Abstract
Single-molecule DNA analysis of testicular germ cells isolated by laser capture microdissection from two Huntington disease patients showed that trinucleotide repeat expansion mutations were present before the end of the first meiotic division, and some mutations were present even before meiosis began. Most of the larger Huntington disease mutations were found in the postmeiotic cell population, suggesting that expansions may continue to occur during meiosis and/or after meiosis is complete. Defining the germ-line cell compartments where the trinucleotide repeat expansions occur could help to elucidate the underlying mechanisms of instability.
At least 14 diseases result from expansion in the number of trinucleotide repeats (TNR) in or adjacent to a protein-coding gene (1–3). In most cases, the repeat sequence consists of CAG/CTG triplets. The expanded alleles characteristically undergo further expansion when the disease gene is transmitted from an affected parent to the offspring, resulting in increased disease severity and an earlier age of onset. Based on studies in model systems, a variety of molecular mechanisms have been proposed to explain TNR expansion, including meiotic recombination (4–7), DNA replication slippage (2, 8–14), and DNA damage repair (15–24). Whether one or all of these processes actually contribute to germ-line expansion mutations in humans is not known, although it is widely accepted that secondary structure formation by TNR tracts is a critical step in the mutation process (reviewed in refs. 2 and 25–28).
Defining the germ-line cell compartments where the TNR expansions occur in humans could help reveal the underlying mechanisms. The presence of expansion mutations in spermatogonia would be consistent with a mitotic mutation process mediated by replication slippage or DNA damage repair. Expansions occurring during meiosis might suggest a role for double strand break formation induced by the SPO11 protein (29–31), whereas expansions after meiosis is completed would implicate DNA damage repair.
No direct information on the presence or absence of CAG/CTG expansion mutations in the various human germ-line cell compartments is available. We investigated the germ line of two men who died from Huntington disease (HD). When men transmit the HD mutation, the relative increase in repeat number among their offspring [or in their sperm (32)] is among the highest observed for CAG/CTG tract instability (reviewed in ref. 33). We analyzed testicular germ cells isolated by laser capture microdissection (LCM) (34). We showed that TNR expansion mutations are present before the end of the first meiotic division, and are even present before meiosis begins. Because a greater proportion of the larger human mutations were found in the postmeiotic cell population, we propose that some expansions may continue to occur after the beginning of meiosis. In humans, TNR mutations may arise at different stages of germ-line development, possibly as a result of several different DNA transactions.
Methods
Tissues. Testis tissue was obtained at autopsy 3 h post mortem and kept at the Harvard Brain Tissue Center at –70°C. No semen samples were available from these patients. Informed consent was obtained from all subjects by using a protocol approved by the Institutional Review Board of the College of Physicians and Surgeons of Columbia University.
Staining. Frozen specimens of testicular tissue were fixed in 50% ethanol at –20°C over 48 h. The specimens were then put at room temperature and the ethanol concentration was raised gradually to 100%. The specimens were manually embedded in paraffin and cut into 5-μm-thick sections. For LCM, the sections were mounted on uncoated glass slides, lightly stained with hematoxylin, and kept in a moisture-free environment without a coverslip.
LCM. We used a PixCell II LCM instrument purchased from Arcturus (Mountain View, CA). Microdissections were performed at resolutions of 7.5 μm.
DNA Extraction. The CapSure Transfer Film cap (Arcturus) was placed in a microcentrifuge tube containing 50 μl of extraction buffer (1 mg/ml proteinase K and 1% Tween 20 in 10 mM Tris·HCl/1 mM EDTA buffer, pH 8.0). The tube was inverted so the fluid was in contact with the surface of the cap. The mixture was incubated overnight in a humidified incubator at 42°C, and the liquid was collected by centrifugation (2,000 × g).
Single-Molecule PCR. One-microliter samples containing an estimated 0.4 target HD molecule were amplified by using two rounds of nested PCR. The 25-μl PCR mixture consisted of 1× FailSafe PCR PreMix K buffer (50 mM Tris·HCl, pH 9.3 at 22°C/50 mM KCl/200 μM each dNTP/1.0 mM MgCl2 and FailSafe PCR Enhancer with betaine; Epicentre Technologies, Madison, WI), 0.875 unit of FailSafe PCR enzyme mix, and 10 pmol each of forward primer (5′-GCGACCCTGGAAAAGCTGATGA-3′) and reverse primer (5′-TGAGGCAGCAGCGGCTGT-3′). First-round PCRs were performed at 98°C for 2 min, followed by 27 cycles of 98°C for 1 min and 64°C for 2 min 30 s. A final extension was performed at 72°C for 10 min. Two microliters of the first-round PCR product, containing the same buffer and enzyme, was further amplified in the second round by using 2 pmol each of forward primer (5′-CCTTCGAGTCCCTCAAGTCCTTC-3′) and reverse primer (5′-CGGCTGAGGAAGCTGAGGAG-3′). The reverse primer used in the second round was labeled at its 5′ end with the Beckman CEQ WellRED Dye D3 (Beckman Coulter). The second round amplifications were performed at 98°C for 2 min, followed by 29 cycles of 98°C for 1 min and 67°C for 2 min. Final extension was performed at 72°C for 10 min. All PCR products with agarose gel mobilities indicating that they were not the WT HD allele were sized on a CEQ2000 (Beckman Coulter) denaturing microcapillary gel electrophoresis system.
Results
In adult patients with HD, variation in disease allele size in blood is severely limited relative to the variation seen in germ cells (35–39). The size of the disease allele in blood can therefore be viewed as indicative of the allele size at conception, and alterations in this size can be taken as evidence of mutation. We studied the instability of an expanded HD allele in postmeiotic testis cells by using tissue taken at autopsy from a 48-year-old affected individual. According to total blood DNA, this patient was heterozygous for a (CAG/CTG)50 disease allele and a 15-repeat normal allele. Analysis of 80 blood alleles by single-molecule PCR (data not shown) revealed 38 WT and 42 disease alleles (defined throughout this paper as >36 repeats). Among the disease alleles, 7 were 49 repeats, 37 were 50 repeats and 9 were 51 repeats. A single 41-repeat contraction was also observed.
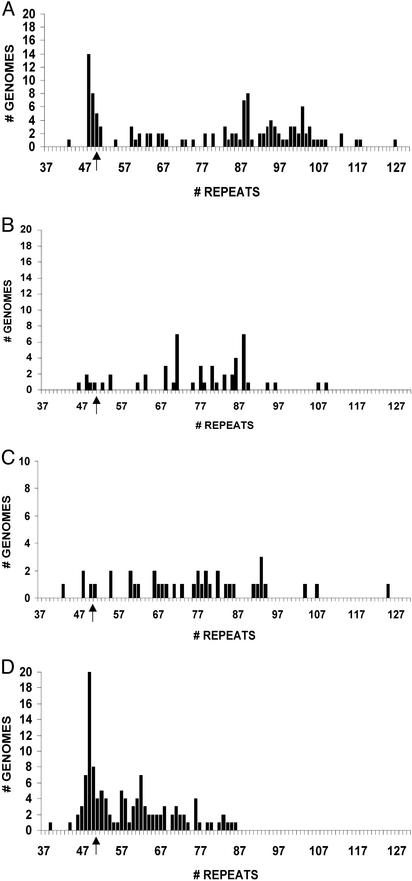
Postmeiotic cells (spermatids and sperm; Fig. 1) were easily identified in histological sections of seminiferous tubules by their characteristic morphology and location close to the lumen of the tubule (40, 41). After LCM, the cells were lysed and diluted to less than one haploid genome equivalent per sample, and the HD TNR tract was amplified by using single-molecule PCR. Among the 292 alleles that could be amplified, 125 (43%) were disease alleles and 57% were WT alleles. The 125 HD alleles (Fig. 2A; Table 1) had a median size of 87 repeats and gave a continuous size distribution (43–127 repeats) similar to allele sizes found in semen samples from affected sperm donors (32, 39). We were conservative in calculating the expansion mutation frequency (no. of disease alleles with an expansion mutation/total no. of disease alleles), such that only alleles with >52 repeats were scored as new mutations. Among the 125 disease alleles, 75% were expansion mutations.
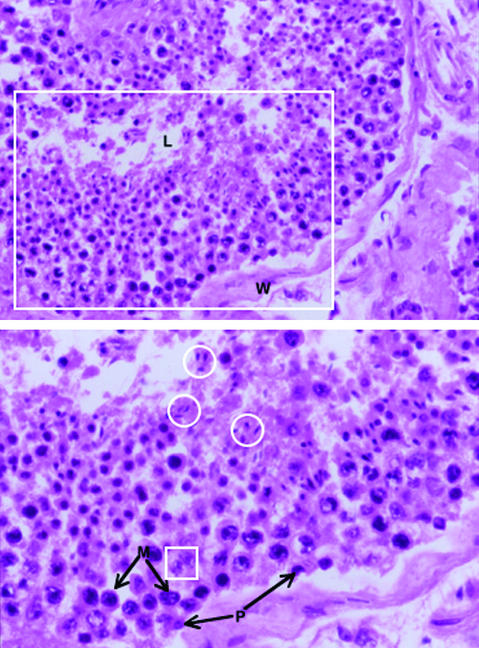
Fig. 1.
Cellular composition of seminiferous tubules. A portion of a seminiferous tubule is shown in Upper. The most immature germ cells (spermatogonia) are located at the periphery, close to the tubular wall (W). Postmeiotic spermatids and maturing spermatozoa are concentrated in the lumen (L). The area in the large rectangle is enlarged in Lower. Note the differences in nuclear size and granularity of the chromatin between premeiotic (P) and meiotic (M) cells. Nuclei from both of these cell types can also be readily distinguished from the small, elongated, homogenously staining nuclei of postmeiotic spermatids and maturing spermatozoa, examples of which are shown within circles. In addition to germ cells, seminiferous tubules contain Sertoli cells, examples of which are shown within the small square. (Magnification: Upper, ×10; Lower, ×20.)
Fig. 2.
Distribution of HD allele sizes from postmeiotic cells of patient 1 (A), mixed premeiotic and meiotic cells of patient 1 (B), premeiotic cells of patient 1 (C), and mixed premeiotic and meiotic cells of patient 2 (D). The arrow indicates the somatic HD allele size that is 50 repeats in both patients.
Table 1. PCR results from single-molecule analysis of premeiotic/meiotic and postmeiotic cell compartments from two testis samples.
|
Premeiotic/meiotic
|
Postmeiotic
|
|||||
|---|---|---|---|---|---|---|
| Sample | Disease alleles | >52 repeats | Total alleles | Disease alleles | >52 repeats | Total alleles |
| Patient 1 | 51 | 45 | 134 | 125 | 94 | 292 |
| Patient 1* | 39 | 34 | 131 | |||
| Patient 2 | 120 | 69 | 388 | |||
A dissection that aimed to include only premeiotic cells.
Synchronization of cell division of spermatogonia along the length of seminiferous tubules is limited in humans compared with rodents (40, 41). Human premeiotic and meiotic cells are readily identified based on their size, nuclear staining properties, and close proximity to the basement membrane (42). Seminiferous tubule sections from the same patient were stained with γ-H2AX antibody (data not shown). Antibody staining is detected in most spermatogonia and primary spermatocytes (43–45). By this criteria, >50% of cells immediately within the outer third of the seminiferous tubules were either premeiotic or meiotic. Although postmeiotic cells can be found in proximity to Sertoli cells near the periphery of seminiferous tubules, these cells could be readily distinguished and excluded from our premeiotic/meiotic LCM preparations (Fig. 1). Fifty-one HD alleles were detected in the premeiotic/meiotic cell preparation (median size, 77 repeats; range 46–109 repeats; Fig. 2B). The mutation frequency was 88%.
A more discriminating microdissection was made to isolate only premeiotic cells from the same patient. Such cells are located at the periphery of seminiferous tubules, show nuclei that are usually smaller than those of meiotic cells, and lack the coarse chromatin seen in cells that have initiated meiosis (Fig. 1). We observed 39 HD alleles in premeiotic cells (Table 1) and a mutation frequency of 87%. Although we may not have been able to completely prevent the capture of some cells just entering meiosis, it is highly unlikely that such contaminants would be present at a level approaching 87%. Allele sizes varied from 43 to 125 repeats (median, 77 repeats; Fig. 2C). When the data from all meiotic and premeiotic cells are pooled and compared with postmeiotic cells, there is a significant difference in expansion mutation frequency (79/90 vs. 94/125, P = 0.02). The slightly lower (15%) mutation frequency in postmeiotic cells could be explained if not all premeiotic/meiotic cells that experienced a TNR expansion mutation are capable of completing meiosis and spermiogenesis.
Premeiotic/meiotic cells were also studied from a second deceased individual 41 years of age who was heterozygous for a (CAG/CTG)50 disease allele and a 17-repeat normal allele (patient 2). Compared with patient 1, the seminiferous tubules were characterized by fewer postmeiotic cells, and less than 20% of the cells in the outer third of the seminiferous tubules stained heavily with γ-H2AX antibody (data not shown). Among the 120 disease alleles detected (Table 1), 57% were expansion mutations with a size distribution from 39 to 86 repeats (median, 57 repeats; Fig. 2D). Compared with the first affected individual, the premeiotic/meiotic cells of the second patient had a lower expansion mutation frequency and a narrower size distribution of disease alleles. Inter-individual variation in mutation frequency and size distribution is also seen when sperm samples from HD patients with similar numbers of repeats are compared (32).
Discussion
Our results from two patients show that HD expansion mutations occur before the end of meiosis (Fig. 2 B–D) and are already present in premeiotic cells (Fig. 2C). The latter observation suggests the occurrence of an important premeiotic mutation phase and supports an earlier argument for such a process in humans (32). How premeiotic mutations arise is not known. The first proposal to explain TNR expansions (46–48) involved DNA replication slippage, implying that germ-line expansion mutations arise during premeiotic germ-line cell divisions. Later, supporting evidence in Escherichia coli and yeast showed that instability was influenced by the orientation of the CAG/CTG repeats with respect to an origin of DNA replication, as well as by the repeat's proximity to an origin (2, 8–11). Finally, results from studies on yeast mutants that affect Okazaki strand synthesis (13, 49) were consistent with a cell-division-dependent mutation model in these organisms. On the other hand, studies on primate and human cells have found the relationship between instability and the orientation and proximity of the repeats to an origin of DNA replication to be more complicated (12, 50). In addition, data on mouse models suggest that TNR instability in somatic tissues accumulates with age and may not always be correlated with cell-division history (15–17, 51), thereby suggesting a role for the repair of DNA damage resulting from DNA breaks or gaps. Thus, slippage during cell-division-dependent DNA replication or repair of DNA lesions induced by TNR secondary structures (or both) could contribute to human premeiotic expansion mutations. Whether such mechanisms produce expansions through many independent events, each one adding a small number of repeats, or result in a single (saltatory) large addition, is not known.
Premeiotic expansion events could arise at a variety of different times during development. One possibility is early in development before the germ-line and somatic cell progenitors become distinct from one another (reviewed in ref. 52). Our single-molecule PCR results on blood DNA from patient 1 [as well as data comparing sperm and a greater variety of human tissues by using total DNA (35–38)] would argue against this possibility, because it might have been expected that all tissues would have comparable levels of instability. However, we must be cautious about ruling out expansion mutations before the germ line and soma become distinct. For example, selection against cells carrying large expansions (reviewed in refs. 52 and 53) may exist in some tissues, thereby reducing what would otherwise be significant somatic allele size variation. Premeiotic TNR instability may also occur between the time of primordial germ cell determination and puberty or could be unique to the postpubertal spermatogonia, but no experimental data concerning these possibilities are available. However, the long lifespan and lifelong cell divisions of postpubertal spermatogonial stem cells make them likely targets for age-dependent mutations that arise by DNA damage repair or cell-division-dependent DNA replication errors, respectively. Of course it is also possible that expansion mutations occur at many different stages of development.
Our data on premeiotic cells suggest that events occurring during meiosis or afterward are not required for expansion mutation. However, our results do suggest the possibility of an additional expansion phase after meiosis begins. In patient 1, a total of 215 disease alleles were detected in the premeiotic, premeiotic/meiotic and postmeiotic cell dissections. Data in Fig. 2 A–C show, for example, that the proportion of disease alleles with >80 repeats in the postmeiotic cells (0.57) was significantly greater (P = 0.01) compared with the proportion found in all of the premeiotic and meiotic cells (0.39). Two mutation mechanisms are obvious candidates to contribute to a second expansion phase. Studies in yeast have demonstrated meiosis-dependent TNR expansion events (4, 6, 7) that require SPO11 protein (5) and that are undoubtedly initiated by double strand break formation. Postmeiotic expansions could arise in haploid cells by DNA damage repair of breaks or gaps in TNR tracts (15–18, 51).
Our finding that human germ-line expansions occur before meiosis is completed (Figs. 2 B–D) differs from experiments in male mice that concluded expansion mutations arise only in cells after they have finished meiosis (18). In that study, premeiotic, meiotic, and postmeiotic cells were isolated by flow cytometry from animals carrying a transgene with a (CAG/CTG)117 tract. One possible explanation for the discrepancy with the human data concerns the different germ cell histories in the two species. The genomes of the human spermatogonial stem cells we studied can be estimated (54) to have experienced 771 (patient 1) or 611 (patient 2) cell divisions over 4 decades after zygote formation. In the mouse study, the spermatogonial stem cells are expected to have experienced ≈35 cell divisions (54) over a period of less than 3 months after zygote formation. Thus, fewer opportunities may exist in mouse than in human to accumulate premeiotic expansion mutations, regardless of what the molecular mechanism may be. Another possible difference between the mouse and human studies is that humans possess the complete HD gene and surrounding sequences, whereas the mouse HD transgene contained only a very limited portion of the human gene.
When all of the captured cell populations are considered together, far more WT alleles were amplified than HD alleles (620 vs. 325, respectively). There are at least two possible explanations for this unexpected finding. First, disease alleles may be missing because some of them may expand above the size range that can be efficiently amplified from microdissected tissue specimens. Using the same PCR protocols, we can detect expanded alleles (177 repeats; data not shown) from other DNA sources that are larger than the largest allele in the testis cell preparations (127 repeats). However, the efficiency of amplification is generally reduced as allele size increases. Second, some captured cells may have contained an HD allele with a DNA strand break in the CAG/CTG tract. Such molecules are an expected intermediate in every expansion model (reviewed in refs. 2, 24–27, and 55). If broken but expanded strands are not ligated to adjacent sequences on the same strand, they cannot be amplified by using PCR primers flanking the repeats. In vitro, the ligation of single (CTG)n strands to adjacent sequences becomes difficult, even when a single strand flap is no more than 10 repeats long, presumably because of secondary structure formation (56). Thus, the inability to find large HD alleles at the expected frequency (or even to find them at all in any particular cell population) may reflect an ascertainment bias against detecting certain intermediates in the mutation process, rather than suggesting their absence.
Acknowledgments
This work could not have been carried out without the support of the Venezuela Huntington Disease Project. A complete listing of the members follows. Columbia University: Nancy Wexler, Judith Lorimer, Julie Porter, Simone Roberts, Karen Marder, Carol Moskowitz, and Graciela Penchaszadeh; Argentina: Fidela Gomez; University of Alabama at Birmingham: Leon Dure; Children's Hospital of Los Angeles: S. Robert Snodgrass; Hospital Virgen del Camino, Spain: Maria Ramos; Indiana University: P. Michael Conneally and Jacqueline Gray; Harvard University: Jang-Ho Cha, James Gusella, Steven Hersch, Zane Hollingsworth, Marcy MacDonald, John B. Penney (deceased), Diana Rosas, and Anne Young; MacNeal Hospital, Illinois: Theresa Stillings; Massachusetts Institute of Technology: Michael Andresen and David Housman; Miami Children's Hospital: Gustavo J. Rey; Mt. Sinai Medical Center, Miami: Amarilis Acevedo-Cruz; New York University: Jose Alvir; North Shore University Hospital: Andrew Feigin; San Diego Public Health: Leticia Acosta; University of California, Irvine: Leslie Thompson; University of Iowa: Jane Paulsen; University of Oxford Wellcome Trust Centre for Human Genetics: Lon Cardon, Stacey Cherney, and Javier Gayan; University of Rochester: Ira Shoulson; University of Southern California: Norman Arnheim; University of Tampa: Juan Sanchez Ramos; University of Texas M. D. Anderson Cancer Center: Jacqueline Bickham; University of Ulm, Germany: Bernhard Landwehrmeyer; and University of Zulia, Venezuela: Margot de Young, Ernesto Bonilla, and Americo Negrette. We also acknowledge grants from the National Institute of General Medical Sciences (to N.A.), the National Institute of Neurological Disorders and Stroke, the W. M. Keck Foundation, and the Hereditary Disease Foundation (to N.S.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HD, Huntington disease; TNR, trinucleotide repeats; LCM, laser capture microdissection.
References
- 1.Cummings, C. J. & Zoghbi, H. Y. (2000) Hum. Mol. Genet. 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 2.Wells, R. D., Warren, S. T. & Sarmiento, M. (1998) Genetic Instabilities and Hereditary Neurological Diseases (Academic, San Diego).
- 3.Ashley, C. T., Jr., & Warren, S. T. (1995) Annu. Rev. Genet. 29, 703–728. [DOI] [PubMed] [Google Scholar]
- 4.Jankowski, C., Nasar, F. & Nag, D. K. (2000) Proc. Natl. Acad. Sci. USA 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jankowski, C. & Nag, D. K. (2002) Mol. Genet. Genomics 267, 64–70. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, H., Sears, D. D., Zenvirth, D., Hieter, P. & Simchen, G. (1999) Mol. Cell. Biol. 19, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schweitzer, J. K., Reinke, S. S. & Livingston, D. M. (2001) Genetics 159, 1861–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenreich, C. H., Stavenhagen, J. B. & Zakian, V. A. (1997) Mol. Cell. Biol. 17, 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang, S., Jaworski, A., Ohshima, K. & Wells, R. D. (1995) Nat. Genet. 10, 213–218. [DOI] [PubMed] [Google Scholar]
- 10.Maurer, D. J., O'Callaghan, B. L. & Livingston, D. M. (1996) Mol. Cell. Biol. 16, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miret, J. J., Pessoa-Brandao, L. & Lahue, R. S. (1998) Proc. Natl. Acad. Sci. USA 95, 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleary, J. D., Nichol, K., Wang, Y. H. & Pearson, C. E. (2002) Nat. Genet. 31, 37–46. [DOI] [PubMed] [Google Scholar]
- 13.Schweitzer, J. K. & Livingston, D. M. (1998) Hum. Mol. Genet. 7, 69–74. [DOI] [PubMed] [Google Scholar]
- 14.Freudenreich, C. H., Kantrow, S. M. & Zakian, V. A. (1998) Science 279, 853–856. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, L. & Shelbourne, P. F. (2000) Hum. Mol. Genet. 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 16.Lia, A. S., Seznec, H., Hofmann-Radvanyi, H., Radvanyi, F., Duros, C., Saquet, C., Blanche, M., Junien, C. & Gourdon, G. (1998) Hum. Mol. Genet. 7, 1285–1291. [DOI] [PubMed] [Google Scholar]
- 17.Fortune, T. M., Vassilopoulos, C., Coolbaugh, M. I., Siciliano, M. J. & Monckton, D. G. (2000) Hum. Mol. Genet. 9, 439–445. [DOI] [PubMed] [Google Scholar]
- 18.Kovtun, I. V. & McMurray, C. T. (2001) Nat. Genet. 27, 407–411. [DOI] [PubMed] [Google Scholar]
- 19.Manley, K., Shirley, T. L., Flaherty, L. & Messer, A. (1999) Nat. Genet. 23, 471–473. [DOI] [PubMed] [Google Scholar]
- 20.Lobachev, K. S., Gordenin, D. A. & Resnick, M. A. (2002) Cell 108, 183–193. [DOI] [PubMed] [Google Scholar]
- 21.van den Broek, W. J., Nelen, M. R., Wansink, D. G., Coerwinkel, M. M., te Riele, H., Groenen, P. J. & Wieringa, B. (2002) Hum. Mol. Genet. 11, 191–198. [DOI] [PubMed] [Google Scholar]
- 22.Richard, G. F., Goellner, G. M., McMurray, C. T. & Haber, J. E. (2000) EMBO J. 19, 2381–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasar, F., Jankowski, C. & Nag, D. K. (2000) Mol. Cell. Biol. 20, 3449–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakupciak, J. P. & Wells, R. D. (2000) IUBMB Life 50, 355–359. [DOI] [PubMed] [Google Scholar]
- 25.Sinden, R. R. (1999) Am. J. Hum. Genet. 64, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurray, C. T. (1995) Chromosoma 104, 2–13. [DOI] [PubMed] [Google Scholar]
- 28.Mitas, M. (1997) Nucleic Acids Res. 25, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. (2000) Mol. Cell 6, 989–698. [DOI] [PubMed] [Google Scholar]
- 30.Keeney, S., Giroux, C. N. & Kleckner, N. (1997) Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 31.Romanienko, P. J. & Camerini-Otero, R. D. (2000) Mol. Cell 6, 975–987. [DOI] [PubMed] [Google Scholar]
- 32.Leeflang, E. P., Tavare, S., Marjoram, P., Neal, C. O., Srinidhi, J., MacFarlane, H., MacDonald, M. E., Gusella, J. F., de Young, M., Wexler, N. S. & Arnheim, N. (1999) Hum. Mol. Genet. 8, 173–183. [DOI] [PubMed] [Google Scholar]
- 33.Brock, G. J., Anderson, N. H. & Monckton, D. G. (1999) Hum. Mol. Genet. 8, 1061–1067. [DOI] [PubMed] [Google Scholar]
- 34.Bonner, R. F., Emmert-Buck, M., Cole, K., Pohida, T., Chuaqui, R., Goldstein, S. & Liotta, L. A. (1997) Science 278, 1481–1483. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald, M. E., Barnes, G., Srinidhi, J., Duyao, M. P., Ambrose, C. M., Myers, R. H., Gray, J., Conneally, P. M., Young, A., Penney, J., et al. (1993) J. Med. Genet. 30, 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Telenius, H., Kremer, B., Goldberg, Y. P., Theilmann, J., Andrew, S. E., Zeisler, J., Adam, S., Greenberg, C., Ives, E. J., Clarke, L. A., et al. (1994) Nat. Genet. 6, 409–414. [DOI] [PubMed] [Google Scholar]
- 37.Giovannone, B., Sabbadini, G., Di Maio, L., Calabrese, O., Castaldo, I., Frontali, M., Novelleto, A. & Squitieri, F. (1997) Hum. Mutat. 10, 458–464. [DOI] [PubMed] [Google Scholar]
- 38.Duyao, M., Ambrose, C., Myers, R., Novelletto, A., Persichetti, F., Frontali, M., Folstein, S., Ross, C., Franz, M., Abbott, M., et al. (1993) Nat. Genet. 4, 387–392. [DOI] [PubMed] [Google Scholar]
- 39.Leeflang, E. P., Zhang, L., Tavare, S., Hubert, R., Srinidhi, J., MacDonald, M. E., Myers, R. H., de Young, M., Wexler, N. S., Gusella, J. F., et al. (1995) Hum. Mol. Genet. 4, 1519–1526. [DOI] [PubMed] [Google Scholar]
- 40.Russell, L. D., Ettlin, R. A., SinaHikim, A. P. & Clegg, E. D. (1990) Histological and Histopathological Evaluation of the Testis (Cache River, Clearwater, FL).
- 41.Johnson, L., McKenzie, K. S. & Snell, J. R. (1996) Tissue Cell 28, 127–136. [DOI] [PubMed] [Google Scholar]
- 42.Nistal Martín de Serrano, M. & Paniagua Gómez-Alvarez, R. (1984) Testicular and Epididymal Pathology (Thieme-Stratton, New York).
- 43.Mahadevaiah, S. K., Turner, J. M., Baudat, F., Rogakou, E. P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W. M. & Burgoyne, P. S. (2001) Nat. Genet. 27, 271–276. [DOI] [PubMed] [Google Scholar]
- 44.Hamer, G., Roepers-Gajadien, H. L., van Duyn-Goedhart, A., Gademan, I. S., Kal, H. B., van Buul, P. P. & De Rooij, D. G. (2003) Biol. Reprod. 68, 628–634. [DOI] [PubMed] [Google Scholar]
- 45.Celeste, A., Petersen, S., Romanienko, P. J., Fernandez-Capetillo, O., Chen, H. T., Sedelnikova, O. A., Reina-San-Martin, B., Coppola, V., Meffre, E., Difilippantonio, M. J., et al. (2002) Science 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichler, E. E., Holden, J. J. A., Popovich, B. W., Reiss, A. L., Snow, K., Thibodeau, S. N., Richards, C. S., Ward, P. A. & Nelson, D. L. (1994) Nat. Genet. 8, 88–94. [DOI] [PubMed] [Google Scholar]
- 47.Kunst, C. B. & Warren, S. T. (1994) Cell 77, 853–861. [DOI] [PubMed] [Google Scholar]
- 48.Richards, R. I. & Sutherland, G. R. (1994) Nat. Genet. 6, 114–116. [DOI] [PubMed] [Google Scholar]
- 49.Ireland, M. J., Reinke, S. S. & Livingston, D. M. (2000) Genetics 155, 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nenguke, T., Aladjem, M. I., Gusella, J. F., Wexler, N. S. & Arnheim, N. (2003) Hum. Mol. Genet. 12, 1021–1028. [DOI] [PubMed] [Google Scholar]
- 51.Gomes-Pereira, M., Fortune, M. T. & Monckton, D. G. (2001) Hum. Mol. Genet. 10, 845–854. [DOI] [PubMed] [Google Scholar]
- 52.Moutou, C., Vincent, M. C., Biancalana, V. & Mandel, J. L. (1997) Hum. Mol. Genet. 6, 971–979. [DOI] [PubMed] [Google Scholar]
- 53.Ashley-Koch, A. E., Robinson, H., Glicksman, A. E., Nolin, S. L., Schwartz, C. E., Brown, W. T., Turner, G. & Sherman, S. L. (1998) Am. J. Hum. Genet. 63, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drost, J. B. & Lee, W. R. (1995) Environ. Mol. Mutagen. 25, 48–64. [DOI] [PubMed] [Google Scholar]
- 55.Sia, E. A., Jinks-Robertson, S. & Petes, T. D. (1997) Mutat. Res. 383, 61–70. [DOI] [PubMed] [Google Scholar]
- 56.Henricksen, L. A., Veeraraghavan, J., Chafin, D. R. & Bambara, R. A. (2002) J. Biol. Chem. 277, 22361–22369. [DOI] [PubMed] [Google Scholar]