Abstract
β-TrCP1 (also known as Fbw1a or FWD1) is the F-box protein component of an Skp1/Cul1/F-box (SCF)-type ubiquitin ligase complex. Although biochemical studies have suggested that β-TrCP1 targets inhibitory subunit of NF-κB(IκB) proteins and β-catenin for ubiquitylation, the physiological role of β-TrCP1 in mammals has remained unclear. We have now generated mice deficient in β-TrCP1 and shown that the degradation of IκBα and IκBβ is reproducibly, but not completely, impaired in the cells of these animals. The nuclear translocation and DNA-binding activity of NF-κB as well as the ability of this transcription factor to activate a luciferase reporter gene were also inhibited in β-TrCP1–/– cells compared with those apparent in wild-type cells. The subcellular localization of β-catenin was altered markedly in β-TrCP1–/– cells. Furthermore, the rate of proliferation was reduced and both cell size and the percentage of polyploid cells were increased in embryonic fibroblasts derived from β-TrCP1–/– mice pared with the corresponding wild-type cells. These results suggest that β-TrCP1 contributes to, but is not absolutely required for, the degradation of IκB and β-catenin and the consequent regulation of the NF-κB and Wnt signaling pathways, respectively. In addition, they implicate β-TrCP1 in the maintenance of ploidy during cell-cycle progression.
The ubiquitin-proteasome pathway of protein degradation is essential for various important biological processes including cell-cycle progression, gene transcription, and signal transduction (1, 2). The formation of ubiquitin–protein conjugates requires three enzymes that participate in a cascade of ubiquitin transfer reactions: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The specificity of protein ubiquitylation is often determined by E3 enzymes, and proteins polyubiquitylated by these enzymes undergo degradation by the 26S proteasome (3, 4). Skp1/Cul1/F-box (SCF) complexes constitute a class of E3 enzymes that are required for the degradation of many key cellular proteins. These complexes consist of invariable components including Skp1, Cul1, and Rbx1 (also known as ROC1 or Hrt1) as well as variable components known as F-box proteins, the latter of which bind to Skp1 through their F-box domains (5–12). F-box proteins function as receptors for target molecules, many, but not all, of which are phosphorylated. The substrate specificity of SCF complexes thus is thought to be determined by the F-box protein. More than 50 F-box proteins have been identified in mammals to date (13, 14).
In mammals, one of the most extensively studied short-lived regulatory proteins that undergo ubiquitylation is IκBα, an inhibitory protein that associates with NF-κB and thereby prevents this dimeric transcription factor from entering the nucleus (15, 16). Another well characterized protein that undergoes ubiquitylation is β-catenin, which plays an essential role in the Wnt signaling pathway and is an important component of cadherin cell-adhesion complexes. The abundance of β-catenin in the cytoplasm is regulated by ubiquitin-dependent proteolysis (17). The abnormal accumulation of β-catenin as a result of mutations either in the adenomatous polyposis coli tumor suppressor protein or in β-catenin itself is thought to initiate colorectal neoplasia. Both IκBα and β-catenin contain a DSGXXS sequence that includes the phosphorylation site required for ubiquitylation (17, 18). We and others have shown that β-TrCP1 (also known as Fbw1a or FWD1) is a member of the F box- and WD40 repeat-containing family of proteins and specifically recognizes IκBα, IκBβ, IκBε, and β-catenin as substrates only when they are phosphorylated at both serine residues in the conserved DSGXXS motif (18–28). A highly homologous protein, β-TrCP2, also contributes to the ubiquitylation of IκBα (29). Another E3 complex, Siah-SIP-Skp1-Ebi, has also been implicated in the degradation of β-catenin (30). The relative importance of β-TrCP1 in the degradation of IκB proteins and β-catenin thus has been unclear.
To determine the physiological functions of β-TrCP1, we generated mice that lack this protein. Characterization of cells from these animals revealed that the degradation of IκB proteins and β-catenin is impaired substantially but not completely in the absence of β-TrCP1. Furthermore, the β-TrCP1–/– cells exhibit a reduced proliferation rate and abnormal ploidy, suggesting that β-TrCP1 contributes to regulation of the cell cycle.
