Abstract
Previous in vitro studies showed that the bromodomain binds to acetyllysines on histone tails, leading to the proposal that the domain is involved in deciphering the histone code. However, there is little in vivo evidence supporting the binding of bromodomains to acetylated chromatin in the native environment. Brd4 is a member of the BET family that carries two bromodomains. It associates with mitotic chromosomes, a feature characteristic of the family. Here, we studied the interaction of Brd4 with chromatin in living cells by photobleaching. Brd4 was mobile and interacted with chromatin with a rapid “on and off” mode of binding. This interaction required both bromodomains. Indicating a preferential interaction with acetylated chromatin, Brd4 became less mobile upon increased chromatin acetylation caused by a histone deacetylase inhibitor. Providing biochemical support, salt solubility of Brd4 was markedly reduced upon increased histone acetylation. This change also required both bromodomains. In peptide binding assays, Brd4 avidly bound to di- and tetraacetylated histone H4 and diacetylated H3, but weakly or not at all to mono- and unacetylated H3 and H4. By contrast, it did not bind to unacetylated H4 or H3. Further, Brd4 colocalized with acetylated H4 and H3 in noncentromeric regions of mitotic chromosomes. This colocalization also required both bromodomains. These observations indicate that Brd4 specifically recognizes acetylated histone codes, and this recognition is passed onto the chromatin of newly divided cells.
Acetylation of lysines on histone tails is thought to form distinct histone codes that direct molecular processes important for transcription (1, 2). A bromodomain is a motif present in a number of chromatin-modifying proteins including histone acetylases of the GNAT family, CBP/p300, general transcription factors including TAFII250, and chromatin remodeling factors of the SWI/SNF family (3, 4). Structural analyses in vitro have shown that the bromodomain is composed of four α-helices and binds to acetylated lysines on histone H3 and H4, although with relatively low affinity (5–9). Based on these studies, the bromodomain has been proposed to act as a chromatin targeting module, deciphering histone acetylation codes (1, 2, 10). However, despite in vitro evidence, it has not been clear whether bromodomain proteins interact with acetylated chromatin in the native nuclear environment in vivo. Besides the question of in vivo interaction, it has not been clear whether differentially acetylated histones are distinguished by bromodomains. The latter question is of interest in view of the fact that bromodomains of different proteins have considerable structural diversity (3, 4). Furthermore, histone acetylation codes are likely to be diverse and translated into distinct processes, as individual lysines on histone H3 and H4 are acetylated in a highly specific and ordered fashion during transcription (11, 12). In addition to transcription, histone acetylation codes may play a role in cell growth, as H3 and H4 are transiently acetylated during replication (13–15). So far, however, studies on the interaction of bromodomains with acetylated histones have been limited to those of a few select proteins, such as PCAF, GCN5, and TAFII250 (5–7, 9), and more recently yeast Bdf1 and Bdf2 (16, 17). The latter two proteins belong to the BET family, whose members carry two tandem bromodomains (3, 4). Bromodomains of the BET family differ substantially from those of other families in amino acid sequence, although they share conserved hydrophobic residues in the α-helices. This group has been studying Brd4, a mammalian member of the family (18, 19) (see Fig. 2 A for scheme). The family also includes Drosophila fsh (20) and mammalian Brd2 (formally RING3/fsrg1) (21, 22). Like other members (21, 23, 24), Brd4 is involved in cell growth and presumably in transcription (18, 19, 25). The most notable feature of Brd4 is that it associates with mitotic chromosomes (18). The retention on chromosomes during mitosis is likely to be a distinctive feature of the BET family (26), in that proteins of other bromodomain families are displaced from the chromosomes during mitosis (27–29). The displacement of these and other transcription factors is a prominent feature of mitosis, and it coincides with marked hypoacetylation of core histones and general transcriptional repression seen in all higher eukaryotes (29–31).
Fig. 2.
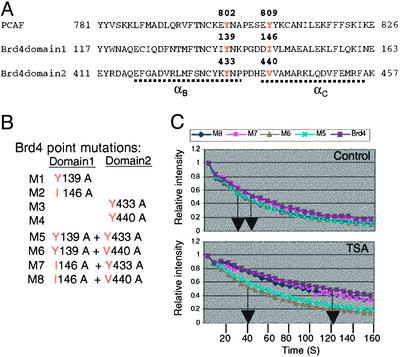
FLIP analysis of bromodomain point mutants. (A) Bromodomain sequence comparison. (B) Bromodomain point mutants tested in this study. (C) FLIP analysis of mutants tested without (control, Upper) or with (Lower) TSA treatment. T1/2 of M5 and M6 was ≈120 s, whereas that of wild-type Brd4 and other mutants was ≈120 s. None of single point mutants showed a discernible difference in the FLIP pattern (data not shown).
Fluorescence photobleaching has been used to visualize real-time movement of a nuclear protein and its chromatin association in living cells (32, 33). These studies revealed that many nuclear proteins, including DNA repair proteins, nuclear receptors, histone H1, a splicing factor, and RNA polymerase I components, are highly mobile within the nucleus and transiently interact with chromatin by a “stop and go” type of binding (34–42). On the other hand, core histones are found to be essentially immobile and remain as a stable component of chromatin (18, 32, 33, 43).
By fluorescence loss in photobleaching (FLIP) experiments, we found that Brd4 interacts with acetylated chromatin in living cells in a bromodomain-dependent manner. These results were supported by subsequent biochemical analyses. Peptide binding experiments revealed that Brd4 interacts with H4 and H3 through acetylated lysine residues, in line with the idea that bromodomains recognize acetylated histone codes. Finally, we show that Brd4 associates with noncentromeric regions of mitotic chromosomes, where some H4 and H3 remain acetylated during mitosis in a bromodomain-dependent manner. Our results indicate that the recognition of acetylated histones by bromodomains persists through mitosis and is passed from one generation of cells to the next.
Materials and Methods
Plasmids and Antibodies. Plasmid vectors for EGFP-Brd4 and deletion mutants have been described (19). EGFP-PCAF and EGFP-Brd2 vectors were constructed by inserting full-length PCAF or Brd2 cDNA into pEGFP-N1 vector. Bromodomain point mutations were generated from full-length Brd4-pEGFPC1 vector by using a mutagenesis kit (Quikchange, Stratagene). Antibodies against Brd4 have been described (18). Rabbit antibody against Brd2 was produced by using recombinant murine Brd2 as an immunogen. Antibodies to acetyl H4 and acetyl H3 histones were obtained from Upstate Biotechnology (Lake Placid, NY); those to transcription factor IIB (TFIIB) and GFP were from Santa Cruz Biotechnology.
Live Cell Microscopy, FLIP Assay, and Immunofluorescence Staining. Murine P19 embryonal carcinoma cells (18, 44, 45) plated at ≈40% confluency on Lab-Tek (Nalge) chambered coverslips were transfected with pEGFP vectors by using Lipofectamine Plus (Invitrogen) for 16–20 h. Some cultures were treated with 50 ng/ml trichostatin A (TSA) for 4 h before harvest. GFP proteins in live cells were detected on a Leica TCS-SP confocal microscope with the 488-nm excitation line of an argon laser. For indirect immunofluorescent staining, cells were fixed in 4% paraformaldehyde, permeabilized in methanol, and incubated for1hwithantibody for Brd4, Brd2 (both 1:250), or acetylated H4 or H3 (1:1,000, Upstate Biotechnology), followed by FITC- or rhodamine-conjugated anti-rabbit antibody. Cells were mounted with gel mount (Biomeda, Foster City, CA), and images were acquired in Leica SP2 confocal microscope. For FLIP experiments, a circular spot 0.5 μm in diameter was repeatedly bleached for 500 ms with intervals of 5 s, and fluorescence intensity was recorded in an area away from the bleached spot (38).
Differential Salt Extraction. Exponentially growing cells (107 cells) transfected with pEGFP-Brd4 or Brd4 deletions were suspended in buffer A, which contains 10 mM Tris·HCl at pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 2.5 mM DTT, and protease inhibitor mixture (Roche Biochemicals), incubated on ice for 10 min, and centrifuged. Nuclear pellets were resuspended in buffer A, and aliquots were then incubated with increasing concentrations of NaCl from 10 mM up to 1 M. Eluted materials were resolved on SDS/4–20% PAGE and immunoblotted.
Peptide Binding Assay. Ten micrograms of nuclear extracts was incubated with 2 μg of biotin-labeled synthetic peptides corresponding to the N-terminal tail of histone H3 and H4 (Upstate Biotechnology) in a buffer containing 50 mM Tris·HCl at pH 7.5, 15 mM MgCl2, 150 mM NaCl, 0.5 mM DTT, and 0.1% Nonidet P-40 for 2 h at 4°C, followed by incubation with 20 μl of M-280 streptavidin beads (Dynal). Beads were washed, and bound materials were eluted with 2× sample buffer. Bound materials were resolved on SDS/PAGE and immunoblotted as above.
Results
Dynamic Interaction of Brd4 with Acetylated Chromatin in Living Cells. Photobleaching techniques offer a powerful means to monitor the mobility of a protein in the nucleus in real time and its interaction with chromatin (32, 33). To visualize the movement of Brd4 in living cells, we used the FLIP assays using P19 embryonal carcinoma cells transiently transfected with Brd4 fused to GFP. The following observations justified the use of GFP-Brd4 for the present study. GFP-Brd4 expressed in these cells was similar to the endogenous protein in terms of intranuclear distribution, mitotic chromosome localization (18), and salt elution profile (see Fig. 3). In addition, GFP-Brd4 caused the same effects as untagged Brd4 on cell-cycle progression from G1 to S and proliferation (19). A small area within the nucleus (Fig. 1A, circle) was repeatedly photobleached, and the loss of fluorescence was monitored in an area outside the bleached spot (Fig. 1 A, rectangle). Loss of GFP-Brd4 fluorescence was compared with that of free GFP, the latter of which distributed throughout the cells, including the cytoplasm. Photobleaching caused an immediate loss of free GFP fluorescence (see 26 s in Fig. 1 A), followed by a further loss that spread to the entire cell by 106 s, indicating the previously established, rapid movement of free GFP. GFP-Brd4 also showed high mobility, as evidenced by a loss of fluorescence in the nucleus over 106 s, although the loss occurred more slowly than that of free GFP. This mobility was a characteristic of living cells because, in fixed cells, fluorescence loss was confined to the bleached spot and did not spread to the rest of nucleus (Fig. 1 A Bottom). Fig. 1B shows the kinetics of fluorescence loss. Free GFP declined rapidly, with the time required for 50% fluorescence loss (T1/2) of ≈20 s, whereas GFP-Brd4 followed slower kinetics with a T1/2 of ≈55 s. These results indicate that the majority of Brd4 is highly mobile in the nucleus and associates with chromatin with a rapid “on and off” interaction, a behavior similar to other nuclear factors (36, 37, 39). In addition, GFP-Brd4 fluorescence did not seem to be entirely lost during this period, as a low level of fluorescence remained even 180 s after bleaching.
Fig. 3.
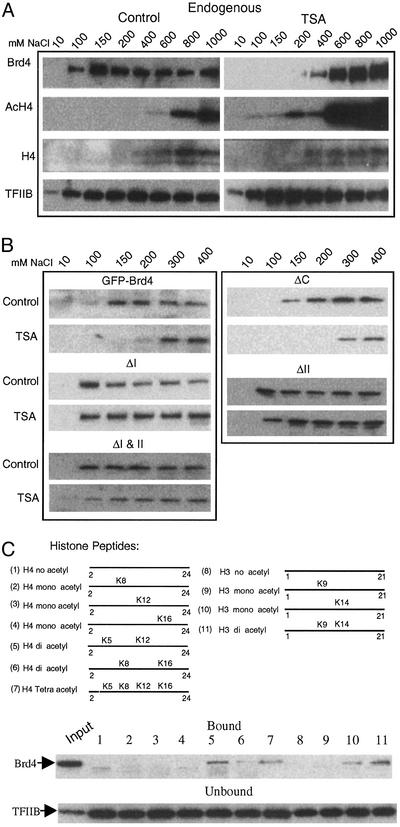
Solubility of Brd4 in differential salt extraction. (A) Endogenous Brd4 was extracted with indicated concentrations of NaCl and detected by immunoblot. (B) Transfected GFP-Brd4 or GFP-Brd4 deletions were extracted and tested as in A. (C) Histone tail peptides tested for Brd4 binding (Top). Bound Brd4 was detected by immunoblot (Middle). TFIIB in unbound materials was detected to verify equal loading (Bottom).
Fig. 1.
FLIP analysis of GFP-Brd4. Murine P19 embryonal carcinoma cells expressing GFP-Brd4 or free GFP were photobleached in the circled area, and loss of fluorescence was monitored in the rectangle. (A) Distribution of GFP proteins before and after photobleaching. Bleaching of free GFP (Top) and GFP-Brd4 was performed with live (Middle) or fixed (Bottom) cells. (Bar indicates 3.6 μm.) (B) Quantification of fluorescence loss in A. Values represent the average of eight independent measurements ± SD. Arrows indicate the time (s) required for 50% fluorescence loss (T1/2). (C) Diagram of Brd4 deletions. Solid blocks (BD1 and BD2) mark bromodomains; shaded blocks mark the ET domain. (D) Distribution of indicated GFP proteins in cells pretreated with 50 ng/ml TSA for 4 h. (E) FLIP patterns for indicated GFP proteins without (control) or with TSA treatment.
We next asked two questions: (i) whether a change in chromatin acetylation affects the mobility of GFP-Brd4 and (ii) whether the bromodomains play a role in controlling the mobility of Brd4. To this end, FLIP analysis was performed with cells treated with a histone deacetylase inhibitor, TSA for 4 h, and with GFP-Brd4 deletions lacking one or both bromodomains (Fig. 1C). TSA treatment increased core histone acetylation in P19 cells (see Fig. 3) (44, 45). In Fig. 1D, TSA reduced the mobility of wild-type GFP-Brd4, as seen by a very slow fluorescence loss outside the bleached spot. Quantification in Fig. 1E showed that GFP-Brd4 fluorescence was lost more slowly in TSA-treated cells with a marked increase in T1/2 (from 60 s to 105 s after TSA), indicating the presence of a relatively immobile component. An increase in the relatively immobile component after TSA treatment was verified by extending FLIP measurements from 120 s to 250 s (not shown). These data indicate that a fraction of Brd4 that stably associated with chromatin was increased by TSA, consistent with an affinity of Brd4 for acetylated chromatin. In contrast, deletion of one bromodomain led to a striking increase in the mobility (Fig. 1D). Quantification in Fig. 1E showed that deletion of either bromodomain (ΔI or ΔII) strongly increased the mobility of Brd4 even in untreated cells. Moreover, TSA failed to alter the mobility of these deletions, resulting in a T1/2 of 25–30 s both with and without TSA. Deletion of both bromodomains (ΔI and II) gave essentially the same outcome as single bromodomain deletions. Conversely, ΔC, lacking the C-terminal domain but retaining both bromodomains, showed a decreased mobility relative to the wild-type Brd4 both with or without TSA, suggesting a negative role of the C-terminal domain in the interaction with chromatin. These results indicate that an increase in core histone acetylation increases Brd4-chromatin association, for which both bromodomains are essential. We also compared the FLIP pattern of Brd4 with that of histone acetylase PCAF (Fig. 1 D and E) (5). The mobility of PCAF was significantly higher than Brd4 in untreated cells (T1/2, 25 s), and TSA treatment did not alter the mobility. On the other hand, Brd2, another BET member (21, 22), showed a FLIP pattern very similar to that of Brd4 (Fig. 1E), supporting the view that interaction with acetylated chromatin is a common function of the BET family.
Bromodomain Mutation Analysis. Previous in vitro studies indicated that the bromodomain binds to acetyllysines through specific hydrophobic residues in the α-helices (6–9, 46). In PCAF, Tyr-809 is shown to be the most critical residue for the interaction, although Tyr-802 also contributes to the binding (5). As seen in Fig. 2A, Tyr-802 in PCAF corresponds to Tyr-139 and Tyr-433 in Brd4, indicating conservation of Tyr in this position. Tyr-809 of PCAF corresponds to Ile-146 and Val-440, indicating substitutions in this position. To begin to identify residues within Brd4 important for the interaction with acetylated chromatin, we tested a series of mutant GFP-Brd4 in FLIP. Mutants M1 through M4 had a point mutation in one of the two bromodomains, whereas M5 through M8 had point mutations in both bromodomains (Fig. 2B). The double bromodomain mutants M5 and M6 showed an increased mobility over wild-type Brd4 (Fig. 2C), particularly noticeable after TSA treatment. However, none of the single domain point mutations showed a detectable change in the FLIP patterns with or without TSA (not shown). Thus, mutations in both domains, but not in one domain, impaired the interaction with acetylated chromatin. On the other hand, the double mutants M7 and M8 exhibited FLIP patterns essentially identical to that of wild-type Brd4, suggesting differential roles of conserved Tyr residues.
TSA Treatment Alters Salt Solubility of Brd4. To explore biochemical evidence for the interaction of Brd4 with acetylated chromatin, we next carried out differential salt extraction experiments. Salt solubility has been used to monitor binding of proteins to the chromatin compartment (47). Nuclear preparations from untreated or TSA-treated cells were extracted with increasing concentrations of NaCl, and each fraction was tested for endogenous Brd4 by immunoblot (Fig. 3A). In untreated cells, Brd4 was first eluted at 100 mM NaCl, followed by peak elution at around 150–200 mM. In TSA-treated cells, Brd4 was not eluted until the NaCl concentration was increased to 200–400 mM, with the peak elution observed at 600 mM, indicating a shift in salt concentration needed for efficient elution of Brd4. The “right” shift in the Brd4 elution profile after TSA treatment was not due to a general change in protein solubility, as the elution profile of TFIIB was not significantly affected by TSA. Further, histone H3 and H4, partially eluted at higher NaCl concentrations, showed greater levels of acetylation after TSA relative to untreated samples, consistent with the effect of TSA (44). These data are in agreement with FLIP results above and indicate that increased histone acetylation stabilizes Brd4 association with chromatin. If the differential salt extraction data above are indeed a biochemical confirmation of FLIP results, one may predict that (i) a deletion of a bromodomain would lower NaCl concentration necessary for elution and (ii) TSA treatment would not change the elution profile of the bromodomain deletions. To test these predictions, we performed differential salt extraction experiments for transfected GFP-Brd4 deletions (Fig. 3B). As expected, the elution profiles of the wild-type GFP-Brd4 were very similar to those of the endogenous Brd4 with or without TSA treatment. In contrast, all three bromodomain deletions (ΔI, ΔII, and ΔI and II) were eluted at a NaCl concentration as low as 100 mM. Further, TSA treatment did not change their elution profiles, indicating that bromodomain deletions reduce the interaction with chromatin and that TSA did not affect the interaction. On the other hand, ΔC was eluted in a manner similar to wild-type Brd4 with and without TSA treatment. These results show that Brd4 preferentially associates with acetylated chromatin, for which both bromodomains are required.
Binding of Brd4 to Acetylated Histone H4 Peptides. To ascertain whether Brd4 distinguishes acetylated from unacetylated chromatin, we tested binding of Brd4 to synthetic histone tail peptides in which lysine residues were differentially acetylated (diagram in Fig. 3C Top). As seen in Fig. 3C Middle, Brd4 avidly bound to diacetylated H4 peptides (acetylated at Lys-5 and Lys-12) and those acetylated at all four Lys resides (tetraacetyl H4). In contrast, Brd4 did not bind to unacetylated H4, nor those acetylated at single resides. Brd4 also bound to diacetylated H3, but it did not bind to unacetylated H3, nor to singly acetylated H3 at Lys-9, although it bound weakly to H3 acetylated at Lys-14. TFIIB, detected in the unbound fraction, confirmed equal loading of extracts (Fig. 3B Bottom). These results indicate that Brd4 recognizes specific patterns of acetyl H4 and H3 and not unacetylated histone counterparts.
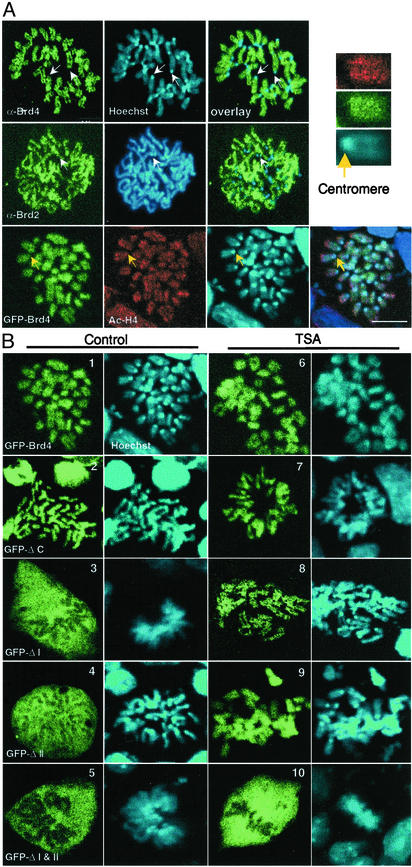
Association of Brd4 with Mitotic Chromosomes Requires Both Bromodomains. Core histones are underacetylated during mitosis, which coincides with the displacement of transcription factors as well as bromodomain proteins from chromosomes (29). Despite the global displacement of proteins from chromosomes, BET family proteins persist on mitotic chromosomes. This displacement is verified in Fig. 4A, where immunostaining localized the endogenous Brd4 as well as Brd2 to the noncentromeric regions of mitotic chromosomes. Little to no residual staining was found in the extrachromosomal regions for these proteins. Fig. 4A also shows that transfected GFP-Brd4 localizes to mitotic chromosomes. Like the endogenous Brd4, it outlined the chromosomal arms except for centromeres. In addition, acetyl H4 staining was detected on the regions overlapping with Brd4 (see enlargement in Fig. 4A). Similarly, acetyl H3 was stained in these regions, as reported (refs. 13 and 29 and data not shown).
Fig. 4.
Chromosomal localization of Brd4 and Brd2. (A) Endogenous Brd4, Brd2, or acetylated H4 was visualized on mitotic chromosomes of P19 cells by indirect immunostaining. DNA was counterstained by Hoechst 33342. Arrows indicate centromeres where staining of Brd4, Brd2, or acetyl H4 was absent. (Bar indicates 8 μm.) (Upper Right) Enlargement of chromosome stained for acetyl H4 (red), Brd4 (green), and DNA (blue). Yellow arrow indicates a centromere. (B) Distributions of transfected GFP-Brd4 and deletions were analyzed on mitotic chromosomes prepared from cells treated with or without TSA.
To assess whether association of Brd4 with chromosomes depends on the bromodomains, we next examined localization of GFP-Brd4 deletions (Fig. 4B Left). Both single bromodomain deletions (ΔI and ΔII) and the double bromodomain deletion (ΔI and II) failed to localize to mitotic chromosomes; they were dispersed in the extrachromosomal space, indicating that both bromodomains are needed for chromosomal localization of Brd4, although ΔC, lacking the C-terminal domain localized to chromosomes, was similar to the wild-type Brd4. Cells treated with TSA can undergo mitosis even though chromatin remains hyperacetylated (29). We studied whether bromodomain deletions localize to chromosomes in TSA-treated cells (Fig. 4B Right). Contrary to what was observed with untreated cells, both of the single bromodomain deletions showed clear chromosomal localization after TSA treatment. However, the double bromodomain deletion remained dispersed in the nonchromosomal area. Thus, single bromodomain deletions regained the capacity to localize to mitotic chromosome after TSA treatment, unlike the lack of recovery of the FLIP mobility by these deletions during interphase.
Discussion
Interaction of Brd4 with Acetylated Chromatin. Photobleaching experiments showed that the majority of Brd4 is mobile in the nucleus and interacts with chromatin with a rapid on and off mode of binding, similar to other nuclear proteins (34–37, 39, 41, 42). This binding required both bromodomains. The observation that deletion of a single bromodomain markedly increased the mobility of Brd4, which could not be altered by TSA, argues that Brd4 preferentially recognizes acetylated chromatin. The observation that deletion of a single bromodomain enhanced the salt solubility of Brd4, which was not affected by TSA, supports this argument. Consistent with this, the two bromodomains did not elicit an additive effect in both FLIP assay and biochemical experiments, indicating that the two domains cooperate with one another and act as one functional motif to recognize acetylated chromatin. That both bromodomains are needed for interaction may not be surprising, given that the first and second bromodomains of Brd4 are somewhat dissimilar in amino acid sequence, despite that each domain is similar to the corresponding domain of other BET proteins (4, 18). Histone peptide binding assays in Fig. 3C confirmed that Brd4 binds only to acetylated H4 and H3, not to unacetylated histones, and indicated that Brd4 recognizes distinct acetylation patterns on H4 and H3. Interestingly, in a similar binding assay, the related protein Brd2 (21, 22) showed binding only to acetylated H4 but not H3 (data not shown). This difference between Brd2 and Brd4 may not be surprising, considering that substantial sequence diversity exists among bromodomains (4), supporting the idea that different bromodomains recognize different sets of acetylated histones as distinct codes. In accordance, TAFII250, another double bromodomain protein, is shown to preferentially bind to acetylated histone H3 on an activated promoter in vivo (11). Further, recent studies (16, 17) indicate that although yeast Bdf1 binds to acetylated H4 and H3, the related Bdf2 interacts with histones with a different specificity. Residue-specific recognition of acetylated histones by different bromodomain proteins likely represents important regulatory processes governing the final outcome of transcription (11, 12, 16, 17). Collectively, these results are consistent with the histone code hypothesis, in which covalent histone modifications are individually deciphered and translated into distinct functions.
Association of Brd4 with Mitotic Chromosomes and Possible Role in the Transmission of Memory Across Cell Division. We show that both the endogenous Brd4 and transfected GFP-Brd4 associate with mitotic chromosomes. Similarly, the endogenous Brd2 localized to chromosomes, as was shown for yeast Bdf1 (26). Thus, association with mitotic chromosomes seems to be a key feature that defines the BET family. Paralleling the results with interphase cells, association with chromosomes required both bromodomains, as deletion of a single bromodomain eliminated the association. These results indicate that the mechanism by which Brd4 associates with mitotic chromosomes is shared by that of the interaction with acetylated chromatin during interphase. In line with this idea, Brd4 localized to the noncentromeric regions of mitotic chromosomes that coincided with acetylated histones. Nevertheless, the observation that a single bromodomain deletion regained the ability to associate with mitotic chromosomes upon TSA treatment, unlike what was observed with interphase cells, suggests that localization of Brd4 to mitotic chromosomes may involve an additional, mitosis-specific mechanism. For example, extensive chromatin packing during mitosis may help stabilize association with Brd4.
That Brd4 and Brd2 remain on chromosomes during mitosis contrasts with the behavior of other bromodomain proteins in that Brg1, Brm, PCAF, GCN5, P300/CBP, and TAFII250 are all displaced from chromosomes during mitosis in mammalian cells (18, 27, 29). This displacement is coincidental with the global hypoacetylation of core histones and general transcription repression seen in higher eukaryotes (29, 30). The large-scale chromatin hypoacetylation and displacement of bromodomain proteins would predict that histone acetylation codes and code-reading activities, if they exist, are largely erased during mitosis in higher eukaryotes. However, some H4 and H3 remain acetylated during mitosis (29). It is possible that histones that remain acetylated during mitosis contribute to the transmission of histone acetylation codes across cell division through the interaction with Brd4. This possibility is worth noting in light of the recent evidence from yeast suggesting that bromodomains of SWI/SNF and histone acetyltransferase contribute to the maintenance of an epigenetic memory for active transcription (48). The mechanism of cellular memory in yeast is, in all likelihood, quite different from that in mammalian cells, as in yeast transcription continues during mitosis (30). Nevertheless, the report may support the role for a bromodomain in the transmission of relevant information across cell division in higher eukaryotes as well.
Together, the double bromodomain of Brd4 is a functional unit that recognizes histone acetylation codes, whose activity persists during mitosis. It is likely that Brd4 and related proteins contribute to the transmission of transcriptional memory from one generation of cells to the next.
Acknowledgments
We thank L. Balsam for participation in the early stage of this project and D. Allis, D. Reinberg, and members of the Laboratory of Molecular Growth Regulation for discussion.
Abbreviations: FLIP, fluorescence loss in photobleaching; TSA, trichostatin A; TFIIB, transcription factor IIB.
References
- 1.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 3.Haynes, S. R., Dollard, C., Winston, F., Beck, S., Trowsdale, J. & Dawid, I. B. (1992) Nucleic Acids Res. 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeanmougin, F., Wurtz, J. M., Le Douarin, B., Chambon, P. & Losson, R. (1997) Trends Biochem. Sci. 22, 151–153. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M. M. (1999) Nature 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 6.Ornaghi, P., Ballario, P., Lena, A. M., Gonzalez, A. & Filetici, P. (1999) J. Mol. Biol. 287, 1–7. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. (2000) Science 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- 8.Hudson, B. P., Martinez-Yamout, M. A., Dyson, H. J. & Wright, P. E. (2000) J. Mol. Biol. 304, 355–370. [DOI] [PubMed] [Google Scholar]
- 9.Owen, D. J., Ornaghi, P., Yang, J. C., Lowe, N., Evans, P. R., Ballario, P., Neuhaus, D., Filetici, P. & Travers, A. A. (2000) EMBO J. 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winston, F. & Allis, C. D. (1999) Nat. Struct. Biol. 6, 601–604. [DOI] [PubMed] [Google Scholar]
- 11.Agalioti, T., Chen, G. & Thanos, D. (2002) Cell 111, 381–392. [DOI] [PubMed] [Google Scholar]
- 12.An, W., Palhan, V. B., Karymov, M. A., Leuba, S. H. & Roeder, R. G. (2002) Mol. Cell 9, 811–821. [DOI] [PubMed] [Google Scholar]
- 13.Jasencakova, Z., Meister, A., Walter, J., Turner, B. M. & Schubert, I. (2000) Plant Cell 12, 2087–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner, B. M. (2000) BioEssays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 15.Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J. & Grunstein, M. (2002) Mol. Cell 10, 1223–1233. [DOI] [PubMed] [Google Scholar]
- 16.Ladurner, A. G., Inouye, C., Jain, R. & Tjian, R. (2003) Mol. Cell 11, 365–376. [DOI] [PubMed] [Google Scholar]
- 17.Matangkasombut, O. & Buratowski, S. (2003) Mol. Cell 11, 353–363. [DOI] [PubMed] [Google Scholar]
- 18.Dey, A., Ellenberg, J., Farina, A., Coleman, A. E., Maruyama, T., Sciortino, S., Lippincott-Schwartz, J. & Ozato, K. (2000) Mol. Cell. Biol. 20, 6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama, T., Farina, A., Dey, A., Cheong, J., Bermudez, V. P., Tamura, T., Sciortino, S., Shuman, J., Hurwitz, J. & Ozato, K. (2002) Mol. Cell. Biol. 22, 6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes, S. R., Mozer, B. A., Bhatia-Dey, N. & Dawid, I. B. (1989) Dev. Biol. 134, 246–257. [DOI] [PubMed] [Google Scholar]
- 21.Denis, G. V. & Green, M. R. (1996) Genes Dev. 10, 261–271. [DOI] [PubMed] [Google Scholar]
- 22.Rhee, K., Brunori, M., Besset, V., Trousdale, R. & Wolgemuth, D. J. (1998) J. Cell Sci. 111, 3541–3550. [DOI] [PubMed] [Google Scholar]
- 23.Denis, G. V., Vaziri, C., Guo, N. & Faller, D. V. (2000) Cell Growth Differ. 11, 417–424. [PMC free article] [PubMed] [Google Scholar]
- 24.Crowley, T. E., Kaine, E. M., Yoshida, M., Nandi, A. & Wolgemuth, D. J. (2002) Mol. Endocrinol. 16, 1727–1737. [DOI] [PubMed] [Google Scholar]
- 25.Houzelstein, D., Bullock, S. L., Lynch, D. E., Grigorieva, E. F., Wilson, V. A. & Beddington, R. S. (2002) Mol. Cell. Biol. 22, 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chua, P. & Roeder, G. S. (1995) Mol. Cell. Biol. 15, 3685–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muchardt, C., Reyes, J. C., Bourachot, B., Leguoy, E. & Yaniv, M. (1996) EMBO J. 15, 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Balbas, M. A., Dey, A., Rabindran, S. K., Ozato, K. & Wu, C. (1995) Cell 83, 29–38. [DOI] [PubMed] [Google Scholar]
- 29.Kruhlak, M. J., Hendzel, M. J., Fischle, W., Bertos, N. R., Hameed, S., Yang, X. J., Verdin, E. & Bazett-Jones, D. P. (2001) J. Biol. Chem. 276, 38307–38319. [DOI] [PubMed] [Google Scholar]
- 30.Gottesfeld, J. M. & Forbes, D. J. (1997) Trends Biochem. Sci. 22, 197–202. [DOI] [PubMed] [Google Scholar]
- 31.Segil, N., Guermah, M., Hoffmann, A., Roeder, R. G. & Heintz, N. (1996) Genes Dev. 10, 2389–2400. [DOI] [PubMed] [Google Scholar]
- 32.Pederson, T. (2001) Cell 104, 635–638. [DOI] [PubMed] [Google Scholar]
- 33.Phair, R. D. & Misteli, T. (2001) Nat. Rev. Mol. Cell Biol. 2, 898–907. [DOI] [PubMed] [Google Scholar]
- 34.Houtsmuller, A. B., Rademakers, S., Nigg, A. L., Hoogstraten, D., Hoeijmakers, J. H. & Vermeulen, W. (1999) Science 284, 958–961. [DOI] [PubMed] [Google Scholar]
- 35.McNally, J. G., Muller, W. G., Walker, D., Wolford, R. & Hager, G. L. (2000) Science 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- 36.Misteli, T., Gunjan, A., Hock, R., Bustin, M. & Brown, D. T. (2000) Nature 408, 877–881. [DOI] [PubMed] [Google Scholar]
- 37.Lever, M. A., Th'ng, J. P., Sun, X. & Hendzel, M. J. (2000) Nature 408, 873–876. [DOI] [PubMed] [Google Scholar]
- 38.Phair, R. D. & Misteli, T. (2000) Nature 404, 604–607. [DOI] [PubMed] [Google Scholar]
- 39.Stenoien, D. L., Patel, K., Mancini, M. G., Dutertre, M., Smith, C. L., O'Malley, B. W. & Mancini, M. A. (2001) Nat. Cell Biol. 3, 15–23. [DOI] [PubMed] [Google Scholar]
- 40.Dou, Y., Bowen, J., Liu, Y. & Gorovsky, M. A. (2002) J. Cell Biol. 158, 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L. I., Phair, R. D. & Misteli, T. (2002) Science 298, 1623–1626. [DOI] [PubMed] [Google Scholar]
- 42.Hoogstraten, D., Nigg, A. L., Heath, H., Mullenders, L. H., van Driel, R., Hoeijmakers, J. H., Vermeulen, W. & Houtsmuller, A. B. (2002) Mol. Cell 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- 43.Ellenberg, J. & Lippincott-Schwartz, J. (1999) Methods 19, 362–372. [DOI] [PubMed] [Google Scholar]
- 44.Minucci, S., Horn, V., Bhattacharyya, N., Russanova, V., Ogryzko, V. V., Gabriele, L., Howard, B. H. & Ozato, K. (1997) Proc. Natl. Acad. Sci. USA 94, 11295–11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefebvre, B., Brand, C., Lefebvre, P. & Ozato, K. (2002) Mol. Cell. Biol. 22, 1446–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howe, L., Auston, D., Grant, P., John, S., Cook, R. G., Workman, J. L. & Pillus, L. (2001) Genes Dev. 15, 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lichota, J. & Grasser, K. D. (2001) Biochemistry 40, 7860–7867. [DOI] [PubMed] [Google Scholar]
- 48.Hassan, A. H., Prochasson, P., Neely, K. E., Galasinski, S. C., Chandy, M., Carrozza, M. J. & Workman, J. L. (2002) Cell 111, 369–379. [DOI] [PubMed] [Google Scholar]