Abstract
Myxococcus xanthus exhibits social behavior and multicellular development. FruA is an essential transcription factor for fruiting body development in M. xanthus. In the present study, the upstream promoter region was found to be necessary for the induction of fruA expression during development. A cis-acting element required for the induction was identified and was located between nucleotides –154 and –107 with respect to the transcription initiation site. In addition, it was found that two binding sites exist within this element of the fruA promoter. By using DNA affinity column chromatography containing the cis-acting element, a fruA promoter-binding protein was purified. The purified protein was shown by N-terminal sequence analysis to be identical to MrpC, a protein identified previously by transposon insertion mutagenesis as an essential locus for fruiting body development [Sun, H. & Shi, W. (2001) J. Bacteriol. 183, 4786–4795]. Furthermore, fruA mRNA was not detectable in the mrpC::km strain, demonstrating that MrpC is essential for fruA expression. Moreover, mutational analysis of the binding sites for MrpC in the fruA promoter indicates that binding of MrpC activates transcription of fruA in vivo. This report provides evidence for a direct molecular interaction involved in temporally regulated gene expression in M. xanthus.
Keywords: FruA, MrpC, cAMP receptor protein, catabolite gene activator protein, transcription activation
Developmental programs in organisms are tightly regulated by temporal and spatial expression of specific genes. Intra- or intercellular signals have important functions in this type of gene regulation. Myxobacteria are among a unique subset of bacteria because they exhibit social behavior and multicellular development (1, 2). The myxobacterium Myxococcus xanthus lives in soil and preys on other bacteria. On nutrient limitation, cells begin to migrate toward aggregation centers by gliding to form fruiting bodies. Inside fruiting bodies, cells differentiate to spores, and a mature fruiting body holds 105 cells. Only 10% of the starved cells become spores. Fruiting body development is mediated by cell–cell interactions, which are coordinated by exchanging intercellular signals. Five intercellular signals (A-, B-, C-, D-, and E-signals) have been known to be involved in development of M. xanthus. In addition, S-signal was shown to be required for the production of three types of cell surface molecules, type IV pili, lipopolysaccharide O-antigen, and fibrils during development (3).
From analysis of protein expression patterns by 2D electrophoresis, it was observed that the expression of certain proteins was activated and the expression of others was repressed in a timely manner during the developmental progression of M. xanthus (4). Some genes turned on in the early stage of development are required for the later developmental genes. Thus, the elucidation of temporally regulated changes of gene expression at the molecular level is fundamentally important to understanding fruiting body development of M. xanthus.
FruA is a protein essential for development in M. xanthus and is involved in regulation of aggregation, fruiting body formation, and sporulation (5, 6). FruA belongs to the response-regulator family of two-component His-Asp phosphorelay systems (7). It is proposed that FruA plays a key role in the C-signal transduction system (6). C-signal is a cell surface-associated protein encoded by the csgA gene and is essential for aggregation, fruiting body formation, and sporulation (8, 9). Analysis of protein expression patterns in the WT, ΔfruA, and ΔcsgA strains during development indicated that developmental genes under the control of FruA can be classified into two groups: C-signal-independent and C-signal-dependent (4). The production of five proteins was found to be fruA-dependent but C-signal-independent, and one protein depended on both fruA and C-signal (4). The induction of fruA expression initiates at ≈6 h and reaches the highest level 12 h after the onset of development (5, 6). This induction depends on A- and E-signals but is independent of C-signal (6). Hence, the FruA-dependent signal transduction system seems to be quite complex.
To understand the signal transduction pathway that includes FruA, we examined how fruA expression is induced during development. The upstream promoter region was shown to be required for the induction of fruA. Using DNA affinity chromatography specific to the fruA promoter, we successfully purified a DNA-binding protein. The purified protein was found to be identical to MrpC, which had been identified previously by transposon insertion mutagenesis as an essential locus for fruiting body development (10). Moreover, we show that fruA fails to be expressed in mrpC::km mutant cells and that the MrpC binding sites are important for the activation of the fruA promoter in vivo. We conclude that MrpC is a direct activator of fruA in the regulatory network that governs M. xanthus development.
Materials and Methods
Bacterial Strains and Growth Conditions. M. xanthus DZF1 (11) was used as a parent strain and grown in CYE medium (10 g/liter casitone/5 g/liter yeast extract/8 mM MgSO4 in 10 mM Mops buffer, pH 7.6) (12) supplemented with kanamycin when necessary. For fruiting body formation, M. xanthus was spotted on clone-fruiting (CF) agar plates (13). Escherichia coli JM83 (14) was used as a recipient strain for transformation, unless otherwise mentioned, and was grown in LB medium (15) supplemented with ampicillin when necessary.
lacZ Fusion Analysis. An integration vector pZKAT for transcriptional lacZ fusion at phage Mx8 attachment site (attB) in the M. xanthus chromosome (16) was constructed by inserting the EcoRI–SmaI fragment containing attP and transcriptional termination signals from pREG1727 (17) into pSI1403Km (18). The fruA promoter regions from nucleotides –185 to +270 and from nucleotides –40 to +270 were amplified by PCR with pMF05 (5) and oligonucleotide primers 5′-TCAAGCTTGTTGACAGACAGCGCCGTGC-3′ (primer a) and 5′-TCGGATCCATCGATACACGAATCGCTGC-3′ (primer b) and 5′-TCAAGCTTCTGGTTCGCGTCTGCGCTTT-3′ (primer c) and primer b (see Fig. 1B for the location; HindIII and BamHI sites are underlined), respectively. PCR products were digested with HindIII and BamHI and cloned into pZKAT. Plasmid DNAs with or without the fruA promoter regions were electroporated into M. xanthus DZF1 as described (19).
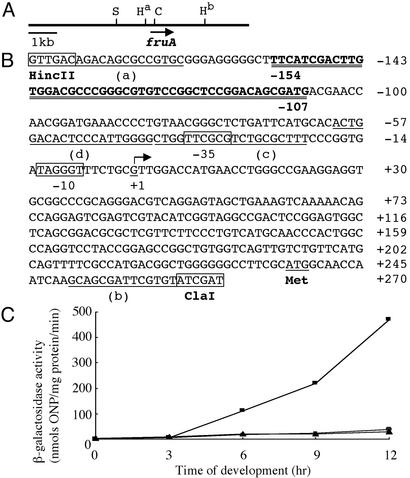
Fig. 1.
fruA expression during development. (A) The map of the 9.5-kbp fragment containing fruA (S, StuI; H, HincII; C, ClaI). (B) The promoter region of fruA. The transcription initiation site is indicated by an arrow. The DNA-binding site identified by footprint analysis shown in Fig. 2B is double-underlined. The sequences corresponding to oligonucleotide primers a–d are underlined. (C) lacZ fusion analysis. The promoter regions from nucleotides –185 to +270 (squares) and from nucleotides –40 to +270 (circles) were fused to lacZ, and β-galactosidase activity was measured during development. Triangles represent pZKAT without the promoter.
For examination of the activity of the intact fruA promoter, the 1.3-kbp StuI–ClaI fragment from pMF03 (ref. 5; Fig. 1A) was used for homologous recombination. The ClaI site of the 1.3-kbp StuI–ClaI (at nucleotide +270, Fig. 1B) fragment was filled in with Klenow fragment of DNA polymerase I and cloned in the SmaIsite of pSI1403Km. The resultant plasmid, pSI1403Km/S-C, was electroporated into M. xanthus DZF1 and integrated at the fruA promoter region.
To examine the effect of deletion of the region from nucleotides –185 to –41 in the context of the intact fruA promoter, the HincIIa (at nucleotide –185; Fig. 1B)–BamHI fragment of pSI1403Km/S-C was replaced with a HindIII–BamHI fragment containing the region from nucleotides –40 to +270 (the HindIII site was filled in with Klenow fragment of DNA polymerase I before ligation). The resultant plasmid, pSI1403Km/S-C(Δ-185-41), was electroporated into M. xanthus DZF1. It should be noted that recombination can occur upstream or downstream of the deletion in the promoter region. Recombination upstream of the deletion was confirmed by PCR amplification.
For mutational analysis, changes of nucleotides in the promoter region from nucleotides –185 to +270 were introduced by site-directed mutagenesis with the overlap extension PCR method (20). Mutant promoters fused to lacZ of pZKAT were introduced into the attB site as described above.
β-Galactosidase activity was measured as described (21).
Preparation of Probes for DNA-Binding Assay and Footprint Analysis. The fruA promoter region from nucleotides –185 to –41 was amplified by PCR using pMF05 (5) and oligonucleotide primers a and 5′-TCGGATCCCCAATGGGAGTGTCCAGT-3′ (primer d; see Fig. 1B for the location). PCR products were digested with HindIII and BamHI and cloned in pUC19 (14). DNA fragments for probes were prepared by digesting the plasmid with HindIII and BamHI and isolating the fragments by PAGE. DNA fragments were then labeled with [α-32P]dCTP by Klenow fragment of DNA polymerase I.
For footprint analysis, a probe corresponding to the region from nucleotides –185 to –41 was prepared as follows. The plasmid described above was digested with BamHI and labeled with [α-32P]dCTP by Klenow fragment of DNA polymerase I. After inactivation of Klenow fragment of DNA polymerase I, the plasmid was digested with HindIII. Therefore, only the strand shown in Fig. 1B was labeled. The labeled DNA fragment was purified by PAGE.
DNA-Binding Assay and Footprint Analysis. DNA-binding reactions were performed at 30°C for 10 min in 10 μl of the reaction mixture containing 10 mM Tris·HCl (pH 8.0), 50 mM KCl, 1 mM DTT, 10 μg/ml BSA, 10% glycerol, and 1 μg of poly(dI-dC)·poly(dI-dC) (Amersham Pharmacia Biotech). The binding patters were analyzed by 5% PAGE, followed by autoradiography. For footprint analysis, after PAGE, bands of interest were excised from the gel and subjected to the treatment of 1,10-phenanthroline-copper as described (22).
Purification of the fruA Promoter-Binding Protein. Cell extract was obtained from M. xanthus DZF1 at 12 h after the initiation of development on CF agar plates. Cells from 1-liter cultures were spotted on CF agar plates. The ammonium sulfate (AS) fraction (40–65%) was prepared as described (23). The AS fraction (2 ml) was applied to a DEAE-Sepharose column (2 ml, Amersham Pharmacia Biotech) equilibrated with TGED buffer (10 mM Tris·HCl, pH 7.9/10% glycerol/0.1 mM EDTA/0.1 mM DTT). The fruA promoter-binding protein (FBP) was eluted with TGED buffer containing 0.1 M NaCl (TGED0.1N) supplemented with protease inhibitors (Complete EDTA-free, Roche Diagnostics) as recommended by the company. FBP was detected by a DNA-binding assay using the promoter region from nucleotides –185 to –41 as a probe. The eluate (2 ml) was applied to a DNA-cellulose column (4 ml, Amersham Pharmacia Biotech) equilibrated with TGED0.1N. FBP was eluted with TGED0.5N. The eluate (8 ml) was diluted with 4 vol of TGED supplemented with 0.1% Nonidet P-40 (TGEDN), mixed with 100 μg of poly(dI-dC)·poly(dI-dC), and incubated on ice for 10 min. The mixture was then applied to a DNA affinity column specific to the fruA promoter. The specific DNA affinity column was prepared as described (24) by using oligonucleotide primers 5′-GATCTTCATCGACTTGTGGACGCCCGGGCGTGTCCGGCTCCGGACAGCGATG-3′ and 5′-GATCCATCGCTGTCCGGAGCCGGACACGCCCGGGCGTCCACAAGTCGATGAA-3′ (the sequences recognized by FBP are underlined) corresponding to the promoter region from nucleotides –154 to –107. The column (2 ml) was equilibrated with TGEDN0.1N. FBP was eluted with TGEDN0.5N. The eluate was diluted with 4 vol of TGEDN, mixed with poly(dI-dC)·poly(dI-dC) and incubated on ice for 10 min. The mixture was again applied to the specific DNA affinity column. FBP was eluted with TGEDN0.5N. These procedures for purification of FBP were performed five times on separate 1-liter cultures.
Purification of MrpC. The mrpC gene (10) was cloned into pET11a (Novagen), and the plasmid was transformed into E. coli BL21(DE3) (25). The mrpC gene was induced by the addition of isopropyl β-d-thiogalactoside (IPTG) at a final concentration of 1 mM in LB medium supplemented with ampicillin at 37°C for 1 h. Cells were harvested by centrifugation and washed with TGED0.1N. Cells were then resuspended in TGED0.1N and disrupted by sonication. After centrifugation, the supernatant was subjected to DNA-cellulose column chromatography followed by DNA affinity column chromatography specific to the fruA promoter as described above.
Results
The Induction of fruA Expression Depends on the Upstream Promoter Region. fruA is expressed exclusively during fruiting body development and its expression reaches the maximum level ≈12 h after the initiation of development (5, 6). To understand how fruA expression is regulated, we first examined fruA expression by using lacZ transcriptional fusions. Because the 1.6-kbp HincII fragment (Fig. 1A) containing the fruA gene is able to complement developmental defects of ΔfruA strains (5, 6), it seems likely that the promoter region up to nucleotide –185 (the HincIIa site) is sufficient for proper regulation of fruA expression. Two promoter fragments extending from nucleotides +270 to –40 and from nucleotides +270 to –185 were fused to lacZ to analyze the importance of the upstream region in the fruA promoter (Fig. 1B). Plasmids containing the two fusions were integrated at the attB site on the chromosome. β-Galactosidase activity was measured during fruiting body formation until 12 h after the initiation of development. Significant induction of β-galactosidase synthesis during development was observed only for the fruA promoter extending to nucleotide –185 (Fig. 1C).
To compare the activity of the promoter extending from nucleotide +270 to nucleotide –185 with the intact fruA promoter on the chromosome, an additional lacZ fusion plasmid was constructed. This plasmid contains the StuI–ClaI fragment (Fig. 1A) fused to lacZ and was integrated by homologous recombination at the fruA gene. The expression level from this fusion was slightly lower than that from the fusion containing the fragment from nucleotide +270 to nucleotide –185 (74% at 12 h after the initiation of development). In addition, to examine the effect of the region from nucleotides –185 to –41 on promoter activity at the native site, the region from nucleotides –185 to –41 was deleted from the StuI–ClaI fragment. The activity of the promoter without the region from nucleotides –185 to –41 was drastically decreased (3.3%). These results indicate that the promoter region up to –185 was sufficient for development-specific fruA expression and that the region from nucleotides –185 to –41 is essential for the induction of fruA expression.
DNA-Binding Activity to the fruA Promoter. We next examined whether a trans-acting factor could be identified because the induction of fruA expression depended on the upstream promoter region. A DNA-binding assay was performed by using a DNA probe containing the promoter region from nucleotides –185 to –41 and AS fractions prepared from vegetative and developmental cells. The cytoplasmic fraction was fractionated by AS precipitation, and the fraction obtained between 40% and 65% was found to contain DNA-binding activity (data not shown). Whereas little DNA-binding activity was detected from vegetative cells (Fig. 2A, lane 2), DNA-binding activity was observed during development (lanes 3–5). Two types of complexes, I and II, were detected (Fig. 2A). These results suggest that an upstream DNA-binding factor might be involved in fruA expression during development.
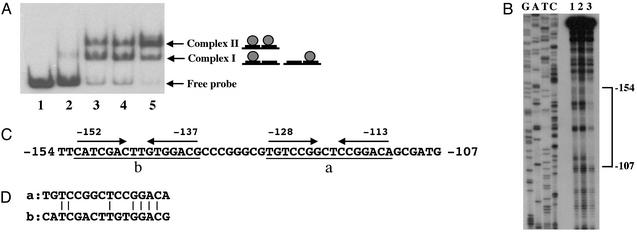
Fig. 2.
Identification of the DNA-binding site. (A) Shown is a DNA-binding assay. The probe contains the promoter region from nucleotides –185 to –41. AS fractions prepared from vegetative (lane 2) and 4-, 8-, and 12-h developmental cells (lanes 3, 4, and 5, respectively) were used for the protein source. Note that the same number of initial cells is harvested at each time point. Lane 1 contains no AS fraction. (B) Shown is footprint analysis. DNA-binding reactions were performed under the same conditions as described for A. After the binding reaction and gel electrophoresis, complex I, complex II, and free probe were excised from the gel and subjected to the treatment with 1,10-phenanthroline-copper. Lane 1, free probe; lane 2, complex I; lane 3, complex II. Lanes G, A, T, and C represent sequence ladders generated by a primer 5′-GATCCCCAGCCCCAATGGGAGTG-3′, which was labeled at the 5′ end with [γ-32P]ATP by T4 polynucleotide kinase and can hybridize just upstream of the –35 region boxed in Fig. 1B. (C) Shown are the sequences of the DNA-binding site. (D) Sequence comparison between regions a and b.
Identification of the DNA-Binding Site in the fruA Promoter. To define the DNA-binding site in the fruA promoter, footprint analysis was performed. After the binding reaction and gel electrophoresis, complex I, complex II, and free probe (Fig. 2A) were excised from the gel and subjected to the treatment with 1,10-phenanthroline-copper (22). The region from nucleotides –154 to –107 was protected in complex II (Fig. 2B). However, no region seemed to be protected in complex I. This finding might result from weak or unstable interactions between the DNA probe and the protein and from dissociation in complex I during the footprint reaction. Because the protected region is relatively long and two types of complexes were observed in the DNA-binding assay, it seems that there are at least two binding sites in the region from nucleotides –154 to –107. Region a, from nucleotides –128 to –113, contains palindromic sequences (Fig. 2C). Additionally, region b, from nucleotides –152 to –137, contains palindromic sequences with two mismatches and exhibits sequence homology to region a (Fig. 2D).
When a probe containing the promoter region from nucleotides –100 to –41 was used in the DNA-binding assay, no complex was detectable (data not shown). In addition, when the promoter region from nucleotides –100 to +270 was fused to lacZ, no synthesis of β-galactosidase was observed during development (data not shown). Therefore, the DNA-binding site identified above is an essential cis-acting element for fruA expression.
Purification and Identification of the fruA Promoter-Binding Protein. The FBP was purified from the 40–65% AS fraction from developmental cells. The AS fraction was subjected to chromatography including a DEAE-Sepharose column, a DNA-cellulose column, and a DNA affinity column specific to the fruA promoter from nucleotides –154 to –107. After the second round of DNA affinity column chromatography, a protein with an apparent molecular mass of 29 kDa was obtained (Fig. 3A).
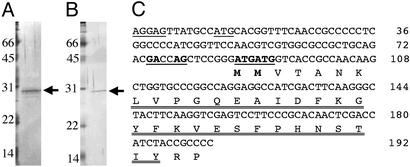
Fig. 3.
Identification of the fruA promoter-binding protein. (A) Shown is purification of FBP from M. xanthus. After the second round of DNA affinity column chromatography, the sample was applied to SDS/15% PAGE and visualized by silver staining. (B) Purification of MrpC2 from E. coli. After the second round of DNA affinity column chromatography, the sample was applied to SDS/15% PAGE and visualized by silver staining. (C) Shown are the sequences of the upstream region of mrpC and N-terminal end of MrpC. The previously assigned ribosome-binding site (nucleotides 1–5) and the initiation codon (nucleotides 13–15) are underlined (10). The N-terminal sequence determined for FBP is double-underlined. The newly assigned initiation codon and the ribosome-binding site are indicated by bold letters and underlined.
To identify the gene encoding FBP, the N-terminal sequence of the purified protein was determined to be LVPGQEAIDFKGYFKVESFPHNSTIY. Comparison of this sequence with the GenBank database (www.ncbi.nlm.nih.gov/blast) revealed it to be identical to amino acid sequences in MrpC (amino acids 8–33; Fig. 3C). MrpC was previously identified by transposon insertion mutagenesis as an essential locus for fruiting body development (10). The initiation codon for MrpC was previously assigned to an upstream ATG (nucleotides 13–15, Fig. 3C) by Sun and Shi (10) based on the proximity of a potential ribosome-binding site, AGGAG, located at nucleotides 1–5 (Fig. 3C). However, N-terminal sequence analysis as described above indicates that either ATG (nucleotides 88–90) or ATG (nucleotides 91–93) is likely to be the initiation codon. This assignment is also supported by sequence similarity to cAMP receptor protein [CRP; also known as catabolite gene activator protein (CAP)] family of transcriptional regulators, and the similarity starts after methionine (ATG at nucleotides 91–93; ref. 10; see Discussion). To test which of the possible start codons is the actual start codon, plasmids were constructed in which MrpC protein was synthesized from an ectopic promoter in E. coli. When MrpC was synthesized from ATG at nucleotides 13–15, MrpC protein (MrpC1) was synthesized with a molecular mass larger than that observed in M. xanthus (data not shown). However, when MrpC was synthesized from ATG at nucleotides 88–90, MrpC protein (MrpC2) was synthesized with a molecular mass similar to that observed in M. xanthus (Fig. 3B). These data suggest that the start codon in mrpC is located either at nucleotides 88–90 or at nucleotides 91–93. It should be noted that the purified protein might be partially degraded during purification procedures because the N-terminal residues (MMVTANK) were not identified by sequence analysis and minor bands were detected on SDS/PAGE (Fig. 3A).
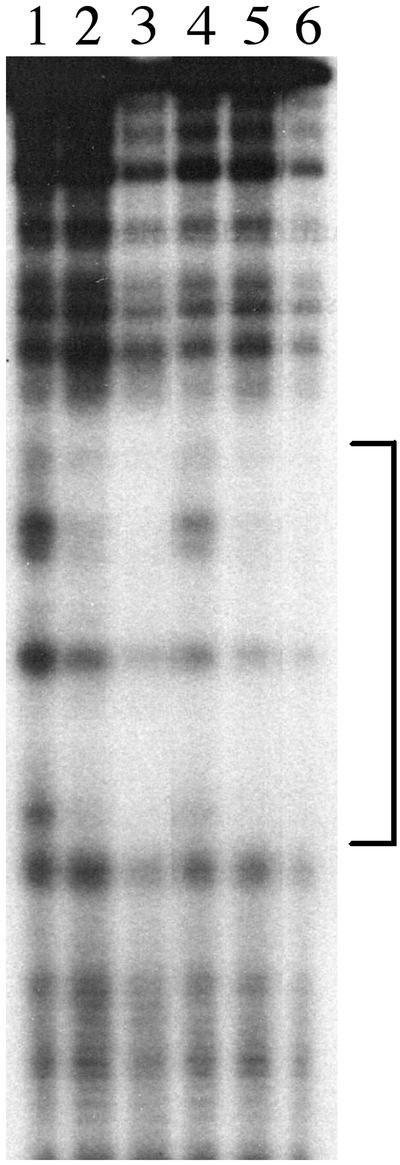
DNA-binding assays and footprint analysis were performed with MrpC2 purified from E. coli to confirm that MrpC2 exhibits the same DNA-binding activity as FBP from M. xanthus. Both FBP and MrpC2 formed complex I and II with a probe containing the fruA promoter region from nucleotides –185 to –41 in a DNA binding assay (data not shown). Then, footprint analysis was performed. After the binding reaction and gel electrophoresis, complex I, complex II, and free probe were excised from the gel and subjected to the treatment with 1,10-phenanthroline-copper. Footprint analysis (Fig. 4) demonstrated that both FBP from M. xanthus (lane 3) and MrpC2 (lane 5) recognized the same sequences in complex II as determined by using the AS fraction (lane 6). In the case of complex I, the same region was found to be partially protected by FBP from M. xanthus (lane 2) and MrpC2 (lane 4), whereas no protected region was observed for complex I formed in the presence of the AS fraction (Fig. 2B, lane 2). This difference may be due to the presence of unknown factors in the AS fraction that affect the stability of complex I. These results indicate that FBP from M. xanthus and MrpC2 from E. coli exhibit indistinguishable binding to the fruA promoter.
Fig. 4.
Footprint analysis. MrpC2 (20 ng) purified from E. coli, FBP (20 ng) from M. xanthus, and the AS fraction (200 μg) were used. The DNA-binding reactions were carried out in a 100-μl volume, and, after gel electrophoresis, complex I, complex II, and free probe were excised from the gel and subjected to the treatment of 1,10-phenanthroline-copper. Lane 1, free probe; lane 2, complex I with FBP; lane 3, complex II with FBP; lane 4, complex I with MrpC2; lane 5, complex II with MrpC2; lane 6, complex II with the AS fraction.
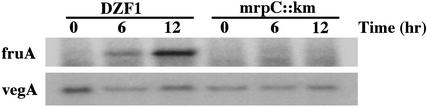
To test the dependence of fruA expression on MrpC, fruA expression was examined by primer extension analysis. Total RNA was prepared from M. xanthus DZF1 and an mrpC::km strain. The mrpC::km strain was constructed by the insertion of a kanamycin-resistance gene at the MscI site of the mrpC gene of DZF1, and the proper insertion was confirmed by Southern blot analysis (data not shown). Developmental defects of the mrpC::km strain were verified by spotting on CF agar plates. fruA mRNA was induced in DZF1 during development, but not in mrpC::km (Fig. 5). As a control, vegA expression was tested for both strains. The vegA gene is known to be expressed during vegetative growth as well as during development (26), and in vitro transcription of vegA can be initiated by RNA polymerase containing SigA, the major housekeeping sigma factor, without an additional factor (27). vegA expression was not affected by the absence of MrpC (Fig. 5). Taken together with the DNA-binding assays, these results strongly suggest that MrpC functions as a trans-acting factor required for the specific induction of fruA expression during development.
Fig. 5.
Expression of fruA in M. xanthus DZF1 and mrpC::km. fruA expression was examined by primer extension analysis as described (18). Total RNA was prepared from DZF1 and mrpC::km during vegetative growth (0 h) and fruiting body development (6 and 12 h). As a control, vegA expression was examined. Oligonucleotide primers 5′-TTGACTTTCAGCTACTCCTGACG-3′ and 5′-GCTTTATCCACGGACATT-3′ were used for fruA and vegA, respectively.
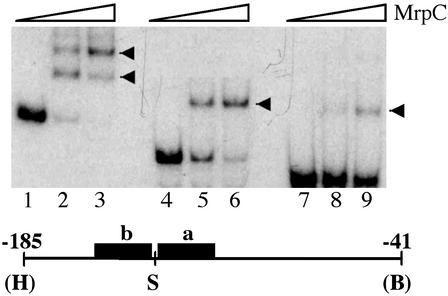
Analysis of the DNA-Binding Site. To further characterize the DNA-binding site for MrpC in the fruA promoter, DNA-binding assays were carried out with purified MrpC2 and various probes. First, we constructed three kinds of probes containing the promoter region including both regions a and b (probe ab), only region a (probe a), or only region b (probe b) to determine whether two binding sites exist as proposed above. With probe ab, two complexes were detected (Fig. 6, lanes 2 and 3). In contrast, only one complex was observed with probe a and probe b (lanes 5 and 6 for probe a, lanes 8 and 9 for probe b). Furthermore, it seems that the affinity of MrpC is higher for probe ab than for probe a or probe b because fewer complexes were formed with probes a and b. In addition, the binding affinity for region a seems to be higher than for region b because more complexes were formed with probe a than with probe b. This result may reflect the fact that region a contains palindromic sequences with no mismatches and that region b contains two mismatches within otherwise palindromic sequences (Fig. 2C).
Fig. 6.
DNA-binding analysis. (Upper) Three kinds of probes (10 fmol), an HindIII (H)–BamHI (B) fragment containing both regions a and b (lanes 1–3), an SmaI (S)–BamHI fragment containing region a (lanes 4–6), or an HindIII–SmaI fragment containing region b (lanes 7–9) were used for DNA-binding reactions in a 10-μl volume. Note that the HindIII and BamHI sites are from the cloning vector. No protein was added for lanes 1, 4, and 7. Purified MrpC2 was added for lanes 2, 5, and 8 (1 ng) and lanes 3, 6, and 9 (2 ng). MrpC–DNA complexes are indicated by arrowheads. (Lower) The fruA promoter region from nucleotides –185 to –41 is shown.
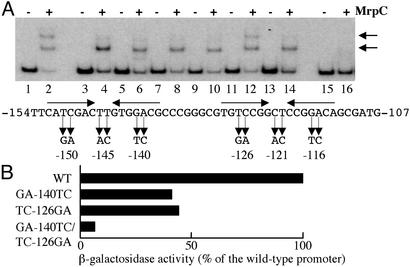
Next, we performed mutational analysis in the binding sites by changing nucleotides at various positions (Fig. 7A). With the probe containing the WT sequences, two types of complexes were observed (Fig. 7A, lane 2). In contrast, only one complex was detected with probes containing mutations in one of the two inverted repeat sequences (lanes 4, 8, 10, and 14). The mutations TT-145AC between the inverted repeat sequences also affected the DNA-binding activity of MrpC (lane 6). In contrast, the mutations CT-121AC did not prevent formation of the two complexes (lane 12). In addition, little complex was detected with the probe containing mutations GA-140TC and TC-126GA in both sites (lane 16). These results demonstrate that the nucleotides at certain positions in the palindromic sequences are critical for the DNA-binding activity of MrpC.
Fig. 7.
Mutational analysis. (A) DNA-binding assay. Probes (10 fmol) contain the promoter region from nucleotides –185 to –41 without mutations (lanes 1 and 2) and with mutations TC-150GA (lanes 3 and 4), TT-145AC (lanes 5 and 6), GA-140TC (lanes 7 and 8), TC-126GA (lanes 9 and 10), CT-121AC (lanes 11 and 12), GA-116TC (lanes 13 and 14), and GA-140TC/TC-126GA (lanes 15 and 16). Purified MrpC2 (1 ng) was added to DNA-binding reaction mixtures in a 10-μl volume for lanes 2, 4, 6, 8, 10, 12, 14, and 16; no protein was added for lanes 1, 3, 5, 7, 9, 11, 13, and 15. MrpC-DNA complexes are indicated by arrows. Changes of sequences are shown below DNA-binding patterns. (B) lacZ fusion analysis. The promoter region from nucleotides –185 to +270 containing the WT or mutant sequences was fused to lacZ, and β-galactosidase activity was measured 12 h after the initiation of development. The activity is shown as a percentage of the activity of the WT promoter.
The effects of mutations GA-140TC, TC-126GA, and GA-140TC/TC-126GA on fruA expression were examined by lacZ fusion analysis (Fig. 7B). The promoters containing mutations GA-140TC or TC-126GA in one of two sites showed 40% and 44% of the activity of the WT promoter, respectively. In addition, the promoter containing mutations GA-140TC/TC-126GA exhibited only 6.0% of the activity of the WT promoter. Therefore, these nucleotides were indispensable for the induction of fruA expression in vivo.
Discussion
We have identified a cis-acting element required for the induction of fruA expression and purified a FBP by using a DNA affinity column specific to the cis-acting element. N-terminal sequence analysis has revealed the purified protein to be identical to MrpC, which had been previously identified as an essential locus for fruiting body development via transposon insertion mutagenesis (10). Furthermore, we show that fruA is not expressed in the mrpC::km strain. Taken together with in vitro DNA-binding assay with mutant promoters and lacZ fusion analyses, we conclude that MrpC directly activates transcription of fruA by binding to the fruA promoter.
MrpC belongs to the CRP [also known as catabolite gene activator protein (CAP)] family of transcriptional regulators (10). The CRP transcriptional regulators contain the cyclic nucleotide (cNMP) binding domain and the DNA-binding domain (28–30). CRP from E. coli (EcCRP) has been shown to require cAMP for specific DNA binding. In contrast, MrpC did not seem to require any cNMPs for specific DNA binding, suggesting that cNMP is not an effector for MrpC. It is possible, however, that purified MrpC formed complexes with cNMP. The N-terminal MrpC sequence exhibits similarity to the cNMP binding domain of EcCRP, including the residues contacting the ribose of cAMP (31). However, the residues contacting the cyclic phosphate and purine ring of cAMP (31) are not conserved in MrpC. Hence, another type of nucleotide may serve as an effector for MrpC. (p)ppGpp, for example, may be a possible candidate, because (p)ppGpp plays essential roles in early development in M. xanthus (32).
EcCRP recognizes 22-bp, 2-fold-symmetric sequences. The DNA-binding site for MrpC seems to span from nucleotides –154 to –107 as judged from footprint analysis, and contains two palindromic sequences. DNA-binding assays with various probes, including mutations, demonstrated that two binding sites exist in this region, and palindromic sequences are critical for the DNA-binding activity of MrpC (Fig. 7A). Whereas region a seemed to be the stronger site than region b (Fig. 6), both regions seemed to be bound by MrpC in complex I (Fig. 4). It may be possible that MrpC requires certain length extended from recognition sequences. It is possible that MrpC favors binding to region a in the experiment shown in Fig. 6 because probe a extends 5 bp beyond the palindromic sequence whereas probe b extends only 3 bp beyond it.
As expected, fruA mRNA was not detected in mrpC::km. It is reasonable to speculate that developmental defects in ΔmrpC (10) are due to the absence of FruA, although it is not known whether MrpC regulates other genes. The ΔfruA and ΔmrpC strains exhibit common characteristics; both strains lack the expression of tps, devTRS, and Ω4500 genes, and methylation of FrzCD is reduced in both strains (5, 6, 33). Therefore, it is likely that MrpC directly activates transcription of fruA during development. However, the involvement of other factors in fruA expression cannot be excluded at this time.
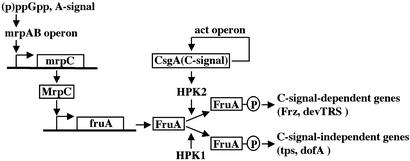
To summarize the FruA-dependent signal transduction system, a model is proposed (Fig. 8). (p)ppGpp and A-signal activate, directly and/or indirectly, the mrpAB operon and mrpC (33). In addition, it is likely that E-signal is also involved in this pathway, because fruA expression is lower in an E-signal mutant (6). mrpAB was identified as an essential locus for development and is located upstream of mrpC (10). It is proposed that the expression of mrpC depends on the mrpAB operon and that MrpC auto-regulates its own gene (10). However, sequences similar to the binding site identified for the fruA promoter are not found in the mrpC promoter region. Therefore, the pathway remains to be verified experimentally in detail.
Fig. 8.
A model for the signal transduction pathway including FruA during fruiting body development of M. xanthus. See Discussion for details.
MrpC then induces fruA expression by binding to the fruA promoter. Because FruA belongs to the response-regulator family and because changes to Ala, Asn, and Gln at Asp at position 59, which is a putative phosphorylation site in FruA, abolish activity of FruA (6), it is reasonable to speculate that FruA is activated via phosphorylation by a histidine kinase. Such a kinase, however, remains to be identified. It is proposed that, in the early stage of development, a small amount of FruA is phosphorylated by an unknown mechanism, resulting in the expression of C-signal-independent genes such as tps and dofA (4). On the other hand, the activity of FruA also seems to be regulated by CsgA (C-signal) (6). C-signal also has been proposed to activate the act operon, whose products regulate the level of CsgA protein (34–37). As the developmental program proceeds and the concentration of C-signal increases, more FruA becomes phosphorylated and induces the expression of C-signal-dependent genes, which leads to FrzCD methylation and devTRS expression (4, 6, 38). It is possible that this regulation of FruA activity may be controlled by two histidine kinases; one (HPK1 in Fig. 8) is activated C-signal-independently and the other (HPK2 in Fig. 8) C-signal-dependently. Such systems, where more than one histidine kinase phosphorylate a single response regulator, are known in other bacteria (7). For instance, histidine kinases KinA, KinB, and KinC are known to phosphorylate a response regulator Spo0F during the regulation of sporulation in Bacillus subtilis (39). Because there are a number of essential genes for development (3), some of which are not shown in Fig. 8, the signal transduction pathway is predicted to be more complicated than the model shown in Fig. 8.
In the present study, we have provided strong evidence that MrpC directly regulates fruA during development. Our findings should expand our understanding of the signal transduction systems operating during development. Further elucidation of the pathway will necessitate the identification of a kinase for FruA and the target genes of FruA.
Acknowledgments
We are grateful to M. Otani for determination of the N-terminal sequence of the purified protein, L. Kroos for plasmid pREG1727, and H. Nariya for construction of the mrpC::km strain. This work was supported by a grant from the Foundation of the University of Medicine and Dentistry of New Jersey.
Abbreviations: AS, ammonium sulfate; FBP, fruA promoter-binding protein; CRP, cAMP receptor protein; CF, clone-fruiting.
References
- 1.Dworkin, M. (1996) Microbiol. Rev. 60, 70–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dworkin, M. & Kaiser, D. (1993) Myxobacteria II (Am. Soc. Microbiol., Washington, DC).
- 3.Shimkets, L. J. (1999) Annu. Rev. Microbiol. 53, 525–549. [DOI] [PubMed] [Google Scholar]
- 4.Horiuchi, T., Taoka, M., Isobe, T., Komano, T. & Inouye, S. (2002) J. Biol. Chem. 277, 26753–26760. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa, M., Fujitani, S., Mao, X., Inouye, S. & Komano, T. (1996) Mol. Microbiol. 22, 757–767. [DOI] [PubMed] [Google Scholar]
- 6.Ellehauge, E., Nørregaard-Madsen, M. & Søgaard-Andersen, L. (1998) Mol. Microbiol. 30, 807–817. [DOI] [PubMed] [Google Scholar]
- 7.Hoch, J. A. & Silhavy, T. J. (1995) Two-Component Signal Transduction (Am. Soc. Microbiol., Washington, DC).
- 8.Kim, S. K. & Kaiser, D. (1990) Cell 61, 19–26. [DOI] [PubMed] [Google Scholar]
- 9.Hagen, T. J. & Shimkets, L. J. (1990) J. Bacteriol. 172, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun, H. & Shi, W. (2001) J. Bacteriol. 183, 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye, M., Inouye, S. & Zusman, D. R. (1979) Dev. Biol. 68, 579–591. [DOI] [PubMed] [Google Scholar]
- 12.Campos, J. M., Geisselsoder, J. & Zusman, D. R. (1978) J. Mol. Biol. 119, 167–178. [DOI] [PubMed] [Google Scholar]
- 13.Hagan, D. C., Bretcher, A. P. & Kaiser, D. (1978) Dev. Biol. 64, 284–296. [DOI] [PubMed] [Google Scholar]
- 14.Vieria, J. & Messing, J. (1982) Gene 19, 259–268. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 16.Stellwag, E., Fink, J. M. & Zissler, J. (1985) Mol. Gen. Genet. 199, 123–132. [DOI] [PubMed] [Google Scholar]
- 17.Fisseha, M., Gloudemans, M., Gill, R. & Kroos, L. (1996) J. Bacteriol. 178, 2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueki, T. & Inouye, S. (1998) Genes Cells 3, 371–385. [DOI] [PubMed] [Google Scholar]
- 19.Kashefi, K. & Hartzell, P. L. (1995) Mol. Microbiol. 15, 483–494. [DOI] [PubMed] [Google Scholar]
- 20.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 22, 51–59. [DOI] [PubMed] [Google Scholar]
- 21.Kroos, L., Kuspa, A. & Kaiser, D. (1986) Dev. Biol. 117, 252–266. [DOI] [PubMed] [Google Scholar]
- 22.Kuwabara, M. D. & Sigman, D. S. (1987) Biochemistry 26, 7234–7238. [DOI] [PubMed] [Google Scholar]
- 23.Ueki, T. & Inouye, S. (2002) J. Biol. Chem. 277, 6170–6177. [DOI] [PubMed] [Google Scholar]
- 24.Kadonaga, J. T. & Tjian, R. (1986) Proc. Natl. Acad. Sci. USA 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990) Methods Enzymol. 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 26.Komano, T., Franceschini, T. & Inouye, S. (1987) J. Mol. Biol. 196, 517–524. [DOI] [PubMed] [Google Scholar]
- 27.Biran, D. & Kroos, L. (1997) Mol. Microbiol. 25, 463–472. [DOI] [PubMed] [Google Scholar]
- 28.Kolb, A., Busby, S., Buc, H., Garges, S. & Adhya, S. (1993) Annu. Rev. Biochem. 62, 749–795. [DOI] [PubMed] [Google Scholar]
- 29.Busby, S. & Ebright, R. H. (1999) J. Mol. Biol. 293, 199–213. [DOI] [PubMed] [Google Scholar]
- 30.Harman, J. G. (2001) Biophys. Biochim. Acta 1547, 1–17. [DOI] [PubMed] [Google Scholar]
- 31.Weber, I. T. & Steitz, T. A. (1987) J. Mol. Biol. 198, 311–326. [DOI] [PubMed] [Google Scholar]
- 32.Singer, M. & Kaiser, D. (1995) Genes Dev. 9, 1633–1644. [DOI] [PubMed] [Google Scholar]
- 33.Sun, H. & Shi, W. (2001) J. Bacteriol. 183, 6733–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S. K. & Kaiser, D. (1991) J. Bacteriol. 173, 1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorski, L., Gronewold, T. & Kaiser, D. (2000) J. Bacteriol. 182, 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gronewold, T. M. A. & Kaiser, D. (2001) Mol. Microbiol. 40, 744–756. [DOI] [PubMed] [Google Scholar]
- 37.Gronewold, T. M. A. & Kaiser, D. (2002) J. Bacteriol. 184, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Søgaard-Andersen, L. & Kaiser, D. (1996) Proc. Natl. Acad. Sci. USA 93, 2675–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perego, M. (1998) Trends Microbiol. 6, 366–370. [DOI] [PubMed] [Google Scholar]