Methods
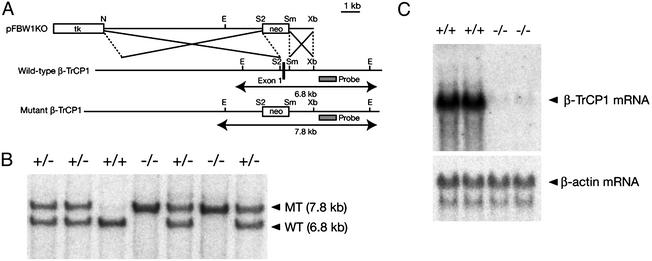
Construction of a Targeting Vector and Generation of β-TrCP1–/– Mice. Cloned DNA corresponding to the β-TrCP1 gene locus was isolated from a 129/Sv mouse genomic library (Stratagene) (31). A targeting vector, pFBW1KO, was constructed by replacing a 530-bp SacII–SmaI fragment containing exon 1 of the β-TrCP1 gene with a phosphoglycerate kinase (PGK)-neo-poly(A) cassette; the vector contained 8.5- and 1.5-kb regions of homology 5′ and 3′ of the neomycin resistance marker, respectively. A PGK-tk-poly(A) cassette was ligated at the 5′ end of the insert. The maintenance, transfection, and selection of embryonic stem cells were performed as described (32). The recombination event was confirmed by Southern blot analysis with a 0.9-kb SacI–SpeI probe that flanked the 3′ homology region (Fig. 1A). The expected sizes of hybridizing fragments after EcoRI digestion are 6.8 and 7.8 kb for the wild-type and mutant alleles, respectively.
Fig. 1.
Targeted disruption of the mouse β-TrCP1 gene. (A) Structures of the targeting vector (pFBW1KO), the mouse β-TrCP1 gene locus, and the mutant allele resulting from homologous recombination. The first exon of the β-TrCP1 gene is depicted by a filled box. A genomic fragment used as a probe for Southern blot analysis is shown as a shaded box, and the expected sizes of the EcoRI fragments of the wild-type and mutant alleles that hybridize with the probe are indicated. neo, neomycin transferase gene linked to the PGK gene promoter; tk, thymidine kinase gene derived from herpes simplex virus linked to the PGK gene promoter. The orientations of both neo and tk are the same as that of the β-TrCP1 gene. Restriction sites: E, EcoRI; N, NotI; S2, SacII; Sm, SmaI; Xb, XbaI. Not all restriction sites are shown. (B) Southern blot analysis of genomic DNA extracted from the tail of mice of the indicated β-TrCP1 genotypes. The DNA was digested with EcoRI and subjected to hybridization with the probe shown in A. The expected sizes of the bands corresponding to wild-type (WT) and mutant (MT) alleles are shown at the right. (C) Northern blot analysis of total RNA isolated from the testis of mice of the indicated β-TrCP1 genotypes. Hybridization was performed with mouse β-TrCP1 and β-actin cDNA probes.
The mutant embryonic stem cells were microinjected into C57BL/6 blastocysts, and the resulting male chimeras were mated with female C57BL/6 mice. The germ-line transmission of the mutant allele was confirmed by Southern blot analysis as described above. Heterozygous offspring were intercrossed to produce homozygous mutant animals. All mice were maintained in a specific pathogen-free animal facility at the Medical Institute of Bioregulation at Kyushu University.
Northern Blot Analysis. Total RNA was isolated from testis by the guanidinium thiocyanate-phenol-chloroform method, and portions (20 μg) were resolved by electrophoresis on a 1% agarose gel containing formaldehyde. The RNA molecules then were transferred to a nylon membrane (Hybond-N, Amersham Pharmacia) and subjected to hybridization with a 32P-labeled probe derived from mouse β-TrCP1 cDNA.
Immunoblot Analysis. Cells were lysed in a solution containing 50 mM Tris·HCl (pH 7.6)/150 mM NaCl/0.5% Triton X-100/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM PMSF/0.4 mM Na3VO4/0.4 mM EDTA/10 mM NaF/10 mM sodium pyrophosphate. Equal amounts of cell lysates (50 μg of protein) were subjected to immunoblot analysis with antibodies to IκBα (C-21, Santa Cruz Biotechnology), IκBβ (S-20, Santa Cruz Biotechnology), or glycogen synthase kinase 3β (7) (Transduction Laboratories, Lexington, KY). The band intensity was measured by LAS-1000 chemiluminescence imager (Fuji) and normalized by the expression of glycogen synthase kinase 3β.
Electrophoretic Mobility-Shift Assay (EMSA) Analysis. EMSA analysis of the DNA-binding activity of NF-κB was performed as described (41) with the exception that thymocytes served as the NF-κB source. Thymocytes were freshly isolated from β-TrCP1+/+ or β-TrCP1–/– mice and stimulated for various times with 300 nM ionomycin (Sigma) and 100 nM phorbol 12,13-dibutyrate (Sigma), after which cell lysates were prepared for use in EMSA analysis.
Preparation of Mouse Embryonic Fibroblasts (MEFs) and Wnt-3a Stimulation. Primary MEFs were isolated on embryonic day 13.5 and cultured as described (32). Conditioned medium containing Wnt-3a was also prepared as described (42). Confluent MEFs in 24-well plates were cultured in 0.5 ml of Wnt-3a-conditioned medium for 2 h, washed twice with DMEM (GIBCO/BRL) supplemented with 10% FBS (GIBCO/BRL), and then cultured for 6 h in DMEM/FBS before analysis.
Luciferase Reporter Assays. MEFs (5 × 105 cells per ml) were transiently transfected with 1 μg of an NF-κB-dependent luciferase reporter construct and 0.5 μg of the pRL-Tk plasmid (Promega) with the use of the FuGENE6 transfection reagent (Roche Applied Science, Indianapolis). After incubation for 48 h, the cells were stimulated with tumor necrosis factor α (TNF-α) (10 ng/ml) for the indicated times. They then were harvested in passive lysis buffer (Promega), and the luciferase activities of cell extracts were measured with the use of the dual luciferase assay system (Promega). The activity of firefly luciferase was normalized relative to that of Renilla luciferase.
Immunofluorescence Microscopy. MEFs were cultured on glass coverslips, fixed with 4% paraformaldehyde in PBS, and incubated with antibodies to p65 (RelA) (SC-109, Santa Cruz Biotechnology) or β-catenin (C19220, Transduction Laboratories) at a concentration of 1 μg/ml in PBS containing 0.1% BSA. Immune complexes were detected with Alexa 546-conjugated goat antibodies to either rabbit or mouse IgG (Molecular Probes), respectively.
Cell-Cycle Analysis. Cell-cycle analysis was performed as described (32).
Results
Generation of β-TrCP1–/– Mice. The mouse β-TrCP1 genomic locus comprises at least 15 exons spanning ≈170 kb on chromosome 19 (31). The targeting construct for the generation of β-TrCP1 knockout mice was designed to delete exon 1 of the β-TrCP1 gene, which includes the translation initiation site (Fig. 1 A). Embryonic stem cells were transfected with the linearized targeting vector, and recombinant clones were selected and injected into C57BL/6 blastocysts. Chimeric males that transmitted the mutant allele to the germ line were mated with C57BL/6 females, and the resulting heterozygotes were intercrossed to yield homozygous mutant mice, which were identified by Southern blot analysis of tail DNA (Fig. 1B). Mating of heterozygotes yielded β-TrCP1+/+, β-TrCP1+/–, and β-TrCP1–/– offspring in numbers that conformed approximately to the expected Mendelian ratio (data not shown). Northern blot analysis confirmed that the β-TrCP1–/– mice did not contain detectable β-TrCP1 mRNA (Fig. 1C).
Both male and female β-TrCP1–/– mice seemed as fertile as their wild-type littermates. Furthermore, we have not detected any health problems in the homozygous mutants up to 16 months of age, suggesting that the absence of β-TrCP1 does not increase the incidence of cancer despite the abnormal accumulation of β-catenin in the nucleus (see below).
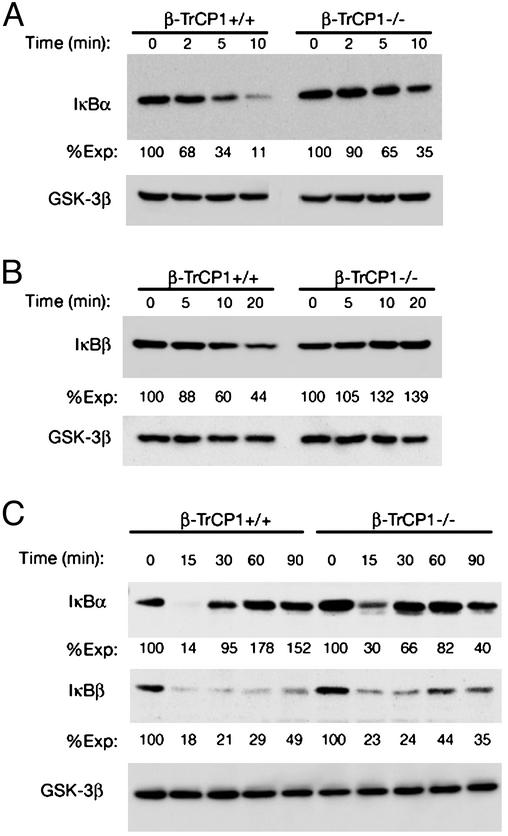
Impaired IκB Degradation in β-TrCP1–/– Cells. Mitogenic stimulation with a phorbol ester and Ca2+ ionophore induces degradation of IκBα and IκBβ in lymphocytes, with the degradation of IκBα occurring before that of IκBβ (21). We compared the degradation of IκBα and IκBβ between lymphocytes isolated from β-TrCP1+/+ mice and those from β-TrCP1–/– mice. The rates of degradation of IκBα (Fig. 2A) and IκBβ (Fig. 2B) were reduced greatly in the β-TrCP1–/– cells compared with those evident in the β-TrCP1+/+ cells. The down-regulation of IκBα and IκBβ induced by TNF-α in MEFs derived from β-TrCP1–/– mice was also inhibited markedly but not completely, relative to that apparent in the wild-type cells at 15 min after stimulation (Fig. 2C). Activation of NF-κB leads to up-regulation of transcription of the IκBα gene, and the consequent increase in the amount of IκBα serves to shut off the activation signal (21). Such an autoregulatory feedback loop is not present in the regulation of the IκBβ gene (21). It is likely that the partial inhibition of NF-κB activation in β-TrCP1–/– cells results in a lesser extent of IκBα up-regulation after its degradation.
Fig. 2.
Impaired degradation of IκBα and IκBβ in cells from β-TrCP1–/– mice. Splenocytes (A), thymocytes (B), and MEFs (C) derived from β-TrCP1–/– mice and their wild-type littermates were stimulated for the indicated times with the combination of 100 nM phorbol 12,13-dibutyrate and 300 nM ionomycin (A and B) or 10 ng/ml TNF-α (C). All cells then were lysed and subjected to immunoblot analysis with antibodies to IκBα, IκBβ, or glycogen synthase kinase 3β (GSK-3β, control) as indicated. The band intensity was measured by an LAS-1000 chemiluminescence imager, normalized by the expression of glycogen synthase kinase 3β, and shown as %Exp (the normalized intensity at 0 min is defined as 100). These data are representative of three or four independent animals.
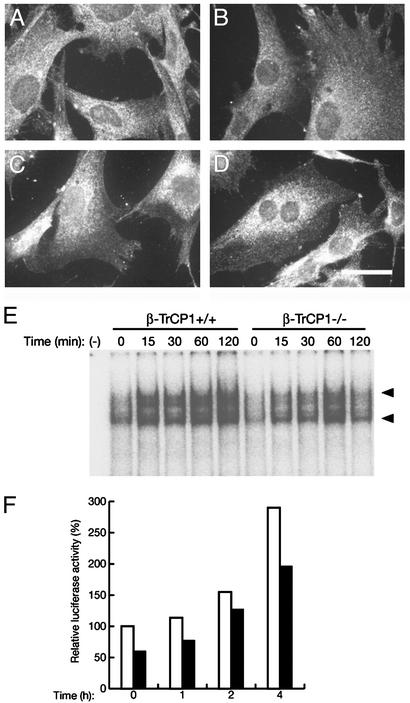
To confirm the impairment in the degradation of IκBs, we examined the nuclear translocation, DNA-binding activity, and transactivating ability of NF-κB. Under resting conditions, NF-κB was localized exclusively in the cytoplasm of both β-TrCP1+/+ and β-TrCP1–/– MEFs (Fig. 3 A and B). After stimulation with TNF-α, however, NF-κB translocated to the nucleus in wild-type MEFs, obscuring the boundary between the nucleus and the cytoplasm (Fig. 3C). The TNF-α-induced translocation of NF-κB to the nucleus appeared less pronounced in β-TrCP1–/– MEFs (Fig. 3D) than in the wild-type cells. EMSA analysis also revealed that the increase in the DNA-binding activity of NF-κB induced by the combination of ionomycin and phorbol 12,13-dibutyrate was slower and not as persistent in β-TrCP1–/– thymocytes compared with that in β-TrCP1+/+ cells, although the maximal activity did not seem to differ substantially between the two genotypes (Fig. 3E). Furthermore, the TNF-α-induced transactivation of a luciferase reporter gene by NF-κB was also inhibited partially in β-TrCP1–/– MEFs compared with that apparent in wild-type cells (Fig. 3F). These various observations thus suggest that NF-κB function is impaired partially in β-TrCP1–/– mice, probably as a result of the reduced rate of IκB degradation.
Fig. 3.
Reduced activity of NF-κB in β-TrCP1–/– cells. (A–D) Nuclear translocation of NF-κB in response to TNF-α. MEFs prepared from β-TrCP1+/+ (A and C) and β-TrCP1–/– (B and D) mice were incubated in the absence (A and B) or presence (C and D) of TNF-α. The cells then were subjected to immunofluorescence analysis with antibodies to the p65 (RelA) subunit of NF-κB. (Scale bar, 20 μm.) (E) DNA-binding activity of NF-κB. Thymocytes prepared from β-TrCP1+/+ and β-TrCP1–/– mice were stimulated for the indicated times with phorbol 12,13-dibutyrate and ionomycin, after which cell lysates were subjected to EMSA analysis with a probe specific for NF-κB. Arrowheads, positions of probe–protein complexes; –, incubation without cell lysate. (F) Activation of a luciferase reporter gene by NF-κB. MEFs derived from β-TrCP1+/+ (open bars) and β-TrCP1–/– (filled bars) mice were cotransfected with an NF-κB-dependent luciferase reporter construct and pRL-Tk. They then were stimulated with TNF-α for the indicated times, after which cell lysates were assayed for luciferase activities. Firefly luciferase activity was normalized by Renilla luciferase activity and then expressed as a percentage of the normalized value for β-TrCP1+/+ cells at time 0. Data are means of triplicates from an experiment that was repeated a total of three times with similar results.
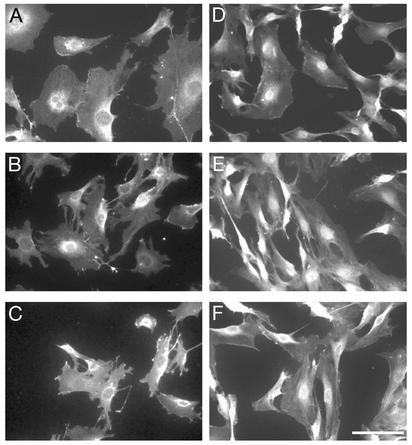
Nuclear Accumulation of β-Catenin in β-TrCP1–/– Cells. We next examined the expression of β-catenin, another target of ubiquitylation by SCFβ-TrCP1, in MEFs from β-TrCP1+/+ and β-TrCP1–/– mice. Although the total abundance of β-catenin in the cells appeared similar in cells of the two genotypes (data not shown), the subcellular localization of this protein differed markedly. Under basal conditions, β-catenin was localized to the cytoplasm of wild-type MEFs, with prominent perinuclear staining being apparent (Fig. 4A). Exposure of the wild-type cells to Wnt-3a for 2 h, however, resulted in translocation of β-catenin to the nucleus (Fig. 4B); the protein again was localized to the cytoplasm 6 h afterwithdrawal of Wnt-3a (Fig. 4C). In contrast, β-catenin was largely restricted to the nucleus of β-TrCP1–/– MEFs regardless of the absence or presence of Wnt-3a (Fig. 4 D and E); it was also retained in the nucleus 6 h after Wnt-3a withdrawal (Fig. 4F). These results suggest that β-TrCP1 contributes substantially to the degradation of nuclear β-catenin.
Fig. 4.
Persistent nuclear accumulation of β-catenin in β-TrCP1–/– MEFs. MEFs derived from β-TrCP1+/+ (A–C) and β-TrCP1–/– (D–F) mice were left untreated (A and D), incubated with Wnt-3a for 2 h (B and E), or cultured in Wnt-3a-free medium for 6 h after Wnt-3a stimulation (C and F). The cells then were subjected to immunofluorescence staining with antibodies to β-catenin. (Scale bar, 20 μm.)
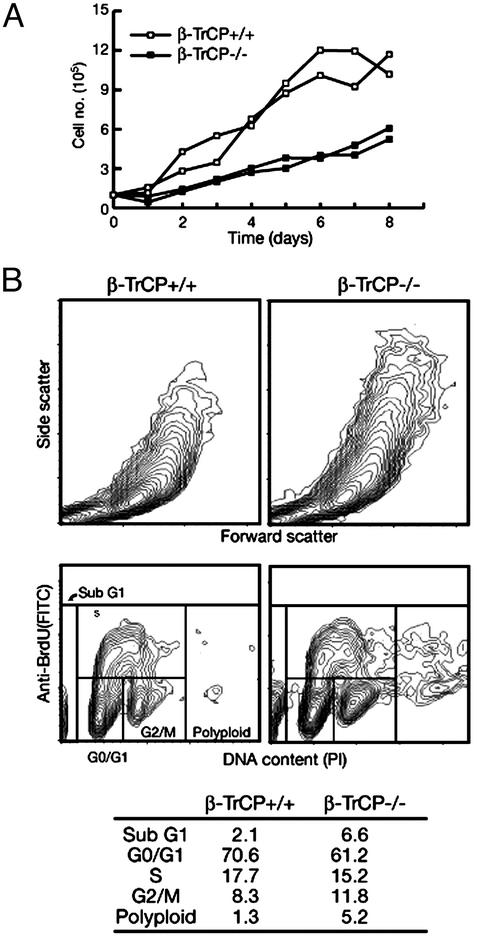
Reduced Growth Rate and Increased Ploidy of β-TrCP1–/– Cells. We also examined the growth properties of β-TrCP1+/+ and β-TrCP1–/– MEFs in culture. The rate of increase in the number of β-TrCP1–/– MEFs was reduced markedly compared with that of β-TrCP1+/+ MEFs (Fig. 5A). The rate of proliferation of lymphocytes derived from β-TrCP1–/– mice in response to various stimulants was also reduced greatly relative to that of those from β-TrCP1+/+ animals (data not shown). Flowcytometric analysis revealed that the size of β-TrCP1–/– MEFs, as estimated from forward and side scatter, was substantially larger than that of β-TrCP1+/+ MEFs (Fig. 5B). In addition, the percentages of both apoptotic (sub-G1 DNA content) and polyploid cells were increased for the mutant MEFs, suggesting that the reduced growth rate of the β-TrCP1–/– cells might be attributable, at least in part, to an increased frequency of apoptosis and polyploidy.
Fig. 5.
Reduced growth rate, increased size, and abnormal ploidy of β-TrCP1–/– cells. (A) Growth curves of β-TrCP1+/+ and β-TrCP1–/– MEFs at passage 6. Data are means of duplicate plates, with each curve corresponding to MEFs from an individual animal. (B) Representative flow-cytometric profiles for nonsynchronized β-TrCP1+/+ (Left) and β-TrCP1–/– (Right) MEFs of three independent embryos. (Upper) Forward and side scatter. (Lower) DNA content, as revealed by staining with propidium iodide (PI), as well as incorporation of bromodeoxyuridine (BrdU), as revealed by staining with FITC-conjugated antibodies to bromodeoxyuridine. The table shows the percentages of cells in each phase of the cell cycle.
Discussion
Degradation by the ubiquitin-proteasome pathway plays a fundamental role in determining the abundance of important regulatory proteins. E3 ubiquitin ligases are thought to determine substrate specificity in this pathway. Structural motifs identified in E3 enzymes include the HECT domain, RING-finger domain, U-box domain, F-box domain, and SOCS-box domain (1, 2, 33). Although database searches reveal numerous cDNAs encoding such domains, a relatively small number of specific targets of E3s has been identified. Gene targeting of E3 enzymes is one approach to elucidation of the enzyme-substrate relations. We generated mice lacking the F-box proteins Skp2 or mCdc4, which target p27 and Notch, respectively, for ubiquitylation, and showed that the degradation of these targets is impaired in the corresponding gene-ablated mice (34) (R. Tsunematsu and K.I.N., unpublished data). However, in some instances the degradation of putative substrates of certain E3s is not affected by E3 deficiency. The degradation of E2F-1, for example, appears normal in Skp2-deficient mice, and that of cyclin E is not altered in mCdc4-deficient mice (our unpublished observations). These results may be explained if other E3 enzymes also contribute to the degradation of such substrates.
We and others have shown by biochemical analyses that β-TrCP1 targets IκB proteins and β-catenin for ubiquitylation-dependent proteolysis only when the DSGXXS motif, which is conserved in IκB and β-catenin family members, is phosphorylated (18–29). If the turnover of these regulators of the NF-κB and Wnt signaling pathways was blocked completely in β-TrCP1-deficient mice, the mutant animals might have been expected to exhibit serious defects early during development. In contrast to the prominent morphological changes apparent in a slimb mutant of Drosophila (Slimb is a Drosophila homolog of β-TrCP1) (35), however, the gross phenotype of β-TrCP1–/– mice appeared normal. Systemic histopathologic examination also did not reveal any substantial abnormalities in the β-TrCP1-deficient mice (data not shown). The most likely explanation for this apparent normality is that β-TrCP2, which is highly related to β-TrCP1 (29), also contributes to the degradation of IκBs and β-catenin and therefore compensates for the loss of β-TrCP1 function. β-TrCP1 and β-TrCP2 form both homodimers and a heterodimer, although only the homodimers recognize IκBα. The disruption of the β-TrCP1 gene thus would result in a loss of function of the β-TrCP1 homodimer and the β-TrCP1–β-TrCP2 heterodimer. Further characterization of the functional differences among both homodimers and the heterodimer awaits the generation of mice deficient in β-TrCP2 and in both β-TrCP1 and β-TrCP2.
Nuclear translocation of β-catenin is thought to trigger cell proliferation through the induction of growth-promoting genes such as those for cyclin D1 (36, 37) and Myc (38). However, despite a prolonged nuclear localization of β-catenin in β-TrCP1–/– MEFs, the growth rate of these cells was reduced substantially compared with that of wild-type MEFs. The increased frequency of both apoptosis and polyploidy associated with this growth retardation may be indicative of a defect in mitosis or in surveillance of genomic integrity in β-TrCP1–/– cells. Although it remains to be determined whether the nuclear accumulation of β-catenin is related to the growth disadvantage of β-TrCP1–/– MEFs, it is possible that the elimination of β-catenin from the nucleus may be necessary for cell-cycle progression or for maintenance of genomic stability. In this regard, a fission yeast mutant lacking Pop1, an F-box protein with WD repeats (similar to β-TrCP1), also exhibits polyploidy (39). The cyclin-dependent kinase inhibitor Rum1 and the S phase regulator Cdc18 accumulate to high levels in this mutant. The maintenance of genome ploidy in fission yeast is controlled by at least two mechanisms, mediated by Cdc2–Cdc13 (B-type cyclin) and by Cdc18. It thus is likely that inhibition of Cdc2–Cdc13 kinase activity as a result of the increased abundance of Rum1 in the pol1 mutant leads to polyploidization through bypass of M phase (40). Given the similarities in molecular structure as well as in the mutant phenotypes of Pop1 and β-TrCP1, the polyploidy of β-TrCP1–/– cells may result from endoreplication, and the increased frequency of apoptosis in these cells may be due to the unbalanced segregation of chromosomes. We have not found evidence of cancer predisposition in the β-TrCP1–/– mice, suggesting that the polyploidy of β-TrCP1–/– cells does not lead directly to tumorigenesis. The possibility remains, however, that the loss of β-TrCP1 is a critical step in the development of certain human cancers.
Acknowledgments
We thank H. Imaki, Y. Yamada, K. Shimoharada, R. Yasukochi, and other laboratory members for technical assistance; M. Pagano for communicating his results before publication; and T. Verjee, C. Sugita, and M. Kimura for help in the preparation of the manuscript. We also thank Ms. Tazim Verjee, editorial associate, for assistance. This work was supported in part by a grant from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations: SCF, Skp1/Cul1/F-box; IκB, inhibitory subunit of NF-κB; PGK, phosphoglycerate kinase; MEF, mouse embryonic fibroblast; TNF-α, tumor necrosis factor α.
References
- 1.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178. [DOI] [PubMed] [Google Scholar]
- 2.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 3.Hershko, A. (1983) Cell 34, 11–12. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover, A., Orian, A. & Schwartz, A. L. (2000) BioEssays 22, 442–451. [DOI] [PubMed] [Google Scholar]
- 5.Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W. & Elledge, S. J. (1996) Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- 6.Feldman, R. M., Correll, C. C., Kaplan, K. B. & Deshaies, R. J. (1997) Cell 91, 221–230. [DOI] [PubMed] [Google Scholar]
- 7.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- 8.Kamura, T., Koepp, D. M., Conrad, M. N., Skowyra, D., Moreland, R. J., Iliopoulos, O., Lane, W. S., Kaelin, W. G., Jr., Elledge, S. J., Conaway, R. C., et al. (1999) Science 284, 657–661. [DOI] [PubMed] [Google Scholar]
- 9.Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J. & Harper, J. W. (1999) Science 284, 662–665. [DOI] [PubMed] [Google Scholar]
- 10.Ohta, T., Michel, J. J., Schottelius, A. J. & Xiong, Y. (1999) Mol. Cell 3, 535–541. [DOI] [PubMed] [Google Scholar]
- 11.Tan, P., Fuchs, S. Y., Chen, A., Wu, K., Gomez, C., Ronai, Z. & Pan, Z. Q. (1999) Mol. Cell 3, 527–533. [DOI] [PubMed] [Google Scholar]
- 12.Seol, J. H., Feldman, R. M., Zachariae, W., Shevchenko, A., Correll, C. C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., et al. (1999) Genes Dev. 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cenciarelli, C., Chiaur, D. S., Guardavaccaro, D., Parks, W., Vidal, M. & Pagano, M. (1999) Curr. Biol. 9, 1177–1179. [DOI] [PubMed] [Google Scholar]
- 14.Winston, J. T., Koepp, D. M., Zhu, C., Elledge, S. J. & Harper, J. W. (1999) Curr. Biol. 9, 1180–1182. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 16.May, M. J. & Ghosh, S. (1998) Immunol. Today 19, 80–88. [DOI] [PubMed] [Google Scholar]
- 17.Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. (1997) EMBO J. 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattori, K., Hatakeyama, S., Shirane, M., Matsumoto, M. & Nakayama, K. I. (1999) J. Biol. Chem. 274, 29641–29647. [DOI] [PubMed] [Google Scholar]
- 19.Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., Andersen, J. S., Mann, M., Mercurio, F. & Ben-Neriah, Y. (1998) Nature 396, 590–594. [DOI] [PubMed] [Google Scholar]
- 20.Hatakeyama, S., Kitagawa, M., Nakayama, K., Shirane, M., Matsumoto, M., Hattori, K., Higashi, H., Nakano, H., Okumura, K., Onoe, K., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 3859–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirane, M., Hatakeyama, S., Hattori, K., Nakayama, K. & Nakayama, K. I. (1999) J. Biol. Chem. 274, 28169–28174. [DOI] [PubMed] [Google Scholar]
- 22.Kroll, M., Margottin, F., Kohl, A., Renard, P., Durand, H., Concordet, J. P., Bachelerie, F., Arenzana-Seisdedos, F. & Benarous, R. (1999) J. Biol. Chem. 274, 7941–7945. [DOI] [PubMed] [Google Scholar]
- 23.Spencer, E., Jiang, J. & Chen, Z. J. (1999) Genes Dev. 13, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston, J. T., Strack, P., Beer-Romero, P., Chu, C. Y., Elledge, S. J. & Harper, J. W. (1999) Genes Dev. 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, C. & Ghosh, S. (1999) J. Biol. Chem. 274, 29591–29594. [DOI] [PubMed] [Google Scholar]
- 26.Hart, M., Concordet, J. P., Lassot, I., Albert, I., del los Santos, R., Durand, H., Perret, C., Rubinfeld, B., Margottin, F., Benarous, R. & Polakis, P. (1999) Curr. Biol. 9, 207–210. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa, M., Hatakeyama, S., Shirane, M., Matsumoto, M., Ishida, N., Hattori, K., Nakamichi, I., Kikuchi, A., Nakayama, K. I. & Nakayama, K. (1999) EMBO J. 18, 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latres, E., Chiaur, D. S. & Pagano, M. (1999) Oncogene 18, 849–854. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, H., Chiba, T., Kobayashi, M., Takeuchi, M., Furuichi, K. & Tanaka, K. (1999) Biochem. Biophys. Res. Commun. 256, 121–126. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzawa, S. I. & Reed, J. C. (2001) Mol. Cell 7, 915–926. [DOI] [PubMed] [Google Scholar]
- 31.Maruyama, S., Hatakeyama, S., Nakayama, K., Ishida, N., Kawakami, K. & Nakayama, K. I. (2001) Genomics 78, 214–222. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama, K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I., Loh, D. Y. & Nakayama, K. I. (1996) Cell 85, 707–720. [DOI] [PubMed] [Google Scholar]
- 33.Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N. & Nakayama, K. I. (2001) J. Biol. Chem. 276, 33111–33120. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama, K., Nagahama, H., Minamishima, Y. A., Matsumoto, M., Nakamichi, I., Kitagawa, K., Shirane, M., Tsunematsu, R., Tsukiyama, T., Ishida, N., et al. (2000) EMBO J. 19, 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, J. & Struhl, G. (1998) Nature 391, 493–496. [DOI] [PubMed] [Google Scholar]
- 36.Tetsu, O. & McCormick, F. (1999) Nature 398, 422–426. [DOI] [PubMed] [Google Scholar]
- 37.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96, 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- 39.Kominami, K. & Toda, T. (1997) Genes Dev. 11, 1548–1560. [DOI] [PubMed] [Google Scholar]
- 40.Correa-Bordes, J. & Nurse, P. (1995) Cell 83, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto, A., Nakayama, K., Imaki, H., Hirose, S., Jiang, Y., Abe, M., Tsukiyama, T., Nagahama, H., Ohno, S., Hatakeyama, S., et al. (2002) Nature 416, 865–869. [DOI] [PubMed] [Google Scholar]
- 42.Shibamoto, S., Higano, K., Takada, R., Ito, F., Takeichi, M. & Takada, S. (1998) Genes Cells 3, 659–670. [DOI] [PubMed] [Google Scholar]