linkers in orange. The DNA is in gray spacefill representation. (C) Cartoon representation of the structure shown in B, using the same color scheme. The chimeric Z-resolvases described here have Zif268 DNA-binding domains in place of the resolvase C-terminal domains (DBD), and a mutant version of the catalytic domain (CAT). The Z-sites have motifs recognized by Zif268 in place of the motifs recognized by the resolvase C-terminal domain (indicated by arrows).
Nearly all applications of site-specific recombination to date have used Cre or FLP, both members of a large family of “tyrosine recombinases” (1). Attempts to alter sequence recognition specificity by these enzymes, by using multiple cycles of mutagenesis and selection, have so far met with limited success (3–5). A more radical approach, substitution of the recombinase's DNA recognition components with those of a completely different protein, is unlikely to be directly applicable to Cre or FLP, because their catalytic and DNA recognition functions are structurally intermingled (6).
A second large family of “serine recombinases” is structurally unrelated to the tyrosine recombinase family and has a different catalytic mechanism (1, 7, 8). The closely related serine recombinases Tn3 resolvase and γδ resolvase are two-domain proteins, in which an N-terminal catalytic domain of ≈140 aa is joined by a short linker sequence to a structurally distinct DNA-binding domain of ≈40 aa. This C-terminal domain confers most of the sequence specificity (9). Resolvase catalyzes recombination at a long (114 bp) sequence, res, that includes three binding sites for resolvase dimers. The phosphodiester linkages that are broken and rejoined during recombination are at the center of binding site I (7).
The crystal structure of a resolvase dimer bound to site I (ref. 10; Fig. 1B) shows that the N- and C-terminal domains are spatially separate and interact with distinct DNA segments. This invites the notion that the DNA sequence specificity of resolvase might be altered, by replacing the C-terminal domain with a domain that binds a different sequence. The chimeric protein might be expected to promote recombination at a site in which two motifs recognized by the new DNA-binding domains are placed in inverted repeat, separated by a suitable central sequence, so that the structural arrangement of the paired catalytic domains of the resolvase dimer is preserved (Fig. 1C). Some “domain-swapping” experiments to this end have been attempted previously, by using domains from pairs of related serine recombinases, but only subtle changes in recognition specificity have been achieved (11–13). Mutagenesis-selection strategies have also been used to alter sequence recognition by intact serine recombinases, with some success (14). For resolvase, attempts to alter sequence recognition are complicated by its requirement for a multipartite recombination site that binds six resolvase subunits.
Recently, multiple mutants of Tn3 resolvase have been isolated that catalyze rapid recombination at a truncated version of res comprising just the 28-bp binding site I (ref. 15; J.H. and W.M.S., unpublished work). Furthermore, unlike wild-type resolvase, activity of these mutants does not depend on the connectivity of the sites, nor on negative supercoiling of the substrate DNA (7). These features make the mutants more suitable for use as genetic tools, and for alteration of sequence recognition. Here, we describe chimeric recombinases with a catalytic domain from one of these “hyperactive” multiple mutant resolvases, fused via a short flexible linker peptide to the DNA-binding domain of the mouse transcription factor Zif268 (16). Recombination catalyzed by these “Z-resolvase” chimeras occurs specifically at “Z-sites” that consist of appropriately spaced pairs of sequence motifs recognized by the Zif268 domain, flanking a central sequence that includes the part of Tn3 res binding site I acted on by the resolvase catalytic domains.
Materials and Methods
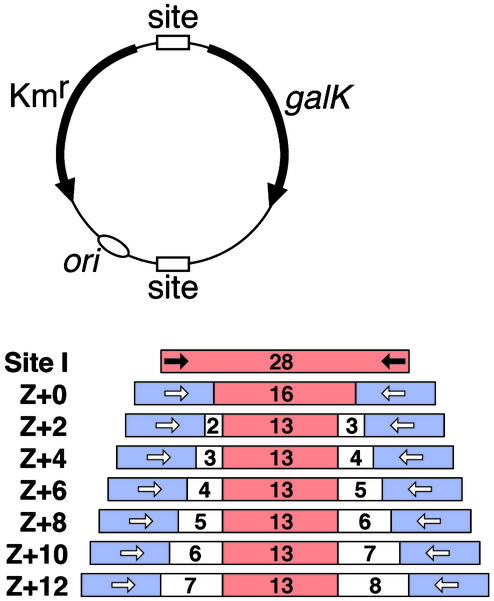
Recombination Substrates. The Z-sites used in the experiments shown in Figs. 3 and 4 were synthesized as oligonucleotides and cloned into appropriate plasmids. The top-strand sequences are of the form —GCGTGGGCG(spacer S1)A ATAT TATAAATT(spacer S2)CGCCCACGC—. The central 13-nt sequence is identical to that of Tn3 res site I. The motifs recognized by Zif268 (16) are bold. The spacer sequences of the Z-sites are as follows. Z+0; S1 = A, S2 = AT. Z+2; S1 = AC, S2 = CAT. Z+4; S1 = ACG, S2 = GCAT. Z+6; S1 = ACGA, S2 = TGCAT. Z+8; S1 = ACGAC, S2 = GTGCAT. Z+10; S1 = ACGACT, S2 = AGTGCAT. Z+12; S1 = ACGACTG, S2 = CATGGCAT. The Z-site-containing test plasmids for the in vivo assays were constructed similarly to pDB35, a plasmid with two copies of Tn3 res site I (Fig. 2), which has been described (15). The substrates used for in vitro Z-site recombination assays were constructed similarly to pOG3, from the cloning vector pMTL23 (17). Full details of the plasmid construction and sequences are available on request.
Fig. 3.
Z-resolvase-mediated recombination in E. coli.(A) Expression plasmids encoding each resolvase variant were introduced into E. coli harboring plasmid substrates with either two copies of site I (Left) or two Z+6 sites (Right). The photographs show colonies on MacConkey indicator plates after 24 h. The percentage of plasmid molecules resolved was determined by isolating plasmid DNA from these cells (see Materials and Methods) and is given in the circle at the lower right of each panel (to the nearest whole number). S10A is a catalytically inactive mutant of Tn3 resolvase (control). NM is the N-terminal, catalytic domain of NM-resolvase (residues 1–148). NM-resolvase (NM-R) and Z-resolvase (Z-R) are described in Materials and Methods. The Z-resolvase used here has the L6 linker (C). (B) Assays of Z-resolvase (L6) activity on substrates containing Z-sites as shown in Fig. 2. The results are presented as in A. (C) Comparison of the activity of Z-resolvases with different linker sequences. The test substrate contained two Z+6 sites. Assays were performed as above; the percentage of resolved substrate molecules in the recovered plasmid DNA is presented on the right.
Fig. 4.
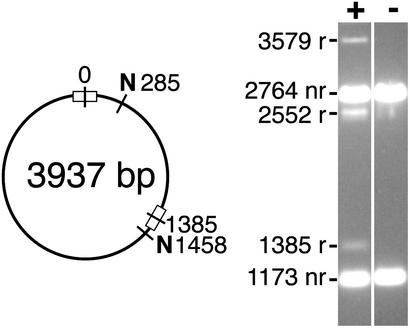
In vitro recombination by Z-resolvase (linker L6). (Left) Structure of the test substrate used, with two Z+6 sites in direct repeat. N denotes sites for the restriction enzyme NruI. (Right) Products after treatment of the substrate with Z-resolvase for 16 h were digested with NruI and separated on a 1.2% agarose gel. nr, nonrecombinant bands; r, recombinant bands. The sizes of the fragments (in base pairs) are indicated. Inversion gives 3,579- and 358-bp bands (the small fragment is not visible on the gel), whereas resolution gives 2,552- and 1,385-bp bands.
Fig. 2.
Structures of the substrates used to assay recombination in Escherichia coli.(Upper) pDB35 (15) contains two copies of site I, which are replaced by Z-sites in the other substrates. Kmr is a gene conferring kanamycin resistance. Recombination between the two sites divides the plasmid into two circles. The galK-containing circle is lost because it does not have an origin of replication or selective marker, making the cells galK–, and giving pale yellow rather than red (galK+) colonies on the indicator plates. (Lower) Z-sites. Tn3 res site I sequence is red, the 9-bp Zif268-binding motif is blue, and the spacer sequence is white. The numbers indicate the lengths of the marked DNA segments (in base pairs). Arrows indicate the inverted repeats bound by recombinase (black for resolvase and unfilled for Z-resolvase).
Chimeric Recombinases. The hyperactive NM-resolvase has the following six differences from the wild-type Tn3 resolvase sequence: R2A E56K G101S D102Y M103I Q105L. Z-resolvase expression plasmids were constructed by cloning a fragment encoding the catalytic domain of NM-resolvase (residues 1–144), and synthetic fragments encoding the linker and the Zif268 DNA-binding domain, into a plasmid similar to pAT5 (15). The amino acid sequence of the 89-residue Zif268 DNA-binding domain was as described (16). Linker peptide sequences were introduced between resolvase residue 144 (arginine) and Zif268 domain residue 2 (glutamate); those tested are shown in Fig. 3C. The sequences of the expression plasmids are available on request. Overexpression and purification of Z-resolvase was by an adaptation of the procedure described for resolvase mutants (15). Briefly, after sonication of the cells and centrifugation, Z-resolvase was redissolved from the pellet in a denaturing buffer containing 6 M urea. After refolding in the presence of 0.02 mM Zn2+, the Z-resolvase was precipitated by dialysis against a low-salt buffer. After redissolution in a buffer containing 6 M urea, it was further purified by cation exchange chromatography and refolded in a buffer containing 2 M NaCl and 0.5 mM Zn2+.
Recombination Assays. The E. coli colony color resolution assay as used in Fig. 3 has been described (15). For quantitation of the extent of resolution (Fig. 3), the cells were washed from the MacConkey agar plates after incubation for 24 h at 37°C. The plasmid DNA was purified and used to transform the E. coli strain DS941 (15). Transformants harboring the test plasmid or its resolution product were selected on MacConkey indicator plates containing kanamycin, and the numbers of white (indicating resolution) and red (unresolved) colonies were recorded (in a total of at least 250 colonies). The purified plasmid DNA was also analyzed by agarose gel electrophoresis (15), before and after digestion with various restriction enzymes, confirming that the recombinant plasmids were of the expected structure (data not shown). The integrity of recombinant Z-sites in product DNA was demonstrated by showing that the DNA was fragmented appropriately by the restriction enzyme PsiI (supplied by New England Biolabs), which cuts at the site TTA/TAA (/, the position of cleavage). This sequence is at the center of the Z-sites (see above). In recombination, resolvase cuts the DNA thus: TTAT/AA (top and bottom strands of this 6-bp segment have the same sequence). The top and bottom strand cuts are therefore staggered by 2 bp.
In vitro assays were as described (15).
Results
The enzyme engineering described here begins with a hyperactive Tn3 resolvase (NM-resolvase) with six substitutions in the N-terminal, catalytic domain (see Materials and Methods). The C-terminal domain is still required for specific binding and catalytic activity at site I; no binding or recombination is observed if all residues after arginine 148 are deleted (Fig. 3A; data not shown). To alter sequence recognition by NM-resolvase, we replaced its C-terminal domain (from residue 145 onwards) with the “zinc finger” DNA recognition domain of Zif268 (16). The two domains were connected by a short linker peptide, several variants of which were tested (Fig. 3C). We also created potential recombination sites (Z-sites) for the chimeric Z-resolvases, by replacing the sequences at the extremities of site I, which are recognized by the resolvase C-terminal domain, with 9-bp motifs recognized by Zif268. The length of DNA between the two 9-bp motifs of the Z-site was varied systematically (Fig. 2).
Combinations of candidate Z-resolvases and Z-sites were tested in vivo by using an E. coli colony color assay (ref. 15; Fig. 2). We observed efficient recombination (resolution) of the test substrate in some cases; observed as white, rather than red, colonies (Fig. 3). The extent of resolution was quantitated by further analysis of plasmid DNA recovered from the cells (see Materials and Methods). We also analyzed recombinant DNA from E. coli by restriction enzyme digests, to show that Z-resolvase-catalyzed recombination occurs between Z-sites, and that the product contains a new Z-site, created by precise joining of two half-sites (see Materials and Methods; data not shown). Z-resolvase was active only at sites with appropriately placed motifs for binding of its Zif268 domain. It was therefore inactive on a substrate that contains two copies of the Tn3-derived site I sequence. Conversely, NM-resolvase efficiently recombined a substrate containing two copies of site I but had no activity on a similar substrate with two Z-sites (Fig. 3A).
The most important factor affecting activity of Z-resolvase was the sequence of the Z-site, and, in particular, the distance between the 9-bp motifs recognized by Zif268 (Fig. 3B). We suspect that the reasons for the observed variation of activity with site length may be complex. When the Zif268 domains are bound too close to the center of the site, they might sterically interfere with structural changes required during catalysis. When the domains are too far away from the center, the stability of the Z-resolvase dimer–DNA complex might be reduced because the interdomain linker is too short. Recombination might be most efficient if the bulkiest parts of the Zif268 domains are on the opposite face of the DNA helix from the catalytic domains (as is the case for the natural resolvase DNA-binding domains; Fig. 1B). The specific spacer sequences chosen (see Materials and Methods) might also affect activity. Rather surprisingly, we found that activity was quite insensitive to variation of the length and sequence of the linker peptide between the Z-resolvase catalytic and Zif268 domains (Fig. 3C). There was no obvious relationship between the linker length and activity on the different Z-sites; all of the Z-resolvases tested were most active on the substrate with Z+6 sites (Fig. 3C; data not shown). Molecular modeling (not shown) indicates that the Z+6 site would place the residues at the ends of the linker closest together, and also places the Zif268 domain away from the catalytic domains, but we cannot yet be sure that these features account for the high recombination activity at this site.
We purified the Z-resolvase that was most active in vivo (Z-R L6; Fig. 3C) and showed that it also catalyzes recombination (resolution and inversion) at Z+6 sites in vitro (Fig. 4). In other reaction conditions, we also observed intermolecular recombination (data not shown). These results conform with previous studies of resolvase-mediated recombination at site I (15, 18). The extent of recombination in vitro was less than that observed in E. coli, for reasons that are as yet unclear. In accord with our observations in vivo, NM-resolvase did not have any Z-site-specific activity in vitro. Likewise, neither Z-resolvase nor the isolated catalytic domain had any detectable recombination activity in vitro at site I or its central 16-bp sequence (data not shown).
Discussion
Our results show that catalysis by the N-terminal domain of a hyperactive mutant resolvase does not require the presence of its proper C-terminal domain; it can be replaced by the DNA-binding domain of the unrelated protein Zif268. The full mechanistic implications of this finding will be discussed elsewhere, but we note here that any specific role for the natural C-terminal domain of resolvase in synapsis of two site Is, or in catalysis of strand exchange, can now be discounted. The successful substitution of the C-terminal domain with a domain that binds DNA much more tightly (19, 20) also suggests that the mechanism of strand exchange does not require dissociation of the domain from the DNA, as is a feature of some models (7). However, it has not yet been proved that Z-resolvases catalyze strand exchange by the same mechanism as wild-type resolvase.
Our results also have implications for potential application of site-specific recombination in genetics, biotechnology, and gene therapy. Specifically, it should be possible to create variant Z-resolvases that promote recombination in vivo at chosen sequences within natural genomes.
Zif268 is a mammalian protein, and several serine recombinases, including γδ resolvase (21), have been shown to be active in mammalian and other eukaryote cells, so we expect that our chimeric proteins will also function in eukaryotic cells. The Zif268 DNA-binding domain is currently the main focus of a field of research whose aim is to create proteins that can bind specifically to any chosen DNA sequence. Already, variants of the Zif268 domain have been created that recognize naturally occurring sequences in the genomes of higher eukaryotes, and these have been used to target chimeric proteins to specific genomic loci (22, 23). It should therefore be quite straightforward to adapt the system described here to promote recombination at many novel sites, by replacing the Zif268 domain of Z-resolvase with one of these variants. In principle, any natural sequence of ≈40 bp could be regarded as a potential Z-site; its ends could be recognized by variant Zif268 domains, and its central sequence could be acted on by the catalytic domain of a hyperactive mutant resolvase. In practice, of course, some sequences will be much more suitable for targeting in this way than others (see below).
We envisage that one could create a recombinase that acts at a naturally occurring sequence, by adopting the following plan. (i) Identify an optimal sequence within a genomic region of interest, with desirable features such as a central sequence with some resemblance to that of Tn3 res site I. (ii) Then, select variant Zif268 domains that bind specifically to motifs that flank the chosen sequence, arranged as in the Z+6 site (Fig. 2). If required, one could create a heterodimeric recombinase, with different Zif268-derived domains to recognize different sequences at the two ends of the site, because resolvase readily forms heterodimers (24). (iii) Likewise, select optimally active variants of the Z-resolvase catalytic domain, by using plasmids analogous to those shown in Fig. 2, but with Z-sites having the central sequence of the chosen target site. The central sequences of the Z-sites used in this study retain at least 13 bp of the original 28-bp Tn3 site I sequence, but current evidence suggests that the identity of only a small subset of these base pairs is important for the interaction of the resolvase N-terminal domain (9, 10, 25). We therefore predict that sequence selectivity of the catalytic domain will not limit the choice of potential sites to an unacceptable extent. (iv) Finally, connect the optimized catalytic and DNA recognition domains with an appropriate linker.
A further general feature of the serine recombinases that should facilitate targeted recombination at novel sequences is that only 2 bp of homology is required at the central “overlap” sequence of two recombining sites, in contrast to the tyrosine recombinases that require 6–8 bp of homology (1, 7, 25).
Many potential uses of these chimeric recombinases can be foreseen, in experimental and applied genetics. For example, as has been noted (26), the integrated DNA copy of a retrovirus such as HIV might be excised and thereby inactivated by recombination between identical sites in the long terminal repeats.
Acknowledgments
We are very grateful to Mary Burke and Adèle McLean for experimental assistance. This work was supported by The Wellcome Trust.
References
- 1.Nash, H. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), 2nd Ed., Vol. 1, pp. 2363–2376. [Google Scholar]
- 2.Gorman, C. & Bullock, C. (2000) Curr. Opin. Biotechnol. 11, 455–460. [DOI] [PubMed] [Google Scholar]
- 3.Nagy, A. (2000) Genesis 26, 99–109. [PubMed] [Google Scholar]
- 4.Buchholz, F. & Stewart, A. F. (2001) Nat. Biotechnol. 19, 1047–1052. [DOI] [PubMed] [Google Scholar]
- 5.Santoro, S. W. & Schultz, P. G. (2002) Proc. Natl. Acad. Sci. USA 99, 4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo, F., Gopaul, D. N. & Van Duyne, G. D. (1997) Nature 389, 40–46. [DOI] [PubMed] [Google Scholar]
- 7.Grindley, N. D. F. (2002) in Mobile DNA II, eds. Craig, N., Craigie, R., Gellert, M. & Lambowitz, A. (Am. Soc. Microbiol., Washington, DC), pp. 272–302.
- 8.Smith, M. C. M. & Thorpe, H. M. (2002) Mol. Microbiol. 44, 299–307. [DOI] [PubMed] [Google Scholar]
- 9.Rimphanitchayakit, V. & Grindley, N. D. F. (1990) EMBO J. 9, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, W. & Steitz, T. A. (1995) Cell 82, 193–207. [DOI] [PubMed] [Google Scholar]
- 11.Avila, P., Ackroyd, A. J. & Halford, S. E. (1990) J. Mol. Biol. 216, 645–655. [DOI] [PubMed] [Google Scholar]
- 12.Grindley, N. D. F. (1993) Science 262, 738–740. [DOI] [PubMed] [Google Scholar]
- 13.Schneider, F., Schwikardi, M., Muskhelishvili, G. & Dröge, P. (2000) J. Mol. Biol. 295, 767–775. [DOI] [PubMed] [Google Scholar]
- 14.Sclimenti, C. R., Thyagarajan, B. & Calos, M. P. (2001) Nucleic Acids Res. 29, 5044–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold, P. H., Boocock, M. R., Grindley, N. D. F., Blake, D. G. & Stark, W. M. (1999) EMBO J. 18, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elrod-Erickson, M., Rould, M. A., Nekludova, L. & Pabo, C. O. (1996) Structure Fold. Des. 4, 1171–1180. [DOI] [PubMed] [Google Scholar]
- 17.Blake, D. G., Boocock, M. R., Sherratt, D. J. & Stark, W. M. (1995) Curr. Biol. 5, 1036–1046. [DOI] [PubMed] [Google Scholar]
- 18.Bednarz, A. L., Boocock, M. R. & Sherratt, D. J. (1990) Genes Dev. 4, 2366–2375. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J.-S. & Pabo, C. O. (1998) Proc. Natl. Acad. Sci. USA 95, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel-Meguid, S. S., Grindley, N. D. F., Templeton, N. S. & Steitz, T. A. (1984) Proc. Natl. Acad. Sci. USA 81, 2001–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwikardi, M. & Dröge, P. (2000) FEBS Lett. 471, 147–150. [DOI] [PubMed] [Google Scholar]
- 22.Isalan, M., Klug, A. & Choo, Y. (2001) Nat. Biotechnol. 19, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibikova, M., Golic, M., Golic, K. & Carroll, D. (2002) Genetics 161, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murley, L. L. & Grindley, N. D. F. (1998) Cell 95, 553–562. [DOI] [PubMed] [Google Scholar]
- 25.Hatfull, G. F., Salvo, J. J., Falvey, E. E., Rimphanitchayakit, V. & Grindley, N. D. F. (1988) Symp. Soc. Gen. Microbiol. 43, 149–181. [Google Scholar]
- 26.Lee, Y. & Park, J. (1998) Biochem. Biophys. Res. Commun. 253, 588–593. [DOI] [PubMed] [Google Scholar